1. Introduction
Skin cancer is one of the most common and aggressive cancers worldwide, leading to significant health deterioration or even loss of life. In the United States alone, it is estimated that over 9500 individuals are diagnosed with skin cancer every day, while more than two individuals lose their lives due to this disease [
1,
2]. Unfortunately, skin cancer is not limited to developed nations, as recent research from Asian countries also reveals its growing incidence and severity as a public health and clinical concern. According to the World Health Organization, reported skin cancer cases result in approximately 853 deaths per year. India faces a similar challenge, with an estimated 1.5 million new instances identified annually. In China, there has been a significant rise in various types of skin cancer, particularly in urban regions. Overall, among all types of cancer affecting Asian countries, skin cancer accounts for approximately 2 to 4 percent of cases, highlighting the significant burden of this disease in the region [
3,
4,
5]. Skin cancer is generally classified into several categories, as illustrated in
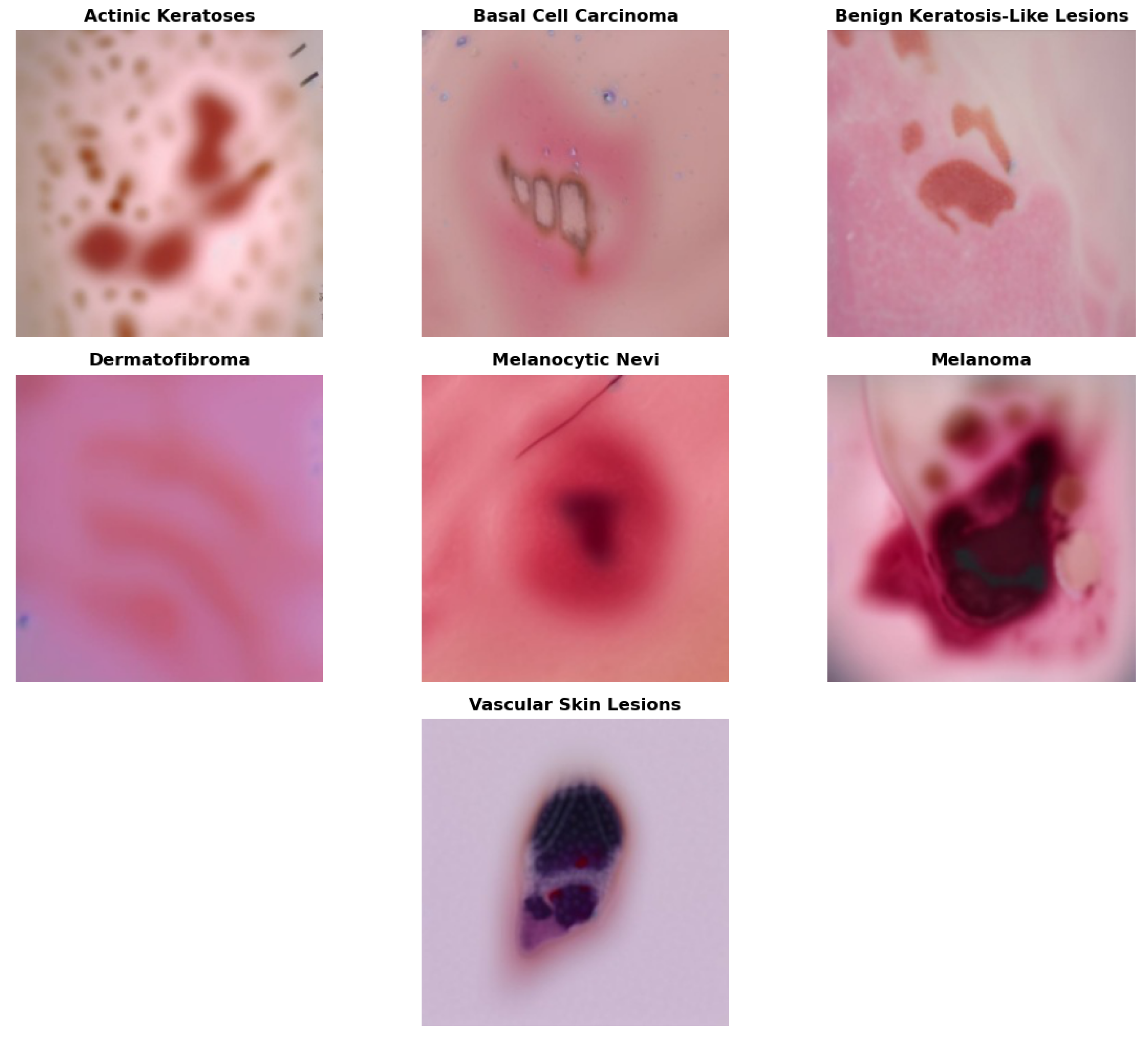
Figure 1.
Skin cancer includes a wide range of malignant pathologies, including dermatofibromas, melanoma, vascular lesions, actinic keratosis, basal cell carcinomas, melanocytic nevi, and benign keratoses. Identifying and preventing these types of skin cancer at an early stage is critical for preserving life. Most people often face challenges in scheduling regular check-ups due to a lack of availability, limited access to healthcare, and individual circumstances. Moreover, the initial undervaluation of skin irregularities can lead to advancement into critical, life-threatening stages [
6,
7,
8].
However, the diagnosis of skin cancer continues to be an essential but challenging task. The advancement of computer-aided techniques for diagnosing skin lesions has become a top priority in recent research. The ABCD rule, which focuses on asymmetry, irregular borders, distinctive color, and dermatological features, is one of the most frequently employed approaches. Dermatologists widely use the ABCD rule to diagnose skin cancer. Nevertheless, it may be challenging to differentiate between malignant and non-cancerous images due to factors such as noise, poor contrast, and uneven boundaries.
1.1. Role of Machine Learning and Deep Learning in the Diagnosis of Skin Cancer
Accurate diagnosis of skin cancer is a crucial area of research, and the potential of machine learning, in particular, offers potential for significant improvements [
9,
10,
11]. The key to successful treatment and increased chances of survival lies in early detection. While conventional diagnostic approaches have long been the standard, the introduction of advanced technologies such as deep learning and transfer learning has opened up new opportunities. These innovative techniques are enhancing both the accuracy and speed of skin cancer diagnosis, representing a significant advancement in medical research and a beacon of hope for the future. Artificial neural networks are used in deep learning, a type of machine learning, to identify feature patterns specific to different kinds of skin lesions [
12,
13,
14].
For instance, a neural network can be trained on a large dataset of skin cancer images. Then, when presented with a new image, it can quickly and accurately identify potential cancerous lesions. Skin cancer can be diagnosed using convolutional neural networks (CNNs), a particular type of neural network that has demonstrated remarkable performance in image-based diagnosis [
15,
16]. CNNs can speed up the diagnostic process and improve accuracy by identifying essential features such as texture, color, and pattern. Additionally, CNNs can be enhanced to better extract relevant information in medical images by incorporating attention mechanisms [
17].
Although there have been advancements in deep neural network architectures and attention mechanisms, skin cancer diagnosis remains a challenging task due to inter-class and intra-class variations. Due to factors such as growth stage, patient demographics, or environmental conditions, lesions of the same type, such as melanoma, can differ significantly in size, color, shape, and texture. This variation makes it difficult for models to generalize across cases within the same class. Another challenge is inter-class similarity, since benign and malignant lesions may be visually similar and it is hard to distinguish between the two with the help of deep learning models as well as medical professionals [
18,
19].
It is this similarity that results in a higher chance of misclassification, especially in borderline cases. Class imbalance is another problem with medical imaging. In the skin cancer datasets, some of the classes are highly represented as compared to others. This also makes it harder to train the models and to generalize, as most available datasets are small and do not represent any varieties of skin or lesion manifestation [
20,
21].
Noise is another source of error that can obscure important lesion features and lower diagnostic accuracy in dermoscopy images and include hair and lighting differences, and imaging artifacts, which contribute to further complexity. Moreover, more sophisticated models that may use transformers or attention mechanisms are more accurate, but are computationally complex and therefore not applicable in real-time clinical use, especially with resource-constrained environments. Lastly, when trained on small or imbalanced datasets, deep learning models are prone to overfitting, resulting in poor generalization to new and unseen cases.
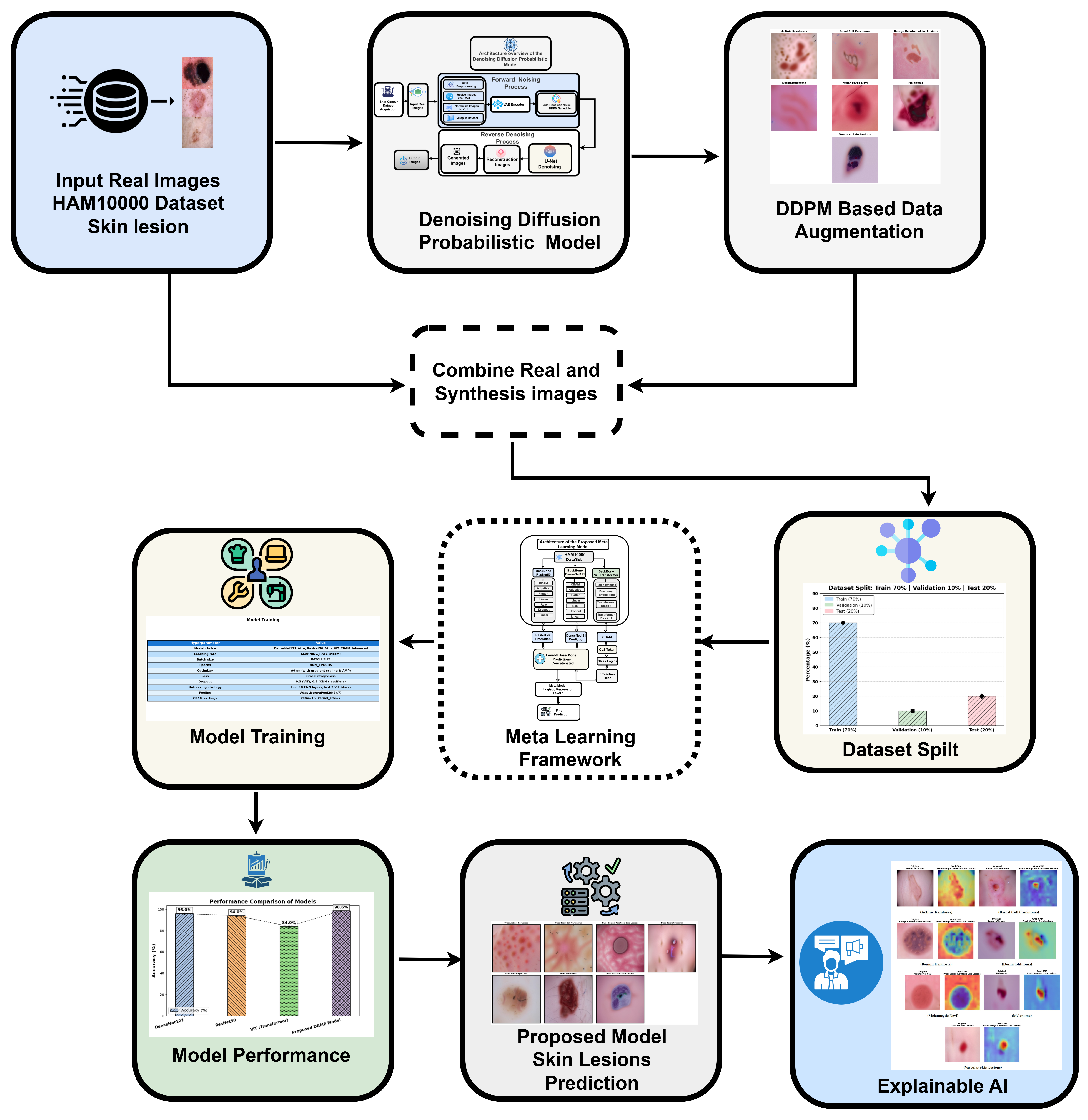
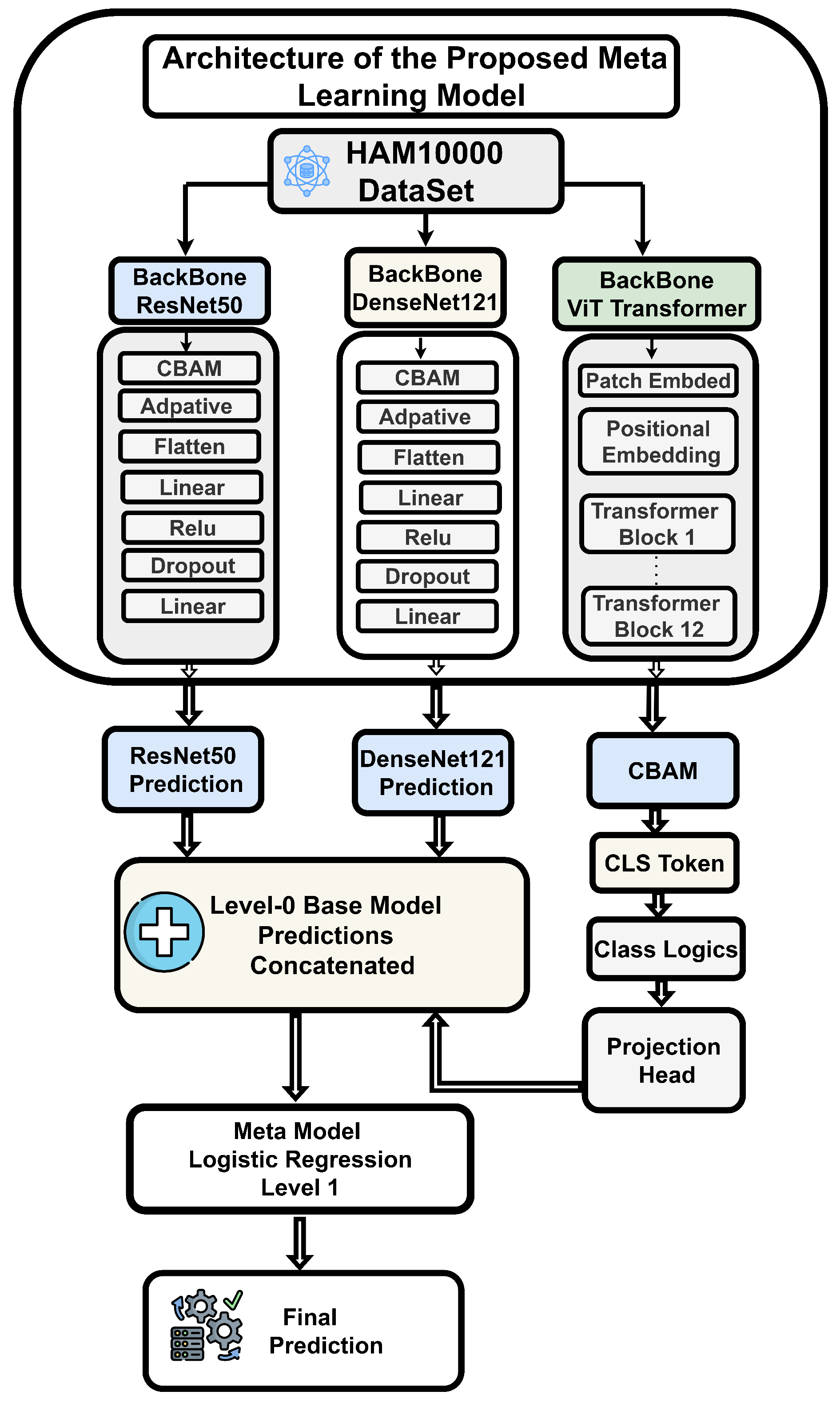
To address these challenges, we proposed the DAME framework (Diffusion-Augmented Meta-Learning Ensemble) as shown in
Figure 2 that integrates the local feature extraction capabilities of ResNet50 and VGG19 with the global context modeling of vision transformers for explainable medical image classification.
1.2. Research Contribution
The main contribution of this research is as follows:
- 1:
Our research proposes the DAME (Diffusion-Augmented Meta-Learning Ensemble) framework, a unified multi-architecture deep learning model that synergistically combines convolutional backbones (ResNet50 and VGG19) with a Vision Transformer (ViT) module.
- 2:
To enhance local and global feature representation, the architecture incorporates the Convolutional Block Attention Module (CBAM), which adaptively refines spatial and channel-wise information.
- 3:
The proposed model is enhanced through the integration of generative modeling, specifically the Denoising Diffusion Probabilistic Model (DDPM), which facilitates robust feature learning under data scarcity, class imbalance, and noise.
- 4:
We incorporated a meta classifier trained on hybrid model predictions to refine the decision boundary further, enabling accurate and generalizable detection of skin cancer metastases.
- 5:
This research introduces a novel approach to enhance black-box model explainability by applying Grad CAM to visualize and highlight regions that impact classification outcomes. The organization of the paper in the subsequent sections is as follows:
2. Related Work
With the widespread application of complex neural network technologies across biomedical domains, healthcare imaging analysis has emerged as a fundamental technique for supporting clinical decision-making. It has also become a primary research focus at the intersection of visual computing and healthcare intelligence. Nevertheless, the complex multidimensional characteristics of clinical images, insufficient data availability, and difficulties in annotation continue to pose persistent challenges to training efficiency and model generalizability. To address these issues, scholars have thoroughly investigated data augmentation strategies, representation learning techniques, and the development of novel classification frameworks.
Table 1 provides a summary of recent studies in skin cancer classification and the identified research gap.
2.1. Medical Image Feature Extraction and Classification
Huang et al. [
22,
23,
24] studied the application of multispectral imaging technology for the identification and classification of skin cancer. They specifically focused on seborrheic keratosis (SK), squamous cell carcinoma (SCC), and basal cell carcinoma (BCC). The experimental observation of the HIS-based system demonstrates a performance enhancement of 7.5% over the conventional RGB-based method. This enhancement is primarily attributed to the increased dataset size employed for training convolutional neural networks; given the computational demands of image processing tasks, larger and heterogeneous datasets are needed to ensure that the CNNs are developed and tested as a whole. There is another important detail that can be improved upon, which is the size of the dataset used to train convolutional neural networks. This development can contribute greatly to the result. Future research should therefore emphasize dataset augmentation and precision enhancement before extending to other architectural aspects.
Yang et al. [
25] proposed a multipurpose convolutional neural network model of multi-class categorization of seven types of skin lesions. Despite their promises, the results of the segmentation are not good regarding their relevance to the work. Moreover, classification accuracy varied across lesion categories, with only two classes achieving satisfactory predictive performance. The validation dataset comprises 7.5% of the total data, which remains reliable as representative samples were ensured during the stratified splitting process. Even though the proposed multipurpose deep neural network was promising in terms of binary cancer detection and lesion segmentation, it is not suitable in complex multi-class classification. The challenges mentioned above are the reasons why the increased research and developments in the field of skin cancer classification are important.
Priyadharshini et al. [
26,
27] introduced an Extreme Learning framework using the Teaching–Learning-Based Optimization (TLBO) approach. The ELM functions as an efficient and precise one-hidden-layer, unidirectional neural network to extract texture features for skin cancer categorization. Simultaneously, the TLBO algorithm enhances model parameters to improve performance. This combination aims to categorize skin lesions as benign or malignant.
In Abhiram et al. [
28], an image classification framework named Deskinned was proposed for the identification of skin lesions. Their model was optimized and assessed using the HAM10000 dataset, and its results were compared against three widely recognized pre-trained frameworks: Inception V3, VGG16, and AlexNet. With a substantially greater precision rate of 97.354%, the study’s findings demonstrate that Deskinned performs better than the other models. Employing data augmentation approaches, they expanded the dataset to 45,756 images to resolve the issue of dataset imbalance in their work. Obviously, overfitting occurs in the training performance from such preprocessing. Their system, however, woudl downsample images to 28 × 28 pixels, thereby losing many features and leading to incorrect classifications in real-world situations. The framework that we have created is able to maintain suitable augmentation and high image resolutions to retain significant features in order to achieve better precision of skin lesion classification.
Arani et al. [
29] developed and evaluated an EfficientNetLite-0 model for mobile-based preliminary skin lesion detection. Their model was benchmarked against MobileNetV2 and ResNet50 architectures to assess performance efficiency. Despite these efforts, most smartphone-based skin cancer detection systems remain constrained by their reliance on cloud computing platforms, which raises concerns regarding diagnostic accuracy, latency, and data privacy. The study’s outcome demonstrated that EfficientNetLite-0 outperforms existing solutions, achieving an impressive accuracy rate of over 94%. The scholars emphasized the limitations of cloud-based platforms in such solutions. However, they proposed that using ensemble methods could address these gaps and even potentially yield enhanced performances. The authors also analyzed the significant implications of assessing the model fusion technique for such implementations. Therefore, our ensemble approach resolves these problems by combining various architectures that utilize on-device computation capabilities. A deep learning-based framework for classifying skin and oral cancer using diagnostic imaging was presented by Raval and Undavia [
30]. They included AlexNet, VGGNet, Inception, ResNet, DenseNet, and Graph Neural Network (GNN). However, specific drawbacks are apparent. The methodological transparency is lacking, as the paper primarily focuses on accuracy without discussing the sensitivity and harmonic means of precision and recall, thereby disregarding the significance of missed detections. Moreover, the dataset splitting of 70:20:15 appears unclear, and the performance of the models for each category has not been thoroughly analyzed. Applying convolutional neural network architectures, especially MobileNet and Xception, improved feature extraction and classification performance. However, it examined only five classes while our study expanded the types of lesions.
Ahammed et al. [
31] developed an AI-driven framework to categorize skin diseases. Through image segmentation and pattern identification techniques, they aim to overcome the limitations associated with manual diagnosis. To improve image resolution and remove artifacts, they utilized digital hair removal and Gaussian-based denoising. The Grab cut segmentation method precisely detects affected lesions, while post-segmentation attribute extraction, using the Grey Level Co-occurrence Matrix and quantitative attributes, captures latent patterns from the segmented images. Evaluation with state-of-the-art machine learning classifiers shows the effectiveness of this framework. However, deeper analysis of research methodological limitations and real-world implementation challenges would improve the research reliability and clinical value. The theme of class imbalance, a common challenge in medical datasets that can result in biased model training, is the focus of research [
32].
Using a variety of parameter configurations, the authors [
33] optimize AlexNet, InceptionV3, and ResNet. They introduced their proposed framework and evaluated its accuracy compared with the SOTA framework. However, the dataset has increased to over 30,000 samples using data augmentation techniques, which raises concerns about overfitting. Furthermore, the outcome remains effective even after implementing the mentioned preprocessing. Despite achieving such high accuracy rates for the proposed framework, the absence of analysis on F1 score, true positive rate (Recall), and incorrect optimistic predictions imposes significant constraints and necessitates further investigation. A resource-efficient neural network-based method for skin cancer classification was proposed by Shinde et al. [
33,
34]. The system demonstrates reliable results by integrating the MobileNet architecture for training and the squeeze-based method for digital hair removal. The accuracy is significantly enhanced by the hair removal method. Its evaluation is limited to the benign and malignant classes, which may reduce its usability for other skin conditions. Our study addresses these limitations. However, its usability for resource-constrained IoT hardware, such as the Raspberry Pi 4, represents a significant advancement in skin cancer classification technology. However, prior to deployment, our work employs the framework of cross domain research [
35] and an optimized image segmentation technique in microscopic images, which uses a deep learning framework in a resource-constrained environment without relying on hardware support.
Moturi et al. [
36] performed an evaluation between two frameworks, MobileNet V2 and a custom CNN, trained and tested on the HAM10000 benchmark. They also introduced a web-based framework for detecting skin lesions. MobileNet V2 achieves an accuracy of 85%, whereas the adapted architecture achieves 95%. However, they emphasized training accuracy instead of validation accuracy. Also, the insufficient analysis of F1 score and Recall metrics further reduces the assessment completeness. A proper evaluation discussion is critical, especially about validation accuracy. Moreover, the paper proposes a web-based framework that returns results based on input images of skin cancer. However, it neglects to address scenarios where non-dermatological images are submitted. Specifying these cases would enhance the completeness of their proposed framework. Convolutional neural network architectures, particularly MobileNet and Xception, have been introduced by Sadik et al. [
37] as a method for designing an expert system that can efficiently identify various skin diseases. The authors pre-train models on the Imagenet benchmark to enhance feature extraction by employing transfer learning. After augmentation and knowledge transfer-based learning, both MobileNet and Xception achieve high performance, demonstrating that comprehensive assessment using key metrics has yielded promising results. The authors further enhance the utility of their findings by implementing a web-based architecture for real-time disease identification.
Riaz et al. [
38,
39] introduced a Hybrid learning framework combining CNN and Local Binary Pattern (LBP) for skin cancer classification, trained and tested on the HAM10000 benchmark. The integration of CNN and LBP demonstrated robust performance, with training accuracy of 98.37% and validation accuracy of 97.32%. This hybrid of handcrafted LBP features and deep convolutional neural networks resulted in enhanced classification of several skin cancer types. Nevertheless, several drawbacks were observed. It was observed that the downsizing of images to 28 × 28 pixels led to information loss, which could have a problem with the accuracy of each type of lesion. Furthermore, the observed discrepancy between training and validation accuracies suggests a risk of overfitting, raising concerns regarding the model’s generalization capability. The absence of a clearly defined train–test–validation split also limits the reproducibility of the reported results. Although the hybrid framework achieved superior performance compared with single-model approaches, further refinement is necessary to minimize misclassification and to ensure stable performance across diverse lesion categories.
2.2. Data Augmentation Methods for Medical Images
Mudassir Saeed et al. [
40,
41] also proposed an enhanced GAN-based augmentation strategy. Their study carefully compared the performance of various generative methods. The CNN-based approach combining multiple convolutional neural networks, including SVM, VGG16, and VGG19 achieved a maximum classification accuracy of 96%.
Wang et al. [
42] introduced a feature extraction network to better identify a narrower region in the latent space where GAN-generated images resemble target domain data. This approach facilitated more effective fine-tuning of pre-trained models on the target distribution.
2.3. Limitations of Existing Approaches
The key limitation of this work is that it is based on binary classification methods, which only distinguish between benign and malignant skin lesions, but ignore the subtle differences between various types of lesions. As a solution to this, we highlight the significance of multi-class classification, which has a high level of precision and recall in all the categories. In prior work, a multi-layer convolutional neural network was developed, accompanied by an additional OpenCV-based chromatic analysis module, to evaluate lesion features. However, validation outcomes were low, and the use of the HAM10000 dataset revealed persistent issues with class imbalance, overfitting, and significant variance between training and validation phases, underscoring the need for more robust validation techniques. Despite the use of rebalancing, preprocessing, as well as augmentation strategies, they only partially helped to improve robustness. The latest reviews also note that real-world medical imaging datasets are also associated with significant obstacles, such as the lack of data, poor image quality, and extreme imbalance of classes. These aspects decrease the accuracy of the model and restrict the generalizability of diagnostic systems. Ordinary augmentation methods, e.g., scaling, rotation, and flipping, may augment apparent variation in the data, but do not create truly new variations, and the problem of data scarcity and imbalance remains unsolved.
To address these shortcomings, the methodology of this paper incorporates diffusion-based generative modeling, that is, applying Denoising Diffusion Probabilistic Models (DDPMs), which have demonstrated outstanding resilience and sample fidelity in image creation. Diffusion enables the augmentation process to produce diverse and natural medical images, thereby improving the stability and diagnostic quality of the classification system. Moreover, the generalized feature representations commonly used in traditional deep learning systems do not tend to represent domain-relevant pathological indicators, which restricts the applicability of such systems to a variety of diseases. We propose a new architecture, the DAME framework, which incorporates disease-aware guidance mechanisms to prioritize clinically significant attributes. Such a design enhances the work of multi-classification, generalization, and offers greater consistency of diagnosis across the different types of skin lesions.
Table 1.
Summary of recent studies on skin cancer classification.
Table 1.
Summary of recent studies on skin cancer classification.
| Reference | Year | Dataset | Classes | Method | Accuracy | Precision | Recall | F1 Score | Pros | Cons |
|---|
| Huang et al. [22] | 2023 | Multispectral/HIS | 3 | YOLO 5 | 79% 78% | 0.888 | 0.758 | 0.792 | HIS-based CNN improved accuracy with larger dataset. | No detailed metrics; computationally intensive; augmentation needed. |
| Priyadharshini et al. [26] | 2023 | DermISdataset | 2 | ELM, TLBO | 93.18% | 89.72% | 92.45% | 91.64% | ELM + TLBO optimization; augmentation improved robustness. | Binary only; dataset imbalance; overfitting risk. |
| Abhiram et al. [28] | 2022 | HAM10000 | 7 | AlexNet, VGG-16, InceptionV3 | 97.35% | 98% | 97% | 97% | Deskinned outperformed InceptionV3, VGG16, AlexNet. | Image reduced to 28 × 28 caused feature loss; overfitting. |
| Arani et al. [29] | 2023 | ISIC 2020 | 2 | EfficientNetLite-0, MobileNet V2, ResNet-50 | 94% | NR | 92.5% | 93% | EfficientNetLite-0 outperformed MobileNetV2 & ResNet50. | Cloud-based dependency; ensemble unexplored. |
| Raval et al. [30] | 2023 | ISIC, HAM10000 | 8 | ResNet, DenseNet | 93% | NR | NR | NR | Applied AlexNet, VGG, ResNet, DenseNet, GNN. | Focused only on accuracy; no F1/recall; unclear splits. |
| Moturi et al. [36] | 2024 | HAM10000 | 7 | MobileNetV2 | 95% | NR | NR | NR | Web-based detection; custom CNN high accuracy. | Training accuracy only; validation overlooked. |
| Sadik et al. [37] | 2023 | Dermnet, HAM10000 | 5 | Inception-ResNet, DenseNet, MobileNet, and Xception | 97% | 97% | 97% | 97% | MobileNet transfer learning; real-time system. | No per-class metrics; accuracy-only focus. |
| Riaz et al. [38] | 2023 | HAM10000 | 7 | CNN, LBP | Train: 98.9% | 98 | 98 | 98 | Hybrid CNN + LBP improved robustness. | Image resized to 28 × 28 lost features; overfitting risk. |
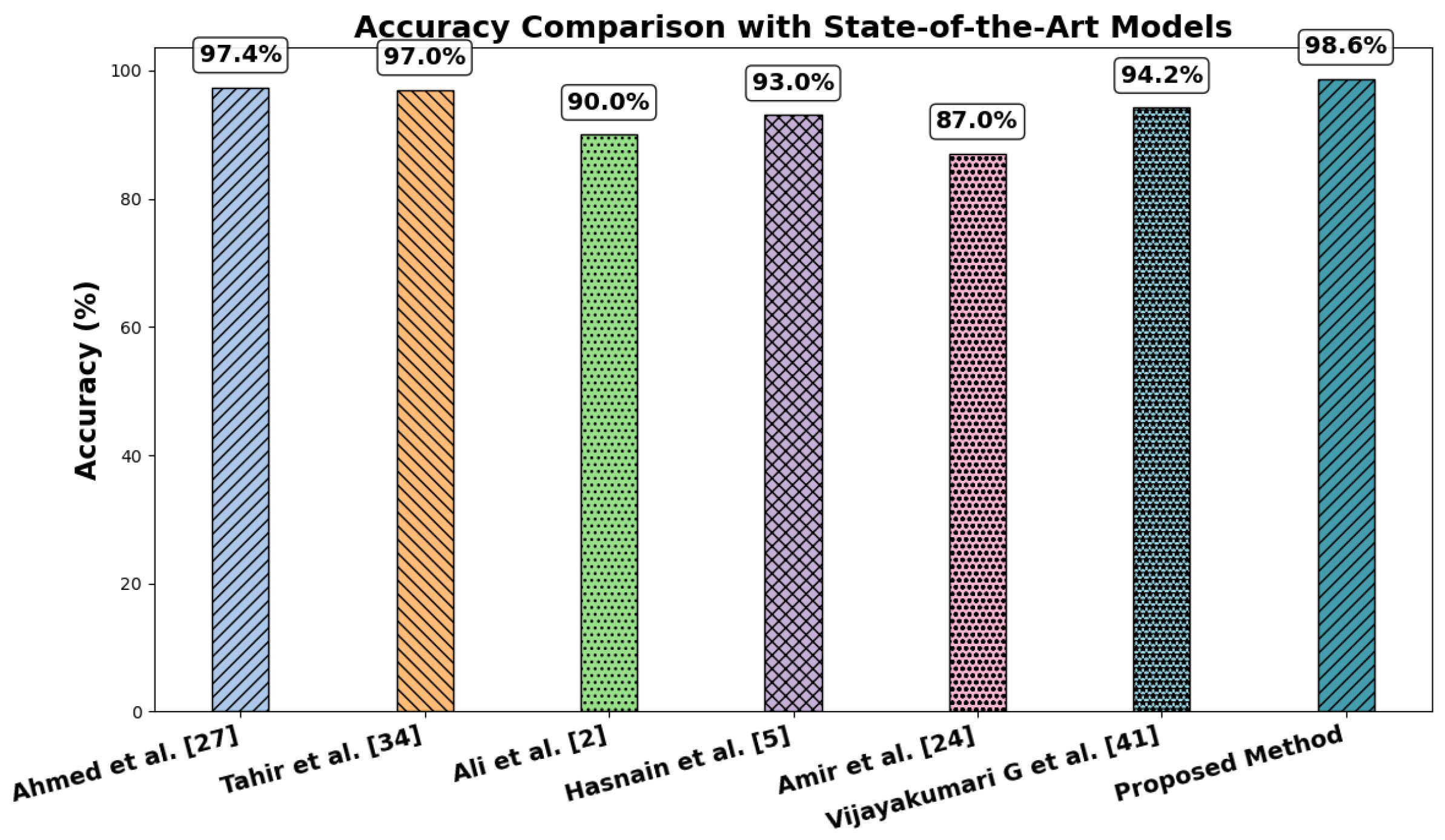
| Ali et al. [2] | 2025 | HAM10000 | 2 | VGG19 | 90.0% | 98 | 98 | 98 | Simple and uniform architecture (stacked 3 × 3 convolutions) makes it easy to implement, understand, and extend for transfer learning. | Very large in terms of parameters (138 M), leading to high memory usage and slower training. |
| Muhammad Hasnain Javid et al. [5] | 2023 | HAM10000 | 2 | ResNet50,
EfficientNet B6, InceptionV3, Xception | 93.0% | 93 | 93 | 93 | Residual connections allow very deep networks to train effectively by solving the vanishing gradient problem, improving accuracy. | Architecture is more complex and harder to customize or modify compared to traditional CNNs. |
| Muhammad Amir Khan et al. [24] | 2025 | HAM10000 | 2 | CNN Adaptive Model | 87.0% | 84 | 91 | 88 | Flexible architecture can be tailored (filters, depth, attention modules, etc.) to fit specific tasks and datasets efficiently. | Requires careful design and tuning; performance may vary widely if not optimized, unlike standardized architectures. |
4. Proposed Methodology
The overall proposed framework for multi-class skin lesion diagnosis is detailed in this section, as shown in
Figure 3. The framework begins with aata acquisition and sampling, followed by training a generative diffusion network to generate synthetic dermoscopic images. Original and generated images are combined to form an augmented dataset, which is subsequently used to train the proposed DAME architecture. The dataset is split into training and testing subsets, enabling performance analysis on both real and synthetic images. Performance is evaluated using standard diagnostic metrics, and expert radiologists verified the accuracy of the predictions. Finally, explainable AI techniques are applied to enhance model interpretability and support reliable clinical diagnosis.
4.1. Material and Methods
Early detection and diagnosis of skin cancer can boost survival rates dramatically. Deep learning-based CAD systems have recently demonstrated promising results in the automatic classification of skin cancer, as well as a meta-learning strategy for skin cancer classification utilizing multiple convolutional neural networks (CNNs). The HAM10000 Dataset [
34] was utilized to assess the performance of our proposed technique. The HAM10000 dataset comprises numerous melanoma and non-melanoma images with varying characteristics, making classification a challenging task. The proposed method enhances skin cancer classification performance, providing a more reliable and accurate diagnosis.
4.1.1. Dataset Description
In order to train, test, and validate our model, we utilized the HAM10000 skin lesion dataset of the International Skin Image Collection (ISIC) database, which is available on Kaggle. The HAM10000 dataset contains images of seven categories of skin lesions, none of which correspond to melanoma-type lesions. Consequently, we have chosen to convert the dataset to a multi-classification type. Secondly, the HAM10000 dataset is extremely unbalanced in terms of class distribution in that only the melanocytic nevi have a total count of over 6705 images, whereas all the other classes have a total of approximately 3310 samples, as shown in
Table 2. Therefore, it is crucial to balance the training set using data augmentation techniques, a necessary step to ensure the model’s accuracy and reliability.
4.1.2. Preprocessing
The images are captured by going through different normalization steps before being fed into the convolutional neural networks with the input medical images. The images are reduced to a normal size of 224 by 224 just before they are sent to convolutional networks. The conversion of the RGB images into the grayscale images is one of the significant stages of the process, as the conversion of images into the grayscale images is one of the most successful methods for identifying the skin lesions because there is no necessity to utilize color information in the original images. The pixel values are summed together to convert the red, green, and blue images to a grayscale image.
4.2. Data Augmentation
In this research, we utilized data augmentation techniques to address the problem of overfitting in our training dataset. Consequently, we implemented various kinds of image transformations, including cropping, random rotations, and splitting, to artificially augment the dataset and collect more information from the existing images. We employed advanced techniques, such as Denoising Diffusion Probabilistic Models, to create new images from existing ones. This approach enabled the augmentation of both the quality and size of the training dataset, resulting in improved efficiency of our deep learning models. Furthermore, data augmentation was essential in reducing overfitting problems in deep learning.
Table 3 shows the augmented images through the diffusion model for each category.
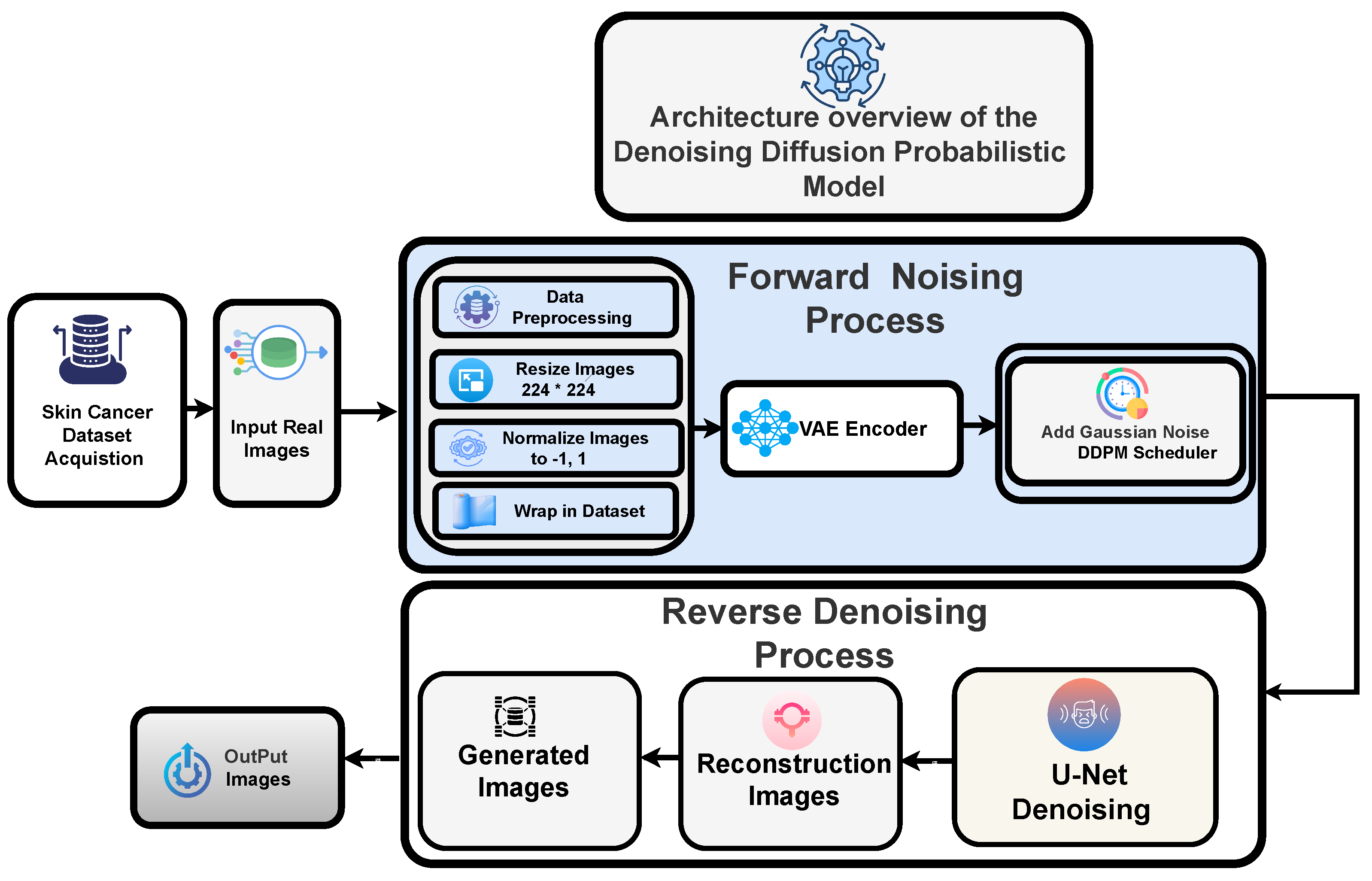
Denoising Diffusion Probabilistic Models
The leading cause of the poor performance of most medical image classification models was attributed not only to the implementation limitations but also to the class imbalance and an insufficient number of samples in the dataset. In addition, we aim to augment the dataset using generative models, specifically diffusion models, as our chosen approach due to their superior performance.
Figure 4 demonstrates the images generated through the diffusion model for each category.
4.3. Forward Noising Process
To generate high-resolution dermatological images conditioned on textual information, we utilized a Denoising Diffusion Probabilistic Model (DDPM) framework constructed using the Diffusers and Transformers libraries. A Variational Autoencoder (VAE) was used to encode dermoscopic images into a latent space after they had been reduced to 512 × 512 pixels, transformed into tensors, and normalized to [−1, 1]. The forward diffusion process was then replicated by gradually adding Gaussian noise to the latent codes in accordance with a specified schedule, with distortion managed by a particular strength parameter. The forward diffusion is a Markov chain that adds Gaussian noise at each step:
Let denote the original data (e.g., an image), and represent the noisy version of the data at step t. The parameter corresponds to the noise schedule, which is a small value that controls the amount of noise added at each step. After many steps T, is nearly pure Gaussian noise.
4.4. Reverse Denoising Process
The reverse procedure cannot be formulated in a closed analytical form, since noise removal, step by step, is mathematically intractable. Hence, it must be approximated using a parameterized function. A deep neural network, most typically a
U-Net topology, is utilized for this operation, owing to its capability to capture fine-grained spatial features through convolutional operations while retaining a broader global structure via skip connections. At each discrete timestep, the network accepts the corrupted input sample
(together with the temporal index
t) and estimates either the injected perturbation
or the corresponding denoised representation
. By recurrently applying these denoising operations along the entire diffusion pathway, The model incrementally reconstructs the underlying data distribution, ultimately converting raw Gaussian perturbations into plausible and high-fidelity outputs.
At each timestep t, let denote the noisy sample. The neural network, typically a U-Net, predicts the noise component as . The noise scaling term is defined as , and the cumulative product of these scaling terms up to timestep t is given by . The variance schedule at each step is represented by , and z denotes the Gaussian noise that is re-sampled at every timestep.
4.5. Training, Validation, and Test Dataset Split
A 70:20:10 training, validation, and testing set proportion was used in order to carefully assess the generalization strength of the models. This controlled subdivision is how the models are trained on a high volume of data with a suitably separate test set to do effective validation. The 70:20:10 split will be used to balance between training stability and overfitting.
4.6. Feature Extraction and Neural Networks
As previously mentioned, we have chosen state-of-the-art feature extraction architectures, including ResNet50, VGG19, and ViT Transformer, because these have been proven to be useful in obtaining meaningful features of images. Optimized structures and weight initializations of these frameworks, which are based on previous tasks and inherent properties of dermoscopic data, are the basis of our dependable skin cancer detection system.
4.7. Classification Model and Fine Tuning
In the meta-learning methodology outlined in this article, as in [
35,
43,
44], several convolutional neural networks (CNNs) are used as baseline models. The models were first trained with the ImageNet dataset, which comprises normal picture datasets, and then extended with the HAM10000 skin cancer dataset, which contains high-resolution dermoscopic images representing diverse types of skin cancer. This two-stage training approach leverages the strengths of transfer learning, preserving low-level feature representations from large-scale data while adapting higher-level parameters to the target domain. The use of meta learning further enhances this process by equipping the models with the capacity to quickly generalize to new classification tasks, leveraging past knowledge and drawing upon experience collected through earlier iterations, retained from the prior training phase. Fine-tuning configurations of the proposed models for classification are summarized in
Table 4. And Algorithm 1 shows skin cancer classification.
4.8. ResNet50
The ResNet50 [
5] model consists of 50 layers, comprising convolutional, pooling, and fully connected layers.The key innovation of ResNet-50 is that it incorporates residual connections, which enable the information flow to bypass specific layers, thereby addressing the problem of vanishing gradients. In our implementation, we utilized the pre-trained backbone, with the vast majority of the blocks frozen. The last 10 blocks, however, were not frozen, allowing us to train the backbone on domain-specific features. To further increase the level of discriminability, a Convolutional Block Attention Module (CBAM) is employed following the final layer of the backbone to reweight informative channels and locations adaptively. These fine tasks are then combined with Adaptive Average Pooling (7 × 7) and presented to a bespoke lesion classifier (Flatten -Linear-ReLU-Dropout-Linear) to identify seven lesion categories.
4.9. DenseNet121
The DenseNet121 [
37] model has been augmented based on its conventional original architecture with an attention mechanism and a task-specific classifier to improve skin cancer classification. The feature extractor is DenseNet-121, which has already been trained on ImageNet. It is also characterized by a highly interconnected structure, whereby all layers can feed on all preceding layers. This form of design enhances gradient propagation, reuses features, and offers various representations that bridge the gap between low- and high-level features. Our deployment process is based on the principle of freezing the backbone of the majority of the visual data and unfreezing the final ten layers to refine on domain-specific data. A 1024-channel activation map of the backbone is then created and refined using the Convolutional Block Attention Module (CBAM). CBAM performs an adaptive recalibration of feature activations by utilizing channel and spatial attention to direct the network’s focus towards disease–discriminative patterns. The optimized features are passed through an Adaptive Average Pooling layer (7 × 7), which normalizes the spatial dimensions across inputs. An adapted classifier head, comprising Flatten, Linear, ReLU, Dropout, and Linear layers, transforms the pooled features into seven output classes. The dropout layer is a regularization technique to reduce overfitting, whereas the dense layers apply non-linear features to make the classes more separable, and CBAM is vital in the optimization of features. In the architecture, transfer learning, attention-based refinement, selective layer unfreezing, and regularized classification are combined to make medical image analysis more stable.
4.10. Vision Transformer Model
Initially, the transformer-based vision model, which was developed to solve natural language processing (NLP) problems, has been shown to be flexible and versatile enough to be adapted to the field of computer vision. It achieves this by considering an image as a sequential patch representation, similar to textual representations in NLP, and, as a result, cross-disciplinary innovation becomes possible. In contrast to convolutional neural networks (CNNs), which acquire multi-scale spatial hierarchies through convolutional layers, the Vision Transformer (ViT) breaks down an input image into a series of fixed-size, non-overlapping patches. All patches are converted into embedded vectors, often in a high-dimensional space, with sufficient representational power. A classification token (CLS) that is learnable is prepended to the patch sequence, and positional embeddings are added to store spatial structure within the sequence.
The ViT CBAM Advanced model, a novel variant built upon the ViT architecture, utilizes the ViT Base patch configuration as its backbone. The pre-trained backbone is first frozen to retain its general-purpose features, while the remaining two transformer encoder blocks are not frozen, allowing task-specific fine-tuning on medical images. Direct access to the CLS token embedding acquired after the final encoder block of the transformer and normalization layer is then provided by removing the original classification head. This embedding, which is normally of size 768, is a small representation of the world around the input image. In order to augment its discriminatory ability, the CLS token is reformulated into a four-dimensional tensor and is subjected to a Convolutional Block Attention Module (CBAM). CBAM, which was originally developed on spatial feature tensors in CNNs, is re-purposed here to enhance the CLS token with the application of both channel and spatial attention to enhance information that is relevant to the class. The attention-enhanced CLS token is then fed through the projection head, and then an advanced classification head comprising Layer Normalization, Linear, GELU, Dropout, and a final Linear layer that generates logits of seven-class classification. The model is the only one to incorporate CBAM into the token-level architecture of ViT, but it is selectively fine-tuned on the last two transformer blocks. This type of integration facilitates the refinement of attention at a fine-grained level in the CLS token space, thereby improving the model’s ability to identify fine semantic details in high-resolution medical images, especially when diagnosing skin lesions.
4.11. Convolutional Block Attention Module (CBAM)
The Convolutional Block Attention Module is a lightweight, effective attention mechanism for feedforward convolutional neural networks. The CBAM attention module infers the attention map along dual dimensions, channels, and space in turn, and then scales features adaptively with the input feature map for context-aware feature enhancement. Because CBAM is a computationally efficient general-purpose module, it can be easily embedded into any convolutional neural network framework with minimal complexity and can be fully trainable end-to-end with the base convolutional neural network.
| Algorithm 1: Proposed Algorithm for Skin Cancer Classification. |
- Input:
Training data with ; Evaluation data D as a set containing elements I within the range with ; Sub-models utilizing CNN and Vision transformer. - Output:
Classification prediction from the meta-learning algorithm.
- 1:
Split dataset into . - 2:
Extract output predictions from sub-models employing CNN models and Vision Transformer model. - 3:
for to S do - 4:
Get output predictions based on . - 5:
. - 6:
Construct a new dataset D with output predictions P and their corresponding target labels Z. - 7:
for to N do - 8:
.
- 9:
Train a meta-model with the newly created dataset D.
- 10:
Validate with ; result = classify . - 11:
Return result.
|
4.12. Proposed Meta-Learning Framework
The design of a meta-learning approach enables adaptive feature selection across multiple convolutional neural network models. The initial step involves generating prediction probabilities for various types of skin lesion images using three fine-tuned CNN sub-models: ResNet50, DenseNet121, and the Vision Transformer model. At level 0, these sub-models are combined to generate an overall prediction for each image. The main idea behind incorporating several models is to improve the overall accuracy of prediction, which is a common practice in ensemble learning, used in deep learning applications, especially with image classification. After the prediction probabilities of the three fine-tuned CNN sub-models are obtained and stacked together at level 0, the next step in the proposed stacked ensemble architecture is to aggregate these predictions and feed it to a meta learner at level 1. The stacked predictions made by the level 0 are used by the meta learner to make the correct classification of the skin lesions. The meta learner, which is a separate model even to logistic regression, is the one that is fed with the joint prediction of the level 0 and creates the final output, giving the proposed image class label, which is the predicted value in the proposed meta-learning model structure as indicated in
Figure 5.
4.13. Employed Meta-Learning Framework and Notations
The notations used in the subsequent equations are explained as follows: M represents the set of sub-models. represents the prediction function of sub-model i, while X and Y represent the input and output, respectively. Additionally, represents the weight assigned to the prediction of sub-model i and represents the optimal model parameters ().
4.14. Prediction Equation
The ensemble prediction is obtained by combining the predictions of all sub-models, weighted according to their respective weights. A commonly used approach is to calculate a weighted average. The combined ensemble prediction, denoted as
, can be represented as follows:
4.15. Weights Optimization for Meta Learning
The weights assigned to each sub-model can be learned or optimized through meta learning to minimize a loss function that quantifies the discrepancy between the ensemble prediction and the desired output
y. Let us denote the loss function as
and the optimal weight as
. The optimization can be formulated as follows:
4.16. Meta-Learning Training Objective
The meta-learning training objective can be formulated as an optimization problem. Given a meta-training dataset
D consisting of multiple tasks
T, the aim is to determine the optimal model parameters:
4.17. Update Rule
The meta-update rule, which updates the model parameters based on the gradients computed from the task loss, can be represented as follows:
4.18. Meta-Model Testing Phase
After the meta-training phase, the learned model can be utilized for meta-testing on new tasks. Given a new task
, the model parameters can be fine-tuned using a small number of task-specific samples to adapt the model. The updated parameters can be obtained through the application of the meta-update rule:
6. Results and Evaluation
The evaluation of the proposed approach included metrics such as accuracy, precision, recall, F1-score, and support.
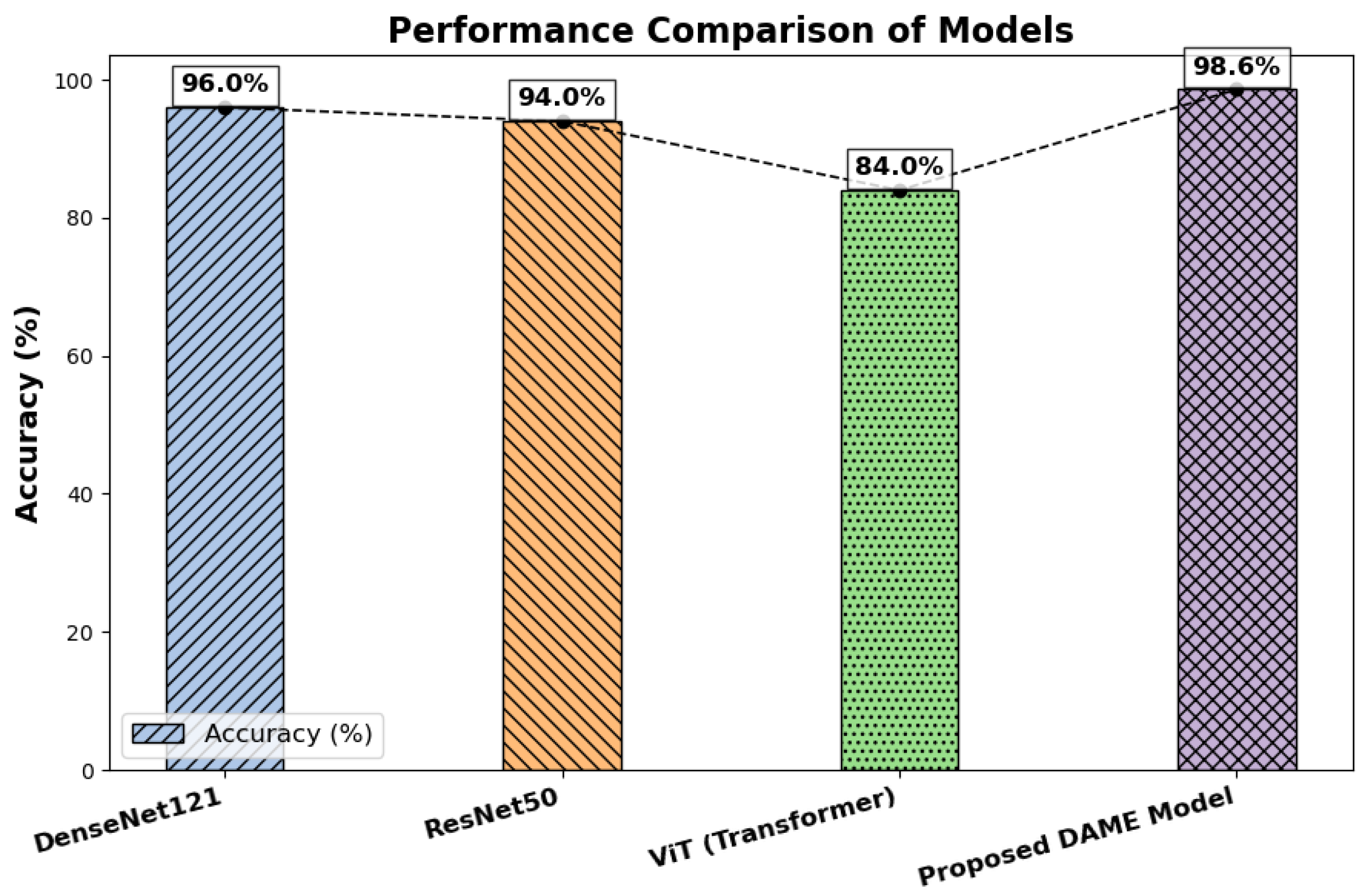
Table 6 presents a summary of the results from comparative classifications using DenseNet121, ResNet50, the Vision Transformer, and the proposed meta-learning-based model. The findings have shown that the meta-learning model consistently outperforms all baseline models in terms of accuracy, precision, recall, and F1-score, with the accuracy rates of DenseNet121, ResNet50, Vit, and the meta-learning model being 96%, 94%, 82%, and 98%, respectively. In contrast, the accuracy of the ensemble technique in the meta-model is significantly higher than that of the individual models. These metrics assess several model performance metrics. While precision compares the proportion of correctly predicted positive cases to the total number of optimistic predictions, accuracy provides a broad measure of how accurately the model makes its predictions. In contrast, recall assesses the ratio of correctly predicted positive instances to the total number of positive samples. Moreover, the F1 score calculates the balanced average of precision and recall using the harmonic mean.
Figure 6 presents a comparison of test accuracy between the baseline models and the proposed model.
6.1. Cross-Validation Results
A paired
t-test was conducted to determine whether the performance differences between the DenseNet121 and ResNet50, ViT, and the proposed DAME model were statistically significant, as shown in
Table 7. The results of the tests indicated that
p = 0.01081 against DenseNet121 ResNet50,
p = 0.02904 against DenseNet121 ViT,
p = 0.09561 against ResNet50 ViT, and
p = 0.09561 against ViT DenseNet121. DenseNet121 was significantly different compared to both ResNet50 and ViT (
p < 0.05), showing that there was a significant difference in the performance of DenseNet121. The significance of the differences between ResNet50 and ViT and DAME was not significant (
p > 0.05), and such results indicate that the levels of their performance were similar. Even though DAME and ViT were comparable in terms of accuracy, DAME produced more reliable results in folds, was also more robust, and represented disease-related features more understandably. Generally, DAME showed robust generalization and practical interpretability, thus making it a robust and useful model to classify skin lesions in conditions of low-resolution imaging, imbalance in classes, and limited data.
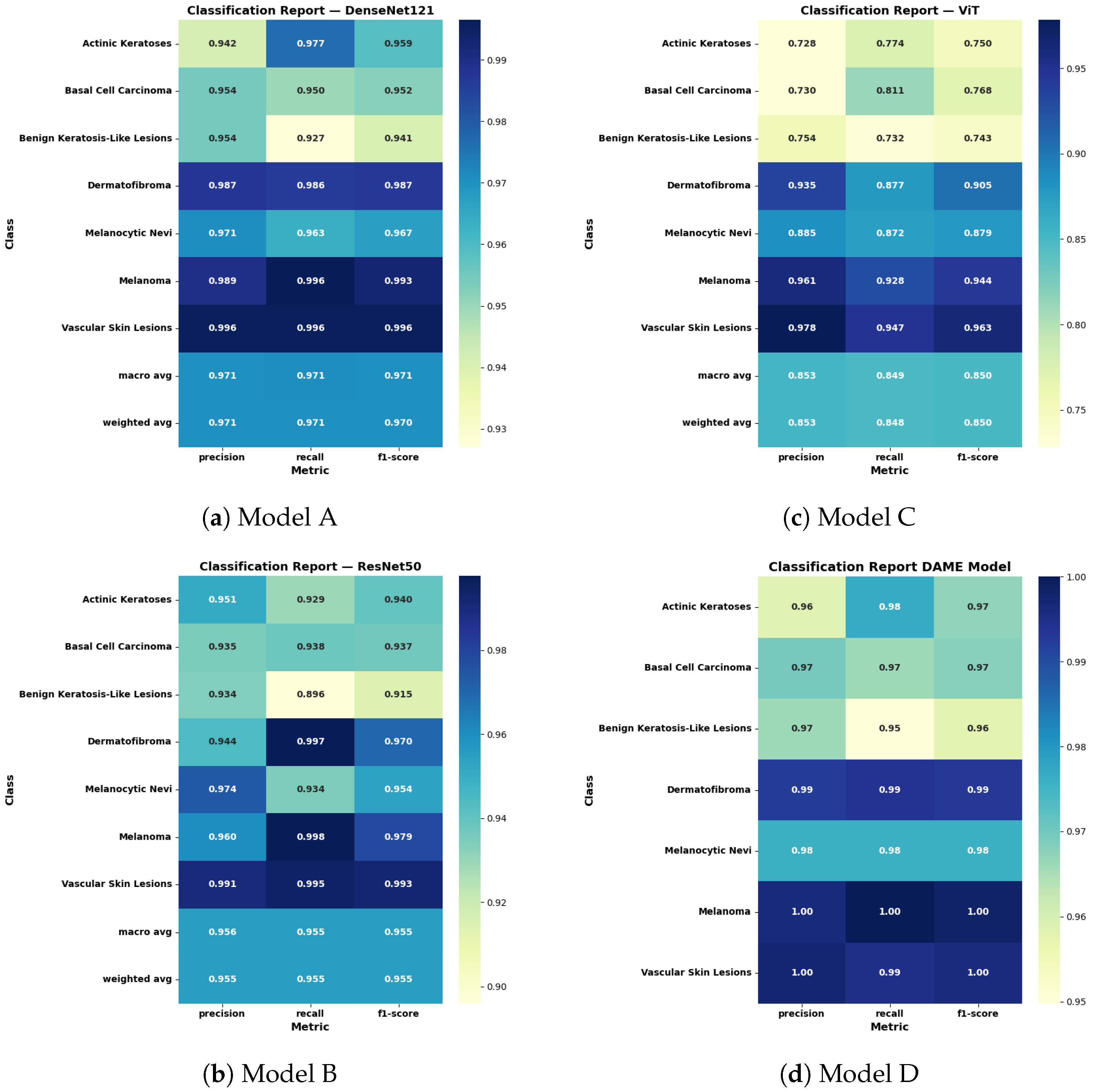
6.2. Classification Report
To comprehensively assess the performance of deep learning architectures in multi-class skin cancer classification, a detailed quantitative evaluation was performed using the classification reports of DenseNet121, ResNet50, the Vision Transformer (ViT), and the proposed Ensemble Model as shown in
Figure 7.
The DenseNet121 architecture that has an attention mechanism has an F1 score of 0.971 on macro and weighted averages, which indicates good generalization and reproducible results between the lesions of various types. However, its sensitivity on benign keratosis-like lesions was relatively minor, implying that it is biased towards under-detection of the lesion.
The ResNet50 model had a slightly lower F1 score of 0.955, but it was highly accurate and precise on specific categories, and especially on melanoma and dermatofibroma, with an F1 score of 0.979 and 0.970, respectively. Nonetheless, it showed weaknesses in identifying basal cell carcinoma and benign lesions, with recall values of 0.937 and 0.915, respectively.
The Vision Transformer achieved comparable accuracy, recording macro and weighted F1-scores of 0.950. It demonstrated stable classification performance for melanoma (F1 score: 0.944) and dermatofibroma (F1 score: 0.905), but showed reduced specificity for actinic keratoses and diminished recall for basal cell carcinoma. These variations are primarily attributed to the transformer’s limited spatial hierarchy and its susceptibility to class imbalance within the dataset.
The proposed DAME model, which combines the predictions of the architectures, significantly outperformed the individual models by achieving a macro and weighted F1-score of 1.00. It had almost perfect recollection of serious types of lesions, including melanoma and vascular skin lesions (1.000, 1.000). It enhanced the accuracy and precision of all other categories, including those that are initially hard to categorize, like benign keratosis-like lesions (precision: 0.97, recall: 0.95). These results may partially reflect dataset-specific characteristics and inherent biases in the HAM10000.
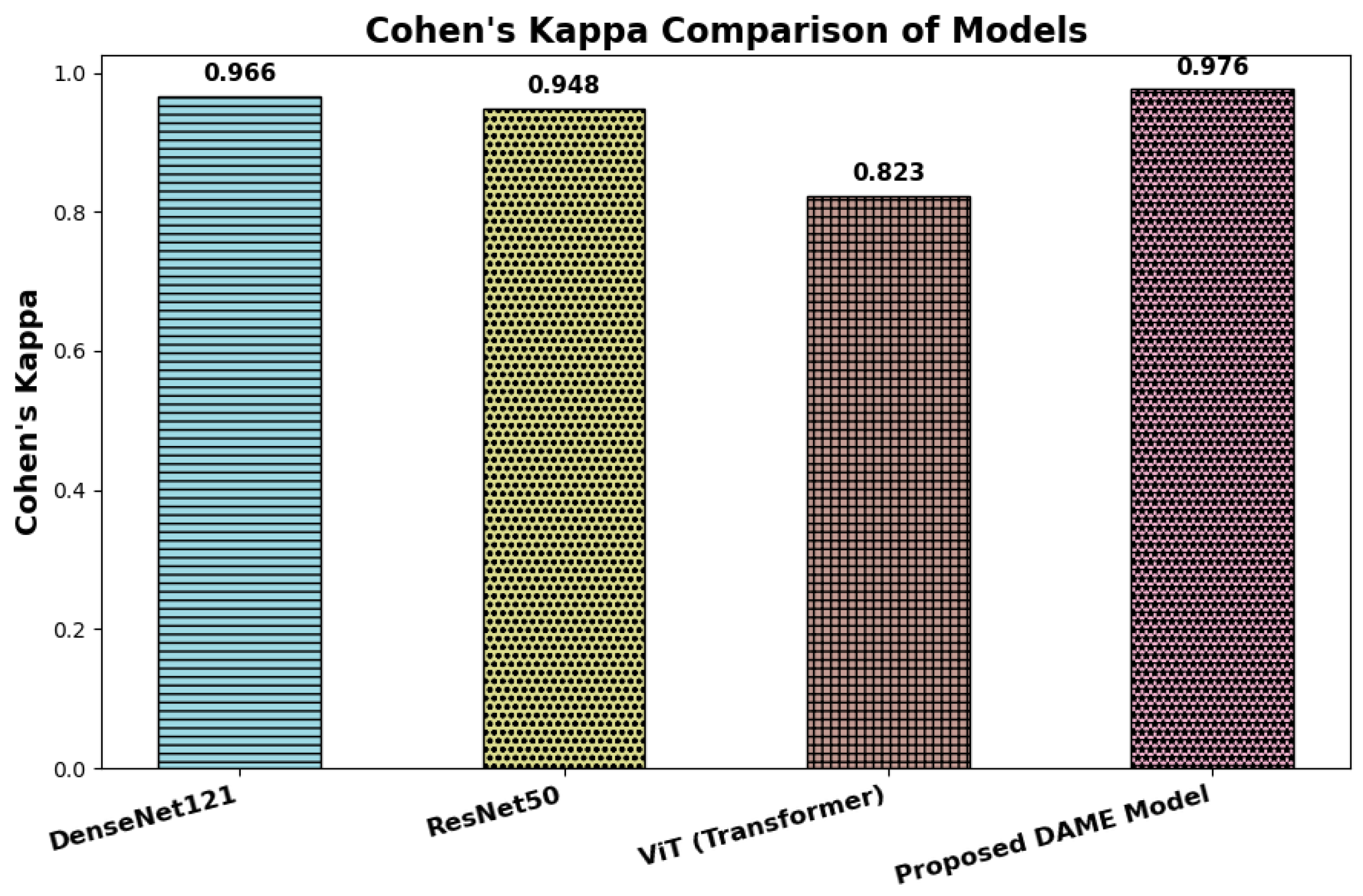
6.3. Cohen’s Kappa
Cohen’s Kappa presents a comparative evaluation of inter-rater agreement across four deep learning architectures: DenseNet121, ResNet50, Vision Transformer (ViT), and the proposed DAME model, based on their respective Cohen’s Kappa (κ) values. Cohen’s Kappa is a robust reliability coefficient that quantifies the degree of agreement between predicted and ground truth labels while accounting for agreement expected by chance. Unlike overall accuracy, Cohen’s Kappa provides a more dependable measure of consistency, particularly in multi-class and imbalanced classification scenarios common in clinical image analysis.
The most valuable model is the DAME architecture, with a value of 0.976, indicating a high degree of consistency in classification performance. It indicates that not only does DAME produce plausible predictions, but it also demonstrates a substantial correlation with the actual class labels, which is crucial in the organization of medical necessity, including dermatological diagnosis. DenseNet121 and ResNet50 follow, with the largest values of 0.966 and 0.948, respectively, indicating a high level of consistency and reliable classification results. The score of Vision Transformer (ViT) of 0.823, however, falls in the category of the nearest-to-perfect agreement, and thus denies stronger levels of alignment. This possible reduction may be explained by the fact that ViT uses more global contextual representations rather than localized characterizations in dermoscopic imaging. In general, the comparison reveals that all four architectures are highly concordant with reference labels; nevertheless, the DMAGE model is more stable and reliable in the classifications. These results confirm the usefulness of its hybrid nature, which combines local feature extraction, multi-resolution representations, and global attention based on transformers to produce better results in medical image interpretation, as shown in
Figure 8.
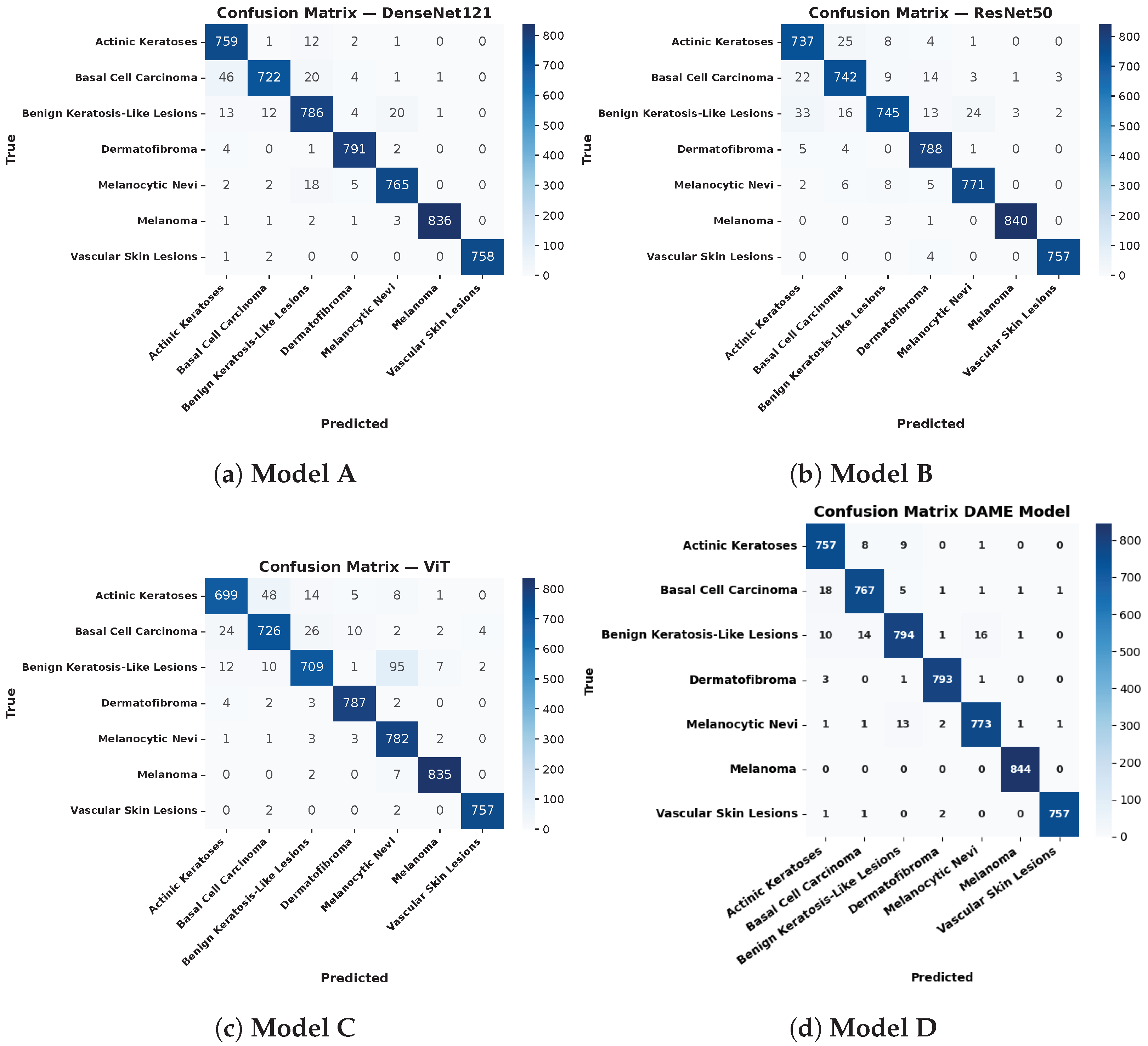
6.4. Confusion Matrix Analysis
The performance of four deep learning models DenseNet121 with the Convolutional Block Attention Module (CBAM), ResNet50 integrated with CBAM, the Vision Transformer (ViT) combined with CBAM, and the proposed DAME configuration was comprehensively evaluated using confusion matrices across seven clinically significant dermatological classes.
The DenseNet121 model, enhanced with CBAM, achieved high diagnostic accuracy in identifying melanoma (836 out of 840 correctly classified) and vascular skin lesions (758 out of 760 correctly classified). Nonetheless, it displayed significant misclassifications among morphologically similar benign classes, especially between Benign Keratosis and Melanocytic Nevi, which suggests considerable overlap in features between the two types of lesions.
The CBAM ResNet50 model showed better inter-class stability in all types of lesions. It was more accurate in classifying Melanoma (840 correctly classified), Benign Keratosis (745 out of 840), and Dermatofibroma (788 out of 800), with misclassification rates on Melanocytic Nevi and basal cell carcinoma (BCC) significantly lower than those obtained with DenseNet121. The CBAM-integrated ViT also achieved competitive performance, correctly classifying Dermatofibroma (787 out of 800) and Vascular Skin Lesions (757 out of 760). Nevertheless, it faced challenges in distinguishing Benign Keratosis from Melanocytic Nevi, primarily due to the transformer’s data-intensive nature and its lack of inherent spatial inductive bias, which makes it more sensitive to intra-class variability.
One of the main findings is that the ensemble model, which combines the predictions of the underlying architectures, yields the best classification results across all lesion types. It showed the improved performance of Melanoma (all 844 samples), Benign Keratosis (794/840), and Actinic Keratoses (757/800). The significant decrease in misclassifications in the confusion matrix has emphasized the usefulness of ensemble learning in reducing the performance shortcomings of single models and improving generalizability. This strategy effectively capitalizes on the synergistic advantage of convolutional and attention-based architectures, which are involved in the robust and accurate classification of skin cancer in terms of multiple classes (which is the objective of the evaluation).
Figure 9 demonstrates the analysis of the confusion matrices of the baseline and DAME proposed models.
Figure 9 shows the evaluation of confusion matrices for the baseline and DAME-proposed models.
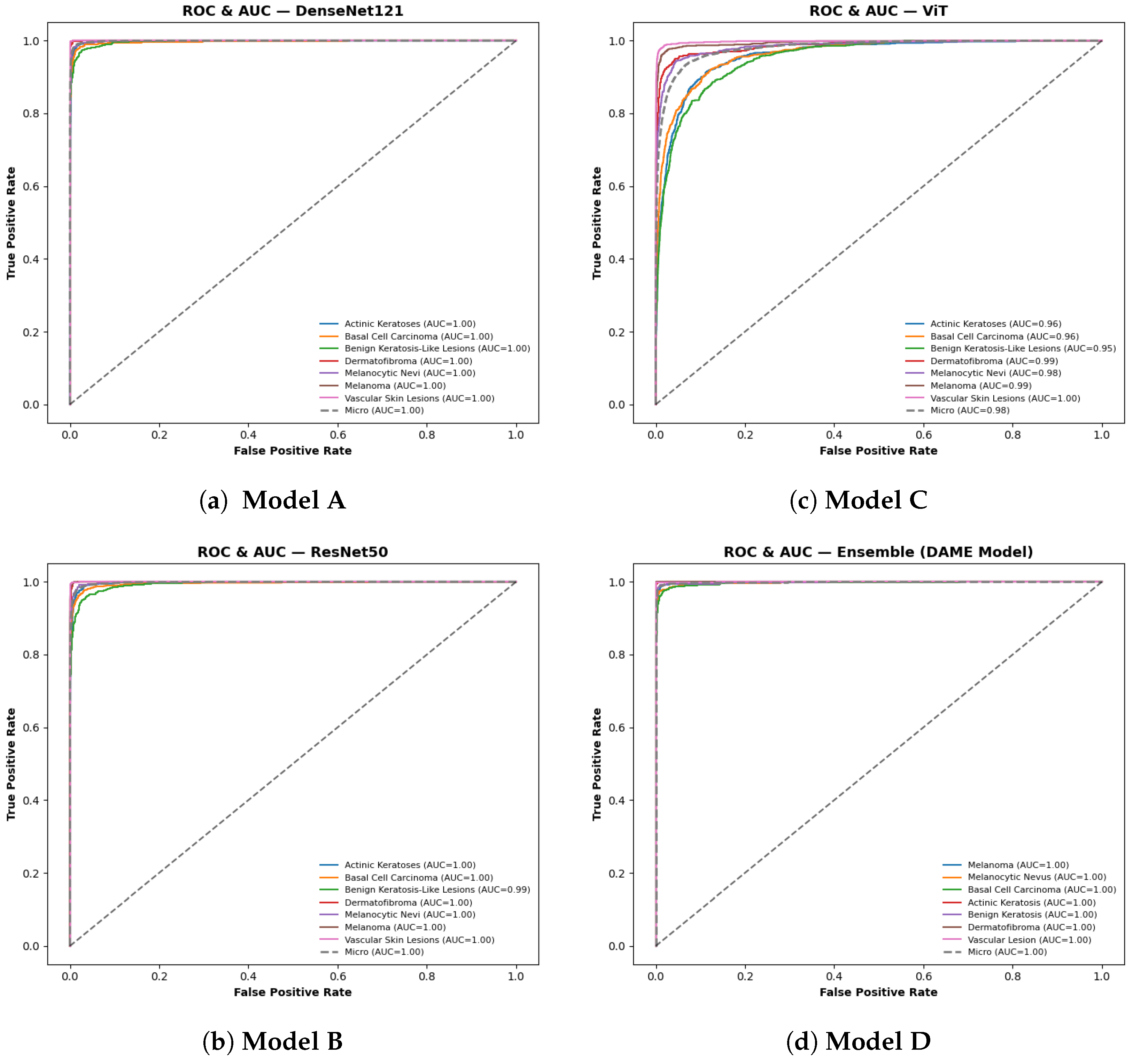
6.5. ROC and AUC Curve Analysis
The ROC AUC evaluation illustrates the models’ validation performance in multi-class dermatological classification. Across all assessments, the macro-averaged ROC curves demonstrated near-perfect class separability, with both DenseNet121 and the proposed DAME ensemble achieving the highest AUC value of 1.00. ResNet50 and the Vision Transformer (ViT) followed closely, with AUC values of 1.00 and 0.98, respectively. The micro-averaged AUC results supported this trend, confirming strong predictive consistency across heterogeneous lesion categories. There was a strong performance in per-class analysis. Dermatofibroma, Melanoma, and Vascular Skin Lesions were able to achieve perfect classification (AUC = 1.000), whereas Actinic Keratoses and basal cell carcinoma had slightly lower but still perfect results of between 0.990 and 0.998.
Figure 10 shows the evaluation of ROC via AUC for the baseline and DAME proposed models.
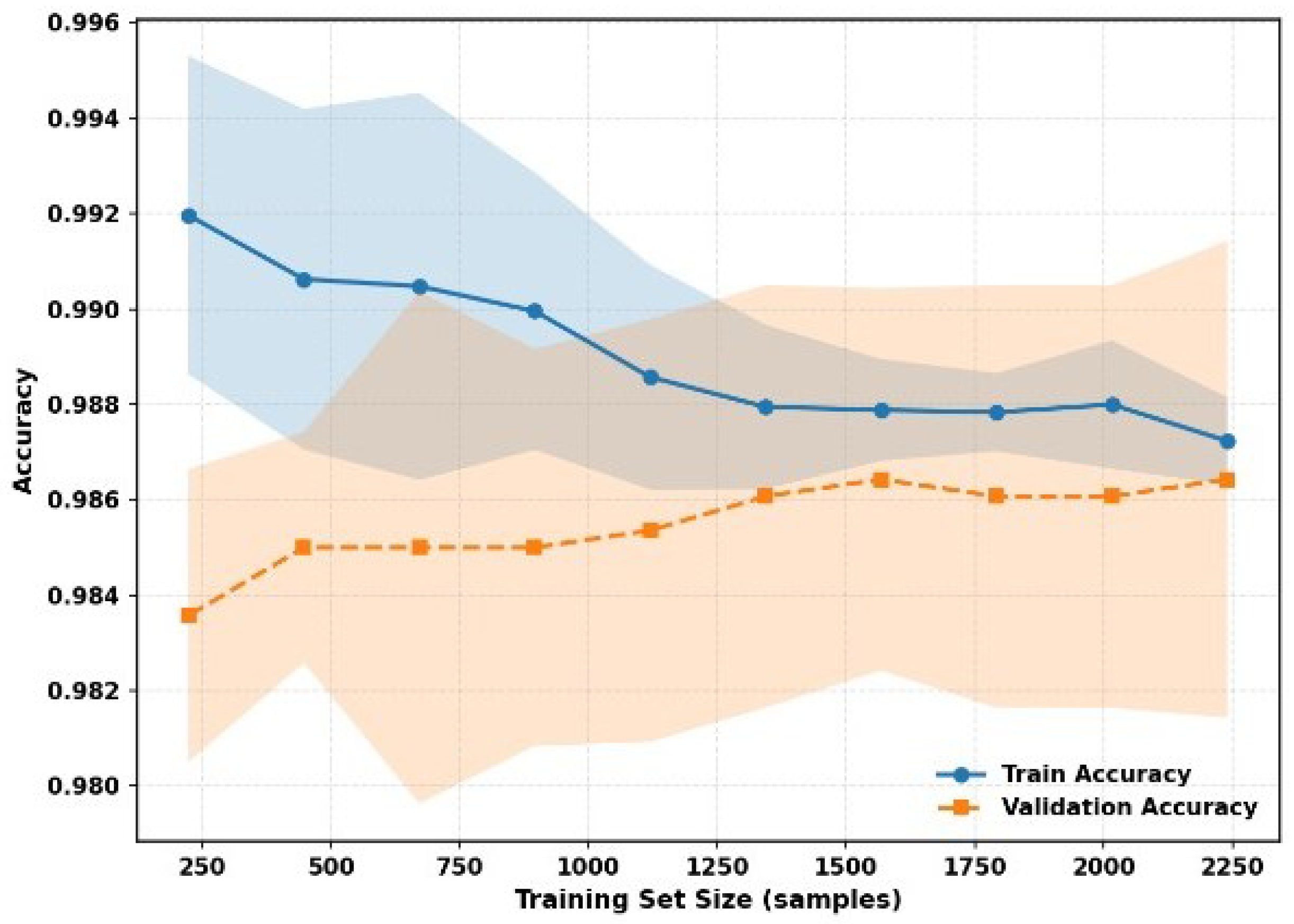
6.6. Training/Validation Accuracy with the DAME Model
The Model Learning Dynamics and Generalization Performance presents a comparative evaluation of training behavior and generalization capability across four architectures: DenseNet121, ResNet50, the Vision Transformer (ViT), and the ensemble-based DAME framework. The assessment includes an analysis of training and validation loss curves, accuracy progression over epochs, and model behavior under varying dataset scales.
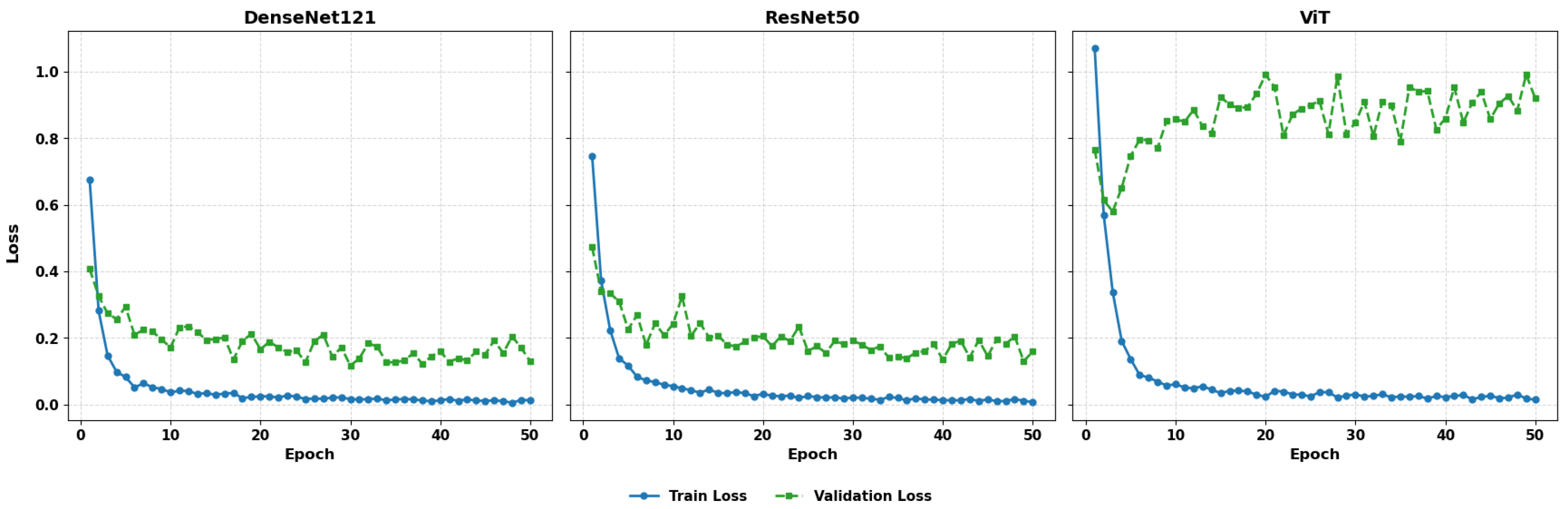
Figure 11 illustrates the comparative training and validation loss plots for DenseNet121, ResNet50, and ViT.
The three models exhibit whipped loss minimization at the initial stages of training indicating the efficient optimization. DenseNet121 has consistent and well synchronized curves of validation loss, showing consistent generalization. ResNet50 also demonstrates successful learning with average changes in the validation loss. Conversely, in contrast to the ViT model, which is able to decrease training loss steadily, its validation curve shows a significant degree of fluctuation, which suggests its susceptibility to noise in the data or lack of an adequate inductive bias. This instability can be a problem to predictive consistency, especially in medical imaging problems that demand fine-grained feature identification.
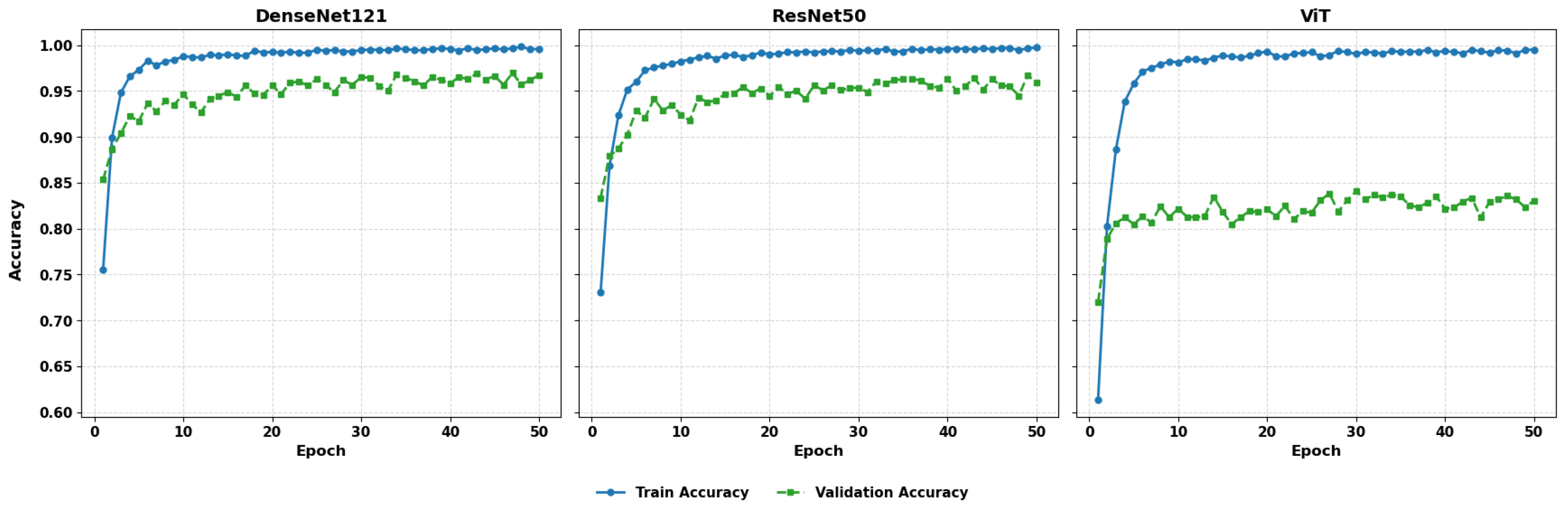
Figure 2 examines the training and validation accuracy of the 50 epochs.
Figure 12 presents the training and validation accuracy with 50 epochs. The DenseNet121, ResNet50, and ViT models demonstrate high training accuracy, which reaches nearly 100% after the initial ten epochs, and their validation accuracy increases to 96–98%. DenseNet121 and ResNet50 have a small margin in terms of training and validation performance, which represents good generalization. On the other hand, ViT has been shown to have greater variability in validation accuracy, although training accuracy is close to perfect, suggesting possible overfitting or a lack of regularization. These findings suggest that, although ViT demonstrates high representational capacity, additional tuning or architectural adaptation may be required to achieve consistent performance.
In
Figure 13, the decreasing distance between the training and validation curves, as well as the less variable tendency in terms of the lower confidence intervals, points to the idea that the DAME framework can be scaled with the increase in the size of the dataset, preserving accuracy and consistency. This strength is due to the fact that it has a hybrid architecture, localized dense features, multi-resolution representations, and global transformer-driven attention, and thus, it can be easily adapted to both small and large data regimes with a strong adaptive capability with minimal overfitting.
Altogether, DenseNet121 and ResNet50 can be characterized by a strong basis, which is equally effective. However, the ViT has lower generalization stability, and the most efficient and sound framework is the DAME. It is a viable validation method, offering scalable performance with balanced learning dynamics, regardless of the dataset size. In general, the DenseNet121 and ResNet50 can be characterized by a strong basis, which is equally effective. However, the ViT has lower generalization stability, and the most efficient and sound framework is the DAME. It is also highly viable in terms of validation and scalable in terms of performance and balanced learning dynamics, regardless of dataset size.
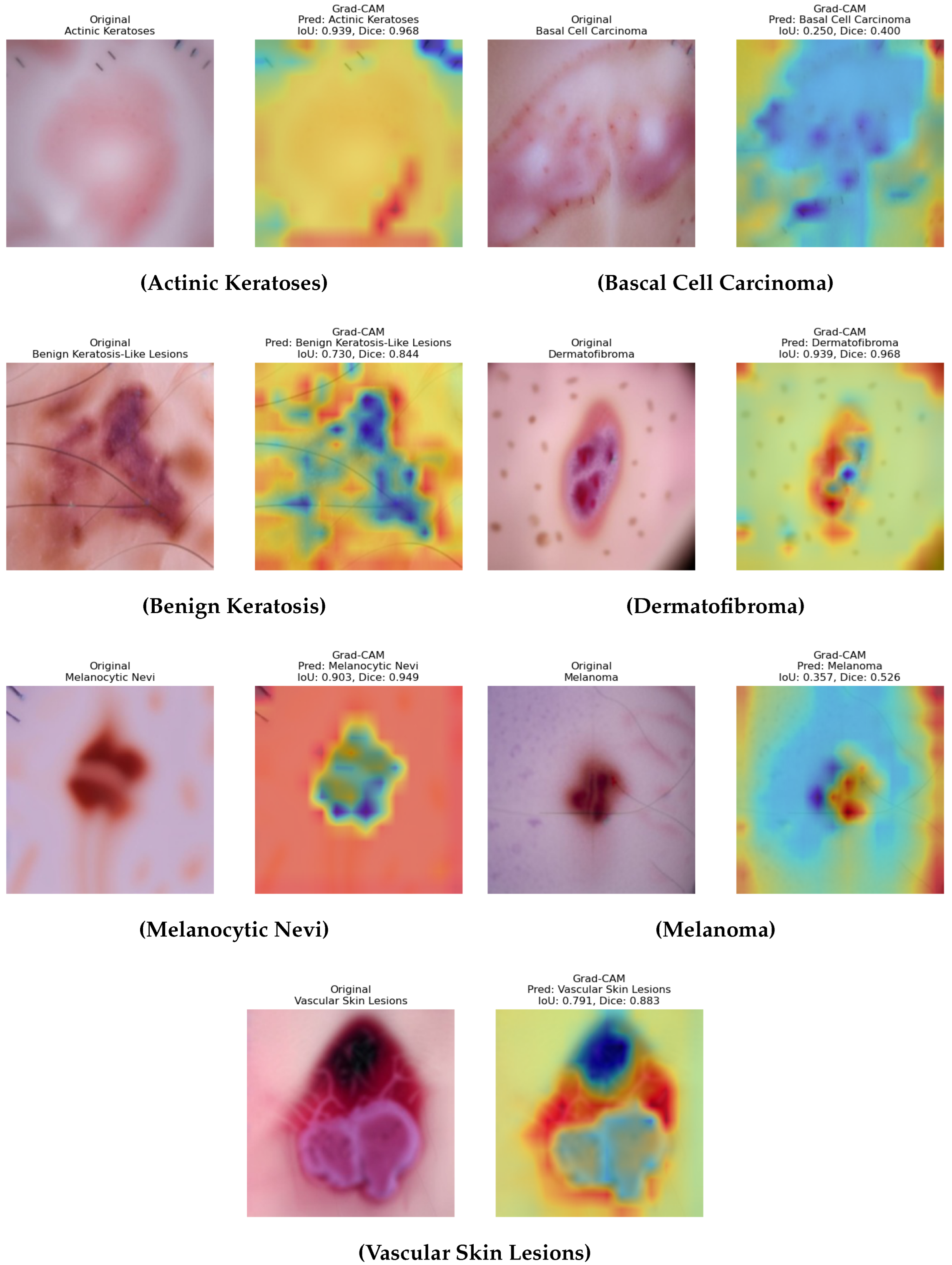
6.7. Explainability Analysis
The interpretability of the DAME framework is demonstrated by comparing original dermoscopic images with their corresponding Grad CAM activation maps, predicted diagnostic categories, and attention weight distributions as shown in
Figure 14. These values represent the contributions derived from the model’s three specialized attention components: the dense connection module, the multi-scale convolutional stream, and the transformer-based global attention unit. This visualization evaluates the extent to which spatially aligned and diagnostically relevant patterns support the model’s predictions. For actinic keratoses and basal cell carcinoma, the model appropriately attends to lesion-specific regions such as clustered pink zones and peripheral asymmetries, with balanced weight distributions that demonstrate integrated feature usage. Similarly, in the case of benign keratosis-like Lesions and dermatofibroma, the model demonstrates roughness areas and nodular states, respectively, which implies that localization of the model and the contribution made by each branch can be interpreted and are proportional. In the Melanocytic Nevi case, the heatmap highlights the central pigment area, and the insignificant noise, like hair, is suppressed, hence showing successful learning of saliency. The melanoma case shows that the weights of attention are skewed. Such an imbalance implies the dependence on high-resolution and global contextual features, which are consistent with the clinical complexity of melanoma. A role of dynamic attention adaptation depending on the type of lesion is also highlighted in the analysis. In the case of vascular skin lesions, the visualization exhibits activation around uniform weights around circular vessel structures, again, with uniform feature integration.
Overall, these visualizations support the explanation of the framework DAME, as they indicate that the diagnostic cues are correlated with the mechanisms of attention. Additionally, the relative stability of the activation maps, along with their morphological properties and the weight capability, highlights the key concepts of explainable AI in medical imaging, as exemplified by the framework’s ability to adjust its focus according to a region-specific shape. The process is especially important in distinguishing the small differences between classes in cases where categories are literally indistinguishable in appearance, such as an abnormal nevus and a small melanoma tumor. In addition to making the concept easier, this adaptive saliency provides greater confidence that it can be used practically in the clinical setting. The DAME methodology uses interpretable visual feedback to fill the gap between reason-based reasoning and inference based on facts. This observation also demonstrates that the model has high potential for generalizing lesions of varying textures, shapes, and pigmentation patterns. The DAME framework addresses the issue of black-box images, which are commonly assumed to be part of deep learning models, by aligning the learned attention maps with interpretable features to those of an expert. It is necessary to mention that the localization of attention with fine spatial fidelity is in agreement with clinical indicators, i.e., asymmetry, irregularity of the border, color heterogeneity, and the most striking indicators in dermatological diagnosis. The hierarchical structure of the model, particularly in complicated cases involving overlapping classes or a question mark, is well incorporated, as it is both globally uniform and locally sensitive. Such insights may not only enhance interpretability but also lay the foundations for a clinician–AI collaboration in diagnostic processes. This leads to an improvement in the technical reliability and ethical accountability of AI-aided medical imaging due to the high explicability of the DAME design.
6.8. Grad CAM Quantitative Evaluation
Quantitative measures were calculated using Grad CAM to measure the level of model focus on meaningful image locations, which consisted of the Mean Intersection over Union (IoU), Mean Dice coefficient, Mean Average Drop, and Average Increase Rate. The results were as under: Mean IoU = 0.701, Mean Dice = 0.791, Mean Average Drop = 0.094, and Average Increase rate = 0.429. The Mean ioU of 0.701 and the Mean Dice of 0.791 show that the Grad CAM heatmaps are nearly similar to the annotated regions of the lesions in the images, and the model can identify the areas that have clinical interest. The insignificance of Mean Average Drop (0.094) implies that the removal of these regions will have a low effect on decrease classification confidence, which means that the attention maps are significant discriminating features. The positive Averages of 0.429 equally demonstrate that confidence in prediction increases with the focus on the areas highlighted. In general, these findings indicate that the Grad CAM visualizations produced by the suggested model are accurate, interpretable, and patient-specifically meaningful, which in turn implies the idea that the model can effectively prioritize areas of interest that are pertinent to the diagnosis in the process of skin lesion classification.
8. Limitations and Future Work
Although the proposed DAME Model has positive outcomes, it has multiple limitations. The existing assessment is limited to distinct image datasets of cancer; thus, additional validation is necessary to determine its applicability to other illnesses, to other imaging scales (including CT and PET), and to multiple-center datasets. Additionally, a multi-module design makes the computation more expensive, which makes the model less suitable in a clinical environment that is limited by resources. Furthermore, the current datasets are clear and well-marked, whereas in reality, hospital data is often noisy, heterogeneous, and ambiguous. Future work will focus on six areas:
(1) Developing both the imaging and modeling aspects of skin cancer diagnosis remains a key research focus. Modern imaging techniques, including multiphoton lifetime tomography, confocal microscopy, optical coherence tomography, and photoacoustic imaging, offer high-resolution structural and biochemical information. (2) Developing lightweight versions of DDPM to make them more efficient to deploy. (3) Generalizing the model to multi-modal and multitask learning conditions to improve generalization. (4) Including real-life clinical feedback into an end-to-end training pipeline to make it more practical. (5) Evaluating the model with real-world clinical datasets that are noisy, inconsistently annotated, have imaging artifacts, and inter-patient variation. (6) Refining domain-adaptive DDPM versions for improved reliability and practical evaluation. These efforts aim to develop more robust models and more accurately determine the utility of such models in clinical practice.
9. Conclusions
In this work, we present a novel framework that addresses some of the most challenging issues in medical image analysis, including class imbalance, data diversity, and the need for high classification accuracy. Our method starts by creating high-quality synthetic medical images using a Denoising Diffusion Probabilistic Model (DDPM). The model gradually transforms noisy input images into realistic ones. When doing this, it helps establish balanced classes and improve generalization by increasing the quantity of training data available, as well as its diversity and quality.
A DAME (Diffusion-Augmented Meta-Learning Ensemble) framework is then included. This component combines meta-learning techniques that adapt to different tasks with the advantages of convolutional networks, which excel at capturing local details. The ensemble is particularly adept at identifying complex medical features across all imaging modalities, as it combines several task-specific learners to produce both fine-grained feature extraction and robust generalization. Our methodology integrates data augmentation, feature extraction, and classification into a straightforward end-to-end process, as opposed to conventional approaches that treat these tasks as separate steps. The proposed DAME framework improves the accuracy and dependability of the final predictions, while the DDPM enriches the dataset with realistic synthetic samples.
Our approach continuously outperforms state-of-the-art techniques in tests, demonstrating improved diagnostic precision, stronger robustness, and more distinct feature separation. As a result, radiologists and pathologists find it to be a valuable tool, particularly for early screening and the identification of complex lesions. Simply put, our study paves the way for more precise, effective, and intelligent healthcare solutions by combining the adaptive learning capabilities of DAME-based Transformer Diffusion-Augmented Meta-Learning Ensemble. Our method advances the development of precise, efficient, and intelligent healthcare solutions.