UNETR++ with Voxel-Focused Attention: Efficient 3D Medical Image Segmentation with Linear-Complexity Transformers
Abstract
1. Introduction
- We identify the deficiency of UNETR++ in its spatial attention design, which relies on dimensionality reduction for computational efficiency but sacrifices feature fidelity.
- We propose a voxel-focused attention (VFA) mechanism that achieves linear complexity without dimensionality reduction, directly computing attention across voxels.
- We integrate VFA into the spatial branch of the EPA block in UNETR++, resulting in an enhanced model that reduces parameters by nearly 50% while maintaining competitive segmentation accuracy.
- We evaluate the proposed method on three widely-used benchmarks (Synapse, ACDC, and BRaTs), demonstrating its effectiveness across diverse medical imaging modalities.
2. Literature Review and Related Work
2.1. Convolution-Based Segmentation Methods
2.2. Transformer-Based Segmentation Methods
2.3. Hybrid Segmentation Methods
3. Methods and Techniques
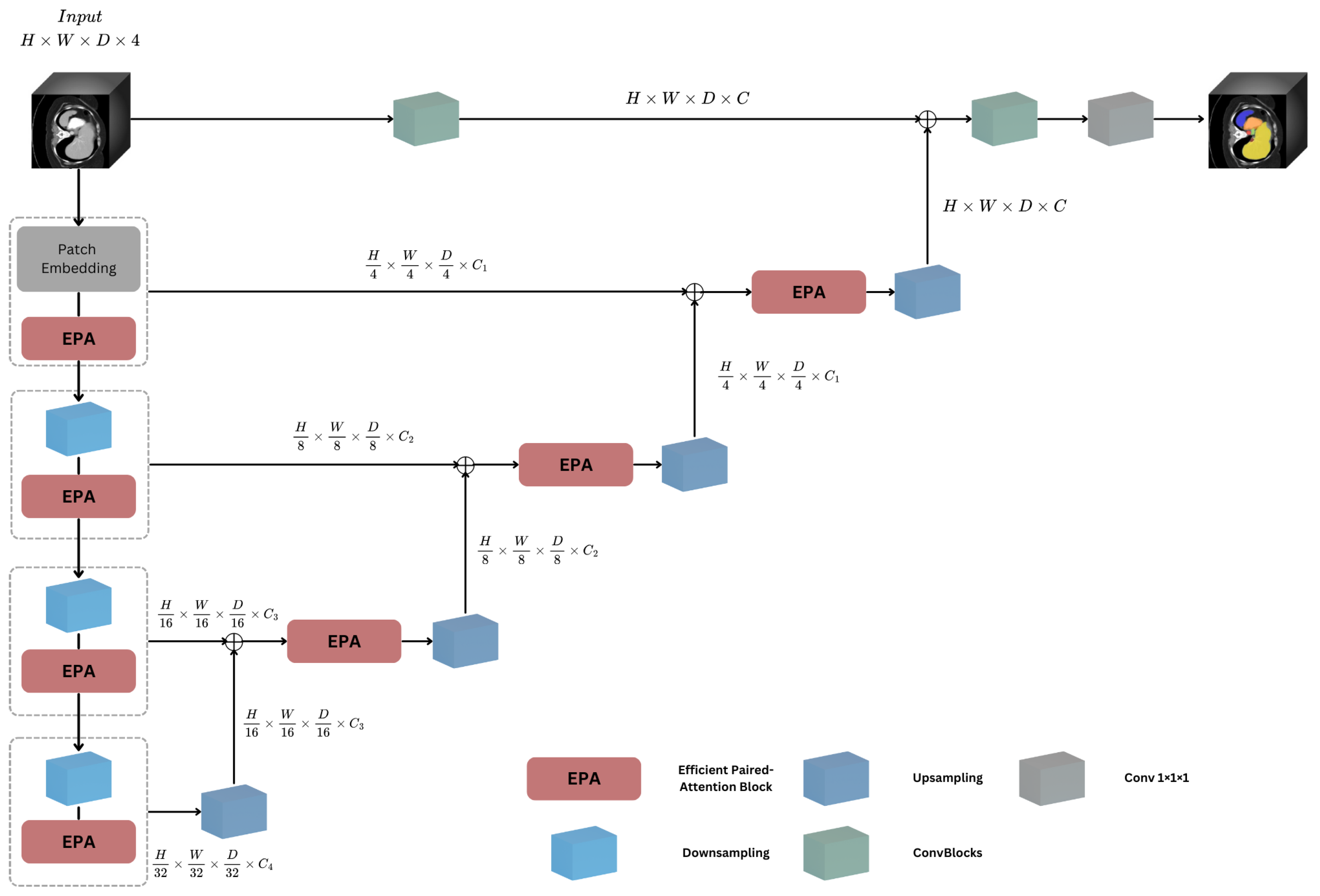
3.1. Overall Architecture
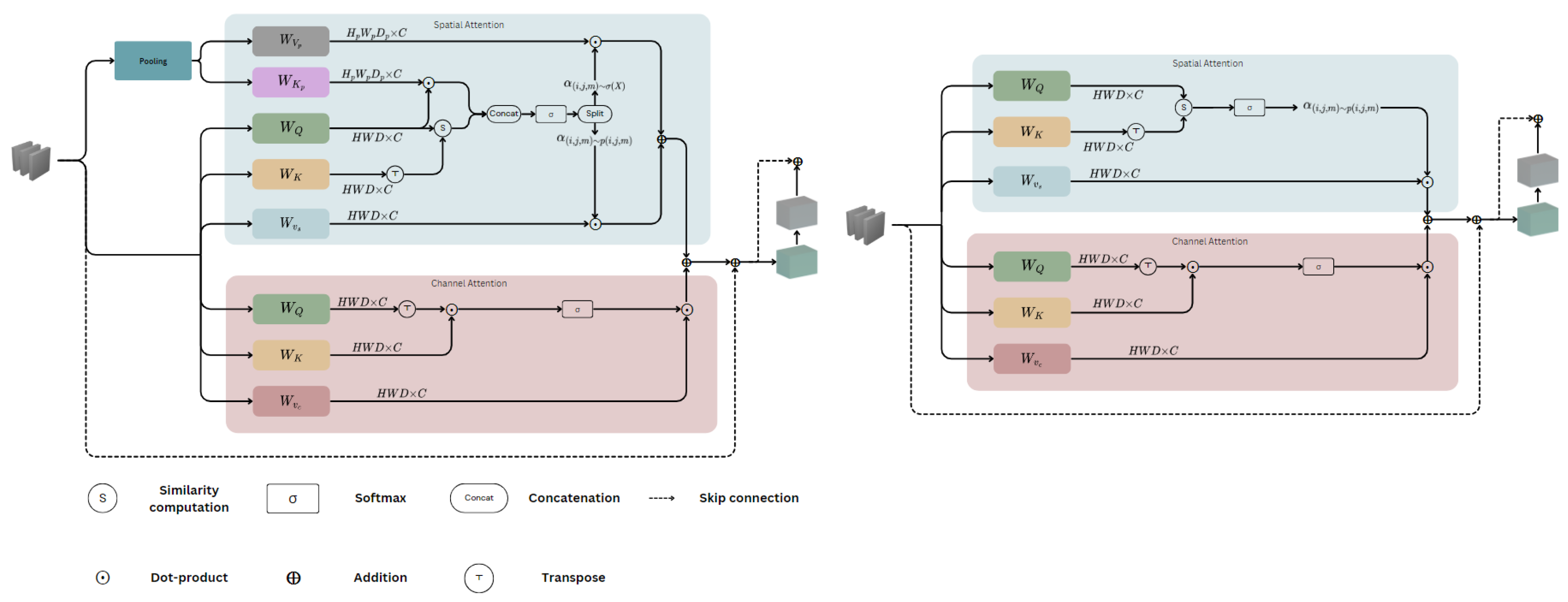
3.2. Voxel-Focused Attention (VFA)
Approaches to VFA
- Approach 1—VFA: This approach considers only the query-centred sliding window voxel-wise attention, without incorporating the pooling attention.
- Approach 2—Full VFA (FVFA): This approach extends the sliding window voxel-wise attention by including the pooling attention operation to capture additional contextual information.
- : Query-key similarity score;
- : Query vector at location ;
- : Query embedding;
- : Key vector at position ;
- : Attention weights;
- d: Dimension of the query and key vectors;
- : Learnable position bias;
- : Value vector.
- : Query-key similarity score;
- : Pooled query-key similarity score;
- : Attention weights;
- QE: Query embedding;
- : Pooled feature map;
- log−CPB: Log-spaced continuous position bias;
- : Relative coordinates;
- T: Learnable tokens.
3.3. Efficient Paired Attention Block
| Algorithm 1 VFA and FVFA UNETR++ Segmentation Pseudocode |
|
4. Experimental Results
4.1. Experimental Setup
- Synapse: This dataset consists of 30 abdominal CT scans with annotations for eight target organs: spleen, right kidney, left kidney, gallbladder, liver, stomach, aorta, and pancreas. Although its sample size is modest, Synapse remains valuable because it is publicly available, well curated, and frequently used for efficiency comparisons, enabling reproducibility and direct benchmarking against a large body of prior work.
- ACDC: The ACDC dataset contains 100 patients’ cardiac MRI images, with annotations for three anatomical structures: right ventricle (RV), left ventricle (LV), and myocardium (MYO).
- BraTS: This dataset includes 484 MRI images with four modalities (FLAIR, T1w, T1gd, and T2w). The dataset is annotated for peritumoral oedema, GD-enhancing tumour, and necrotic/non-enhancing tumour core.
- Synapse: In total, 18 samples were used for training, and 12 samples were used for evaluation.
- ACDC: The dataset was split into a 70:10:20 ratio for training, validation, and testing, respectively.
- BraTS: The data was split into an 80:5:15 ratio for training, validation, and testing, respectively.
- lr: Learning rate at the current epoch;
- initial_lr: Initial learning rate at the start of training;
- epoch_id: The current epoch number;
- max_epoch: The total number of epochs for training;
- 0.9: The exponent that controls the decay rate.
4.2. Comparison with State-of-the-Art Methods
4.2.1. Synapse Dataset
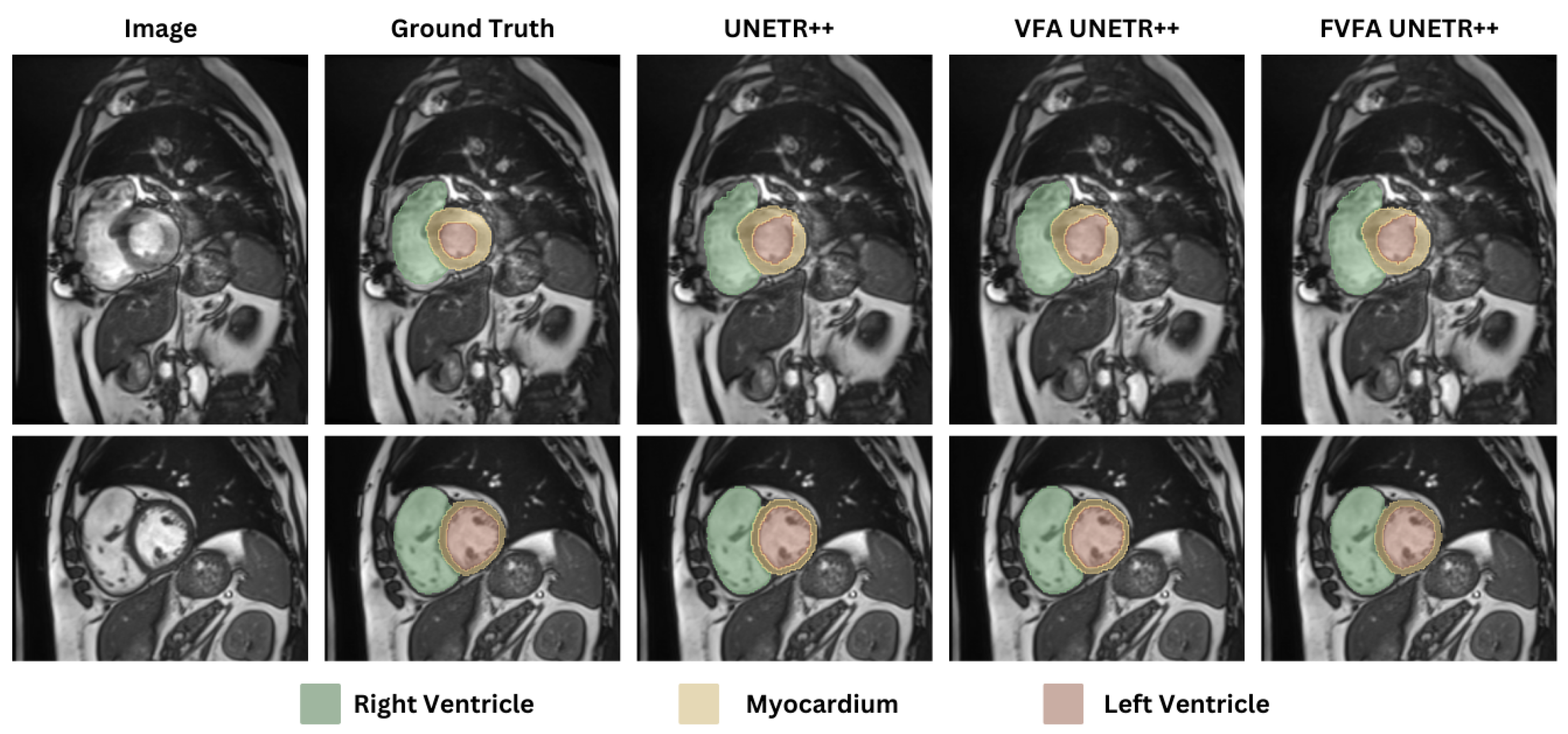
4.2.2. ACDC Dataset
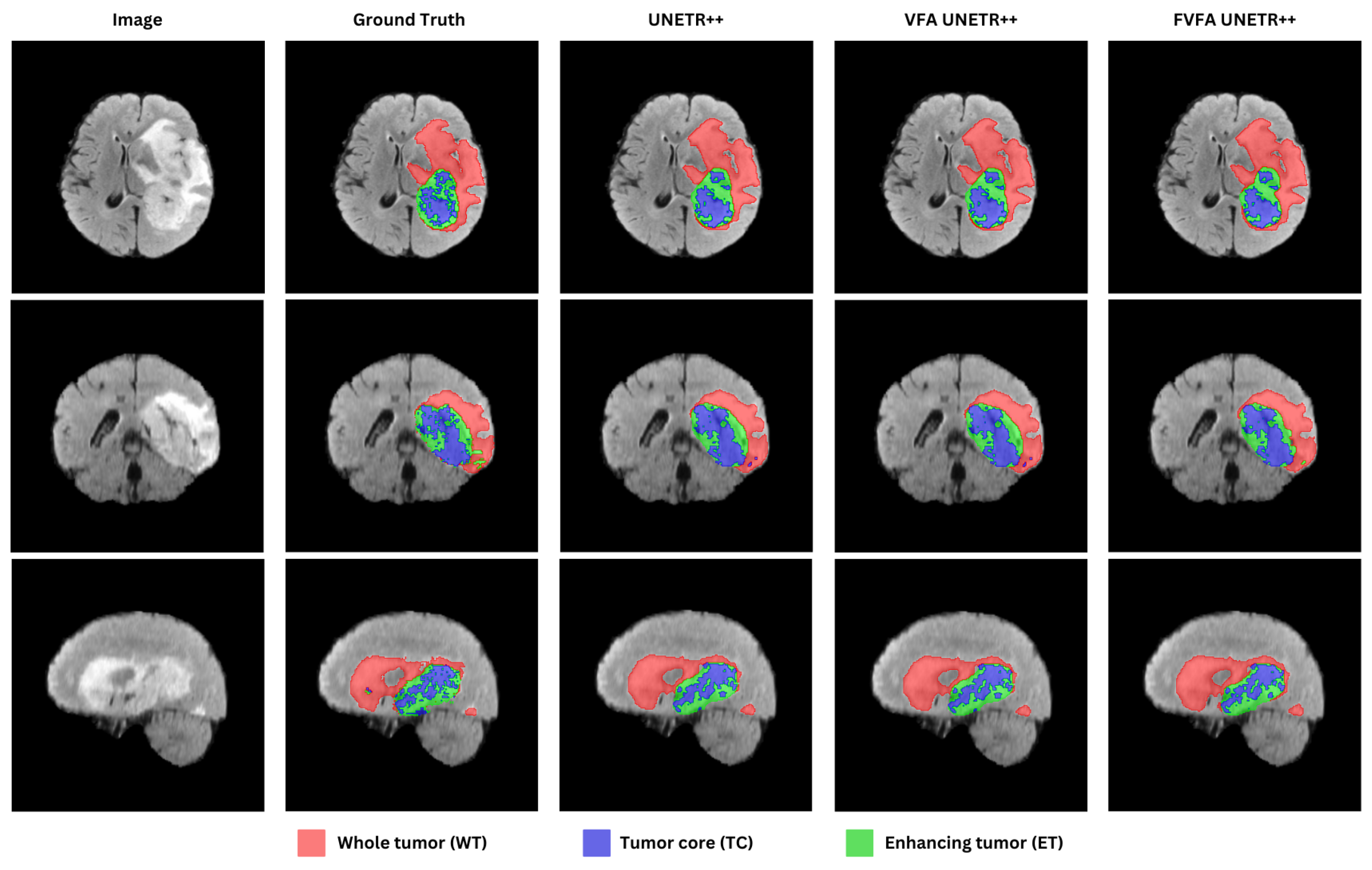
4.2.3. BraTs Dataset
5. Comparison of CUDA C++ and Native Python-Only Implementations
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ViTs | Vision Transformers |
| CNNs | Convolutional Neural Networks |
| VFA | Voxel-Focused Attention |
| FVFA | Full-Voxel-Focused Attention |
| EPA | Efficient Paired Attention |
| DADC | Dual Attention and Depthwise Convolution |
| QLV | Query-Learnable-Value |
| LQE | Learnable Query Embedding |
| DSC | Dice Similarity Coefficient |
| HD95 | Hausdorff Distance 95 |
| Spl | Spleen |
| RKid | Right Kidney |
| LKid | Left Kidney |
| Gal | Gallbladder |
| Liv | Liver |
| Sto | Stomach |
| Aor | Aorta |
| Pan | Pancreas |
| RV | Right Ventricle |
| LV | Left Ventricle |
| MYO | Myocardium |
References
- National Health Service. Magnetic Resonance Imaging (MRI) Scan; Retrieved from Health A to Z; National Health Service: London, UK, 2022.
- National Health Service. CT Scan; Retrieved from Health A to Z; National Health Service: London, UK, 2023.
- Padhi, S.; Rup, S.; Saxena, S.; Mohanty, F. Mammogram Segmentation Methods: A Brief Review. In Proceedings of the 2019 2nd International Conference on Intelligent Communication and Computational Techniques (ICCT), Jaipur, India, 28–29 September 2019; pp. 218–223. [Google Scholar] [CrossRef]
- Doi, K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef]
- Chen, Y.; Zhuang, Z.; Chen, C. Object Detection Method Based on PVTv2. In Proceedings of the 2023 IEEE 3rd International Conference on Electronic Technology, Communication and Information (ICETCI), Changchun, China, 26–28 May 2023; pp. 730–734. [Google Scholar] [CrossRef]
- Wang, W.; Xie, E.; Li, X.; Fan, D.P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. PVT v2: Improved baselines with Pyramid Vision Transformer. Comput. Vis. Media 2022, 8, 415–424. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. arXiv 2021, arXiv:2103.14030. [Google Scholar] [CrossRef]
- Yang, J.; Li, C.; Dai, X.; Gao, J. Focal Modulation Networks. In Advances in Neural Information Processing Systems; Koyejo, S., Mohamed, S., Agarwal, A., Belgrave, D., Cho, K., Oh, A., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2022; Volume 35, pp. 4203–4217. [Google Scholar]
- Dong, X.; Bao, J.; Chen, D.; Zhang, W.; Yu, N.; Yuan, L.; Chen, D.; Guo, B. CSWin Transformer: A General Vision Transformer Backbone with Cross-Shaped Windows. arXiv 2022, arXiv:2107.00652. [Google Scholar] [CrossRef]
- Wang, W.; Dai, J.; Chen, Z.; Huang, Z.; Li, Z.; Zhu, X.; Hu, X.; Lu, T.; Lu, L.; Li, H.; et al. InternImage: Exploring Large-Scale Vision Foundation Models with Deformable Convolutions. arXiv 2023, arXiv:2211.05778. [Google Scholar]
- Shi, D. TransNeXt: Robust Foveal Visual Perception for Vision Transformers. arXiv 2024, arXiv:2311.17132. [Google Scholar]
- Shaker, A.M.; Maaz, M.; Rasheed, H.; Khan, S.; Yang, M.H.; Khan, F.S. UNETR++: Delving into Efficient and Accurate 3D Medical Image Segmentation. IEEE Trans. Med. Imaging 2024, 43, 3377–3390. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2023, arXiv:1706.03762. [Google Scholar]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.A.; Cetin, I.; Lekadir, K.; Camara, O.; Gonzalez Ballester, M.A.; et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [PubMed]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Landman, B.; Xu, Z.; Igelsias, J.; Styner, M.; Langerak, T.; Klein, A. Miccai multi-atlas labeling beyond the cranial vault–workshop and challenge. In Proceedings of the MICCAI Multi-Atlas Labeling Beyond Cranial Vault—Workshop Challenge, Munich, Germany, 5 October 2015; Volume 5, p. 12. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; pp. 234–241. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar] [CrossRef]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.W.; Wu, J. UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation. arXiv 2020, arXiv:2004.08790. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016, Athens, Greece, 17–21 October 2016; Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W., Eds.; pp. 424–432. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. arXiv 2018, arXiv:1807.10165. [Google Scholar]
- Angermann, C.; Haltmeier, M. Random 2.5 d u-net for fully 3d segmentation. In Proceedings of the Machine Learning and Medical Engineering for Cardiovascular Health and Intravascular Imaging and Computer Assisted Stenting: First International Workshop, MLMECH 2019, and 8th Joint International Workshop, CVII-STENT 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, 13 October 2019; Proceedings 1. Springer: Berlin/Heidelberg, Germany, 2019; pp. 158–166. [Google Scholar]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S.; et al. nnU-Net: Self-adapting Framework for U-Net-Based Medical Image Segmentation. arXiv 2018, arXiv:1809.10486. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. arXiv 2022, arXiv:2201.03545. [Google Scholar] [CrossRef]
- Roy, S.; Koehler, G.; Ulrich, C.; Baumgartner, M.; Petersen, J.; Isensee, F.; Jaeger, P.F.; Maier-Hein, K.H. Mednext: Transformer-driven scaling of convnets for medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Daejeon, Republic of Korea, 23–27 September 2023; Springer: Berlin/Heidelberg, Germany, 2023; pp. 405–415. [Google Scholar]
- Li, H.; Nan, Y.; Yang, G. LKAU-Net: 3D Large-Kernel Attention-Based U-Net for Automatic MRI Brain Tumor Segmentation. In Proceedings of the Medical Image Understanding and Analysis; Yang, G., Aviles-Rivero, A., Roberts, M., Schönlieb, C.B., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 313–327. [Google Scholar]
- Guo, M.H.; Liu, Z.N.; Mu, T.J.; Hu, S.M. Beyond self-attention: External attention using two linear layers for visual tasks. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 45, 5436–5447. [Google Scholar] [CrossRef]
- Huang, T.; Huang, L.; You, S.; Wang, F.; Qian, C.; Xu, C. Lightvit: Towards light-weight convolution-free vision transformers. arXiv 2022, arXiv:2207.05557. [Google Scholar]
- Karimi, D.; Vasylechko, S.D.; Gholipour, A. Convolution-Free Medical Image Segmentation Using Transformers. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, Strasbourg, France, 27 September–1 October 2021; de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 78–88. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar] [CrossRef]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. UNETR: Transformers for 3D Medical Image Segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 3–8 January 2022; pp. 574–584. [Google Scholar]
- Zhou, H.Y.; Guo, J.; Zhang, Y.; Yu, L.; Wang, L.; Yu, Y. nnformer: Interleaved transformer for volumetric segmentation. arXiv 2021, arXiv:2109.03201. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2022; pp. 205–218. [Google Scholar]
- Liu, Y.; Zhang, Z.; Yue, J.; Guo, W. SCANeXt: Enhancing 3D medical image segmentation with dual attention network and depth-wise convolution. Heliyon 2024, 10, e26775. [Google Scholar] [CrossRef]
- Huang, X.; Deng, Z.; Li, D.; Yuan, X. Missformer: An effective medical image segmentation transformer. arXiv 2021, arXiv:2109.07162. [Google Scholar] [CrossRef] [PubMed]
- Hatamizadeh, A.; Nath, V.; Tang, Y.; Yang, D.; Roth, H.R.; Xu, D. Swin unetr: Swin transformers for semantic segmentation of brain tumors in mri images. In Proceedings of the International MICCAI Brainlesion Workshop; Springer: Berlin/Heidelberg, Germany, 2021; pp. 272–284. [Google Scholar]
- Rahman, M.M.; Marculescu, R. Medical Image Segmentation via Cascaded Attention Decoding. In Proceedings of the 2023 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 2–7 January 2023; pp. 6211–6220. [Google Scholar] [CrossRef]
- Niknejad, M.; Firouzbakht, M.; Amirmazlaghani, M. Enhancing Precision in Dermoscopic Imaging using TransUNet and CASCADE. In Proceedings of the 2024 32nd International Conference on Electrical Engineering (ICEE), Tehran, Iran, 14–16 May 2024; pp. 1–5. [Google Scholar] [CrossRef]
- Azad, R.; Niggemeier, L.; Huttemann, M.; Kazerouni, A.; Aghdam, E.K.; Velichko, Y.; Bagci, U.; Merhof, D. Beyond Self-Attention: Deformable Large Kernel Attention for Medical Image Segmentation. arXiv 2023, arXiv:2309.00121. [Google Scholar] [CrossRef]
- Rahman, M.M.; Marculescu, R. Multi-scale Hierarchical Vision Transformer with Cascaded Attention Decoding for Medical Image Segmentation. arXiv 2023, arXiv:2303.16892. [Google Scholar] [CrossRef]
- Rahman, M.M.; Munir, M.; Marculescu, R. EMCAD: Efficient Multi-scale Convolutional Attention Decoding for Medical Image Segmentation. arXiv 2024, arXiv:2405.06880. [Google Scholar] [CrossRef]
- Perera, S.; Navard, P.; Yilmaz, A. SegFormer3D: An Efficient Transformer for 3D Medical Image Segmentation. arXiv 2024, arXiv:2404.10156. [Google Scholar]
- Tsai, T.Y.; Yu, A.; Maadugundu, M.S.; Mohima, I.J.; Barsha, U.H.; Chen, M.H.F.; Prabhakaran, B.; Chang, M.C. IntelliCardiac: An Intelligent Platform for Cardiac Image Segmentation and Classification. arXiv 2025, arXiv:2505.03838. [Google Scholar] [CrossRef]
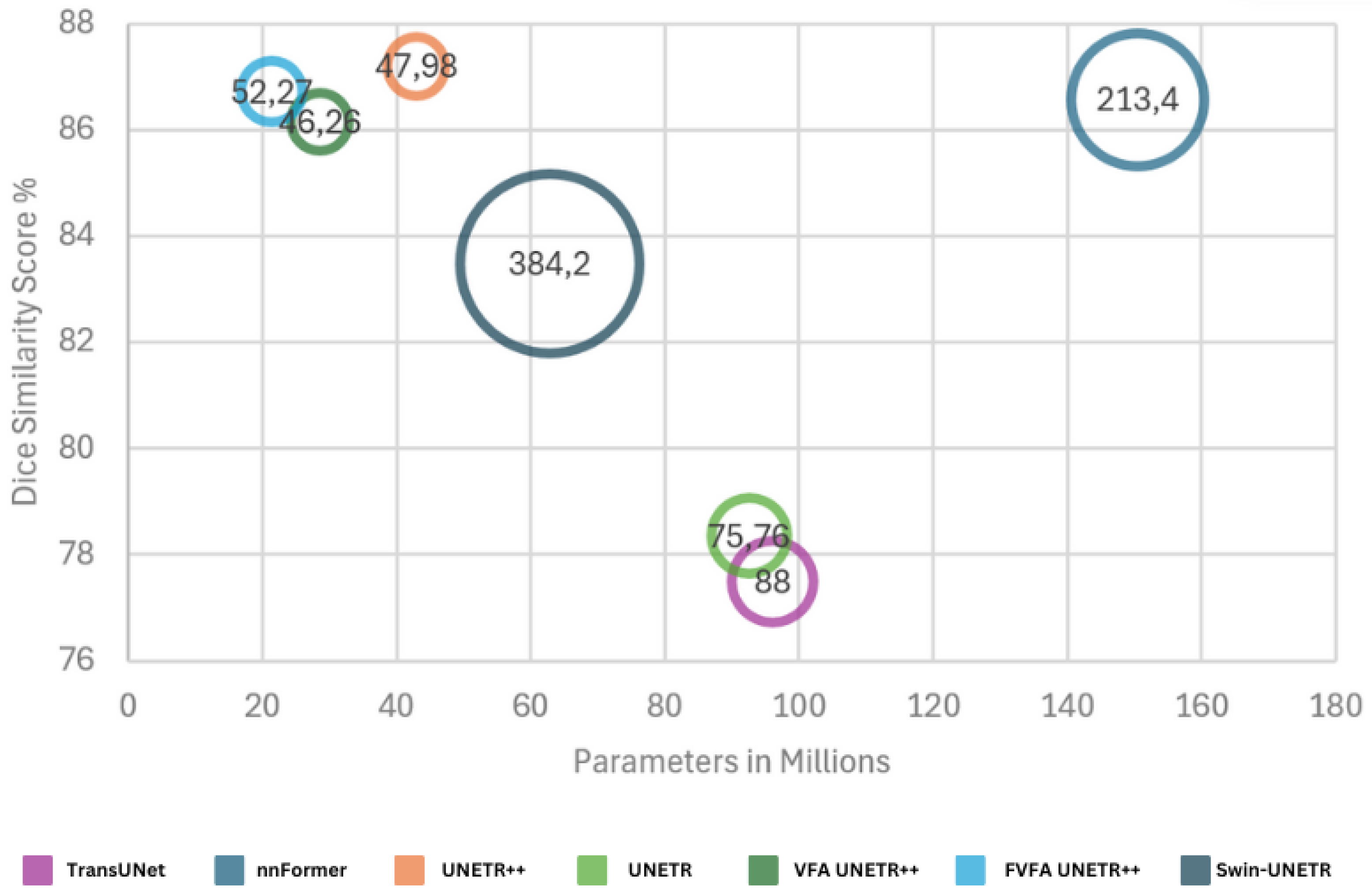


| Methods | Params | FLOPs | Spl | RKid | LKid | Gal | Liv | Sto | Aor | Pan | Average | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HD95 ↓ | DSC ↑ | |||||||||||
| U-Net [17] | - | - | 86.67 | 68.60 | 77.77 | 69.72 | 93.43 | 75.58 | 89.07 | 53.98 | - | 76.85 |
| TransUNet [30] | 96.07 | 88.91 | 85.08 | 77.02 | 81.87 | 63.16 | 94.08 | 75.62 | 87.23 | 55.86 | 31.69 | 77.49 |
| Swin-UNet [33] | - | - | 90.66 | 79.61 | 83.28 | 66.53 | 94.29 | 76.60 | 85.47 | 56.58 | 21.55 | 79.13 |
| UNETR [31] | 92.49 | 75.76 | 85.00 | 84.52 | 85.60 | 56.30 | 94.57 | 78.00 | 89.60 | 60.47 | 18.59 | 78.35 |
| MISSFormer [35] | - | - | 91.92 | 82.00 | 85.21 | 68.65 | 94.41 | 80.81 | 89.06 | 65.67 | 18.20 | 81.96 |
| Swin-UNETR [36] | 62.83 | 384.2 | 95.37 | 86.26 | 86.99 | 66.54 | 95.72 | 77.01 | 91.12 | 68.80 | 10.55 | 83.48 |
| nnFormer [32] | 150.5 | 213.4 | 90.51 | 86.25 | 86.57 | 70.17 | 96.84 | 86.83 | 92.04 | 83.35 | 10.63 | 86.57 |
| UNETR++ [12] | 42.96 | 47.98 | 89.98 | 90.29 | 87.58 | 73.20 | 96.92 | 85.18 | 92.28 | 82.19 | 9.84 | 87.20 |
| SCANeXt [34] | 43.38 | 50.53 | 95.69 | 87.46 | 85.92 | 73.66 | 96.98 | 85.60 | 92.96 | 82.95 | 7.47 | 89.67 |
| PVT-CASCADE [37] | - | - | 83.44 | 80.37 | 82.23 | 70.59 | 94.08 | 83.69 | 83.01 | 64.43 | 20.23 | 81.06 |
| TransCASCADE [38] | - | - | 90.79 | 84.56 | 87.66 | 68.48 | 94.43 | 83.52 | 86.63 | 65.33 | 17.34 | 82.68 |
| 2D D-LKA Net [39] | - | - | 91.22 | 84.92 | 88.38 | 73.79 | 94.88 | 84.94 | 88.34 | 67.71 | 20.04 | 84.27 |
| MERIT-GCASCADE [40] | - | - | 91.92 | 84.83 | 88.01 | 74.81 | 95.38 | 83.63 | 88.05 | 69.73 | 10.38 | 84.54 |
| PVT-EMCAD-B2 [41] | - | - | 92.17 | 84.10 | 88.08 | 68.87 | 95.26 | 83.92 | 88.14 | 68.51 | 15.68 | 83.63 |
| VFA UNETR++ | 28.6 | 46.26 | 88.68 | 87.77 | 87.67 | 69.61 | 97.05 | 84.55 | 92.43 | 81.49 | 10.49 | 86.16 |
| FVFA UNETR++ | 21.42 | 52.27 | 89.56 | 87.39 | 87.33 | 72.96 | 96.71 | 85.56 | 92.56 | 81.68 | 8.84 | 86.72 |
| Methods | RV | Myo | LV | Average (DSC) |
|---|---|---|---|---|
| TransUNet [30] | 88.86 | 84.54 | 95.73 | 89.71 |
| Swin-UNet [33] | 88.55 | 85.62 | 95.83 | 90.00 |
| UNETR [31] | 85.29 | 86.52 | 94.02 | 86.61 |
| MISSFormer [35] | 86.36 | 85.75 | 91.59 | 87.90 |
| nnFormer [32] | 90.94 | 89.58 | 95.65 | 92.06 |
| UNETR++ [12] | 91.89 | 90.61 | 96.00 | 92.83 |
| SegFormer3D [42] | 88.50 | 88.86 | 95.53 | 90.96 |
| IntelliCardiac [43] | 92.27 | 90.33 | 95.09 | 92.56 |
| VFA UNETR++ | 91.45 | 90.59 | 96.10 | 92.71 |
| FVFA UNETR++ | 91.75 | 90.74 | 96.21 | 92.90 |
| Methods | Params (M) | FLOPs |
|---|---|---|
| UNETR++ [12] | 66.8 | 43.71 |
| VFA UNETR++ | 44.36 | 42.4 |
| FVFA UNETR++ | 39.03 | 47.54 |
| Methods | Params | FLOPs | Mem | DSC (%) |
|---|---|---|---|---|
| UNETR [31] | 92.5 | 153.5 | 3.3 | 81.2 |
| SwinUNETR [36] | 62.8 | 572.4 | 19.7 | 81.5 |
| nnFormer [32] | 149.6 | 421.5 | 12.6 | 82.3 |
| UNETR++ [12] | 42.6 | 70.1 | 2.7 | 82.7 |
| VFA UNETR++ | 28.6 | 72.29 | 3.1 | 82.8 |
| FVFA UNETR++ | 21.4 | 78.3 | 5.8 | 82.4 |
| Methods | Training Time | Inference Time | HD95 |
|---|---|---|---|
| UNETR++ [12] | 244.14 | 5.69 | 5.27 |
| VFA UNETR++ | 245.21 | 5.28 | 5.01 |
| FVFA UNETR++ | 248.58 | 7.94 | 5.08 |
| Implementations | Training Time (s) | Inference Time (s) | Mem |
|---|---|---|---|
| Python | 261.15 | 9.06 | 7.5 |
| CUDA | 248.58 | 7.94 | 5.8 |
| Implementations | Training Time (s) | Inference Time (s) | Mem |
|---|---|---|---|
| Python | 248.37 | 6.25 | 4.2 |
| CUDA | 245.21 | 5.28 | 3.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ntanzi, S.; Viriri, S. UNETR++ with Voxel-Focused Attention: Efficient 3D Medical Image Segmentation with Linear-Complexity Transformers. Appl. Sci. 2025, 15, 11034. https://doi.org/10.3390/app152011034
Ntanzi S, Viriri S. UNETR++ with Voxel-Focused Attention: Efficient 3D Medical Image Segmentation with Linear-Complexity Transformers. Applied Sciences. 2025; 15(20):11034. https://doi.org/10.3390/app152011034
Chicago/Turabian StyleNtanzi, Sithembiso, and Serestina Viriri. 2025. "UNETR++ with Voxel-Focused Attention: Efficient 3D Medical Image Segmentation with Linear-Complexity Transformers" Applied Sciences 15, no. 20: 11034. https://doi.org/10.3390/app152011034
APA StyleNtanzi, S., & Viriri, S. (2025). UNETR++ with Voxel-Focused Attention: Efficient 3D Medical Image Segmentation with Linear-Complexity Transformers. Applied Sciences, 15(20), 11034. https://doi.org/10.3390/app152011034







