Balancing Privacy and Utility in Artificial Intelligence-Based Clinical Decision Support: Empirical Evaluation Using De-Identified Electronic Health Record Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. Data Processing and De-Identification
2.3. Data Analysis and Evaluation
2.4. Re-Identification Risk via k-Anonymity Distributions
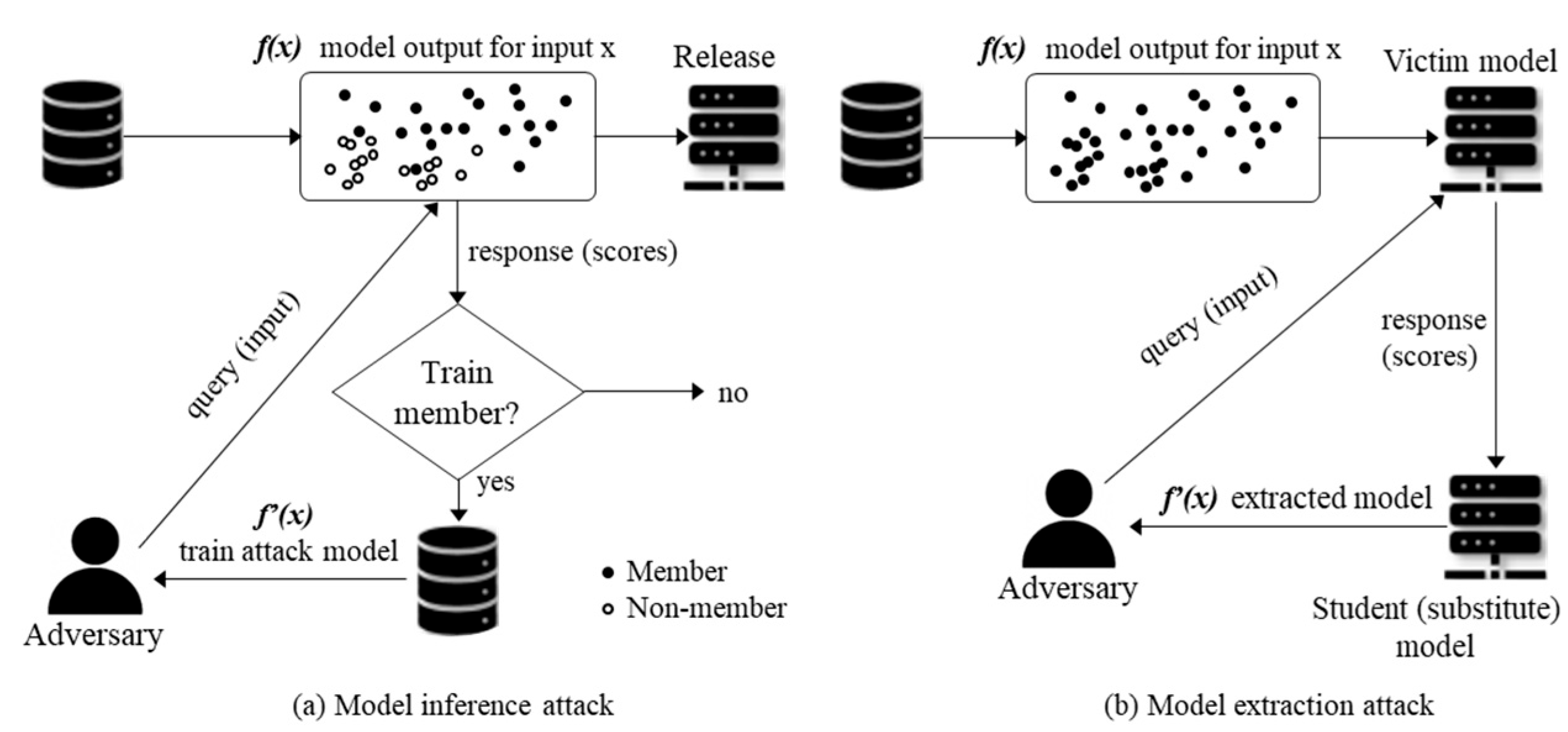
2.5. Membership Inference Attack (MIA)
2.6. Model Extraction Attack (MEA)
2.7. Differential Privacy Stochastic Gradient Descent (DP-SGD)
| Algorithm 1. Privacy-Preserving Training Pipeline |
For clarity, the key training parameters are denoted as follows: b (mini-batch size), q = b/N (sampling rate), C (clipping threshold), σ (noise multiplier), η (learning rate), and E (number of training epochs).
|
| Data Preprocessing (suppression stage): Generalize QIs (e.g., age → 5-year bins, admission date → quarter) Suppress records violating the k ≥ 5 criterion to obtain an anonymized dataset D′ Training (DP-SGD stage): Iteratively sample mini-batches from D′ Clip per-sample gradients by C Add Gaussian noise with multiplier σ Update model parameters using learning rate η. Privacy Accounting: Track the cumulative privacy loss via RDP composition Derive the overall (ε, δ) budget. |
|
2.7.1. Threat Model
2.7.2. Mini-Batch Sampling and Per-Sample Gradient Computation
2.7.3. Gradient Clipping
2.7.4. Noise Addition
2.7.5. Parameter Update
2.7.6. Privacy Accounting
2.7.7. Design Choice and Implementation Details
3. Results
3.1. k-Anonymity Distributions
3.2. Baseline Evaluation
3.3. Evaluation of DP-SGD in Clinical Models
3.3.1. MIAs with DP-SGD
3.3.2. MEAs with DP-SGD
3.3.3. Privacy–Utility Metrics
4. Discussion
4.1. Principal Findings
4.2. Interpretation
4.2.1. k-Anonymity
4.2.2. Membership Inference Attacks
4.2.3. Model Extraction Attacks
4.2.4. Privacy–Utility Trade-Off
4.2.5. Defense with DP-SGD
4.3. Implications for CDSS and Data Sharing
4.4. Strengths
4.5. Limitations
4.6. Future Work
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| EHR | Electronic health records |
| MEA | Model extraction attack |
| MIA | Membership inference attack |
| CDSS | Clinical decision support systems |
| ECE | Expected calibration error |
| DP | Differential privacy |
| DP-SGD | Differentially private stochastic gradient descent |
| KCD | Korean Standard Classification of Diseases |
| ICD | International Classification of Diseases |
| IRB | Institutional review board |
| QI | Quasi-identifier |
| AUROC | Area under the receiver operating characteristic curve |
| AUPRC | Area under the precision–recall curve |
| CI | Confidence interval |
| BCa | Bias-corrected and accelerated |
| EC | Equivalence classes |
| MAE | Mean absolute error |
| RDP | Rényi differential privacy |
| BH | Benjamini–Hochberg |
| FDR | False-discovery rate |
| AI | Artificial intelligence |
| TPR | True positive rate |
| FPR | False positive rate |
References
- Mahmood, S.S.; Levy, D.; Vasan, R.S.; Wang, T.J. The Framingham Heart Study and the epidemiology of cardiovascular disease: A historical perspective. Lancet 2014, 383, 999–1008. [Google Scholar] [CrossRef]
- Andersson, C.; Johnson, A.D.; Benjamin, E.J.; Levy, D.; Vasan, R.S. 70-year legacy of the Framingham Heart Study. Nat. Rev. Cardiol. 2019, 16, 687–698. [Google Scholar] [CrossRef]
- Tsao, C.W.; Vasan, R.S. Cohort Profile: The Framingham Heart Study (FHS): Overview of milestones in cardiovascular epidemiology. Int. J. Epidemiol. 2015, 44, 1800–1813. [Google Scholar] [CrossRef] [PubMed]
- Colditz, G.A.; Hankinson, S.E. The Nurses’ Health Study: Lifestyle and health among women. Nat. Rev. Cancer. 2005, 5, 388–396. [Google Scholar] [CrossRef]
- Colditz, G.A.; Philpott, S.E.; Hankinson, S.E. The Impact of the Nurses’ Health Study on Population Health: Prevention, Translation, and Control. Am. J. Public Health 2016, 106, 1540–1545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yan, C.; Malin, B.A. Membership inference attacks against synthetic health data. J. Biomed. Inform. 2022, 125, 103977. [Google Scholar] [CrossRef]
- Chen, J.; Wang, W.H.; Shi, X. Differential Privacy Protection Against Membership Inference Attack on Machine Learning for Genomic Data. Pac. Symp. Biocomput. 2021, 26, 26–37. [Google Scholar]
- Wang, K.; Wu, J.; Zhu, T.; Ren, W.; Hong, Y. Defense against membership inference attack in graph neural networks through graph perturbation. Int. J. Inf. Secur. 2023, 22, 497–509. [Google Scholar] [CrossRef]
- Cobilean, V.; Mavikumbure, H.S.; Drake, D.; Stuart, M.; Manic, M. Investigating Membership Inference Attacks against CNN Models for BCI Systems. IEEE J. Biomed. Health Inform. 2025; in press. [Google Scholar] [CrossRef]
- Famili, A.; Lao, Y. Deep Neural Network Quantization Framework for Effective Defense against Membership Inference Attacks. Sensors 2023, 23, 7722. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, L.; Wang, Q. MiDA: Membership inference attacks against domain adaptation. ISA Trans. 2023, 141, 103–112. [Google Scholar] [CrossRef]
- Liu, Y.; Luo, J.; Yang, Y.; Wang, X.; Gheisari, M.; Luo, F. ShrewdAttack: Low Cost High Accuracy Model Extraction. Entropy 2023, 25, 282. [Google Scholar] [CrossRef] [PubMed]
- Qayyum, A.; Ijaz, A.; Usama, M.; Iqbal, W.; Qadir, J.; Elkhatib, Y.; Al-Fuqaha, A. Securing Machine Learning in the Cloud: A Systematic Review of Cloud Machine Learning Security. Front. Big Data 2020, 3, 587139. [Google Scholar] [CrossRef]
- Boenisch, F. A Systematic Review on Model Watermarking for Neural Networks. Front. Big Data 2021, 4, 729663. [Google Scholar] [CrossRef]
- Han, D.; Babaei, R.; Zhao, S.; Cheng, S. Exploring the Efficacy of Learning Techniques in Model Extraction Attacks on Image Classifiers: A Comparative Study. Appl. Sci. 2024, 14, 3785. [Google Scholar] [CrossRef]
- Ni, Z.; Zhou, Q. Differential Privacy in Federated Learning: An Evolutionary Game Analysis. Appl. Sci. 2025, 15, 2914. [Google Scholar] [CrossRef]
- Tayyeh, H.K.; AL-Jumaili, A.S.A. Balancing Privacy and Performance: A Differential Privacy Approach in Federated Learning. Computers 2024, 13, 277. [Google Scholar] [CrossRef]
- Zhang, S.; Hagermalm, A.; Slavnic, S.; Schiller, E.M.; Almgren, M. Evaluation of Open-Source Tools for Differential Privacy. Sensors 2023, 23, 6509. [Google Scholar] [CrossRef]
- Namatevs, I.; Sudars, K.; Nikulins, A.; Ozols, K. Privacy Auditing in Differential Private Machine Learning: The Current Trends. Appl. Sci. 2025, 15, 647. [Google Scholar] [CrossRef]
- Chu, Z.; He, J.; Zhang, X.; Zhang, X.; Zhu, N. Differential Privacy High-Dimensional Data Publishing Based on Feature Selection and Clustering. Electronics 2023, 12, 1959. [Google Scholar] [CrossRef]
- Zheng, L.; Cao, Y.; Yoshikawa, M.; Shen, Y.; Rashed, E.A.; Taura, K.; Hanaoka, S.; Zhang, T. Sensitivity-Aware Differential Privacy for Federated Medical Imaging. Sensors 2025, 25, 2847. [Google Scholar] [CrossRef]
- Hernandez-Matamoros, A.; Kikuchi, H. Comparative Analysis of Local Differential Privacy Schemes in Healthcare Datasets. Appl. Sci. 2024, 14, 2864. [Google Scholar] [CrossRef]
- Yu, J.-X.; Xu, Y.-H.; Hua, M.; Yu, G.; Zhou, W. Enhancing Privacy-Preserving Network Trace Synthesis Through Latent Diffusion Models. Information 2025, 16, 686. [Google Scholar] [CrossRef]
- Zhang, Z.; Ma, X.; Ma, J. Local Differential Privacy Based Membership-Privacy-Preserving Federated Learning for Deep-Learning-Driven Remote Sensing. Remote Sens. 2023, 15, 5050. [Google Scholar] [CrossRef]
- Aloqaily, A.; Abdallah, E.E.; Al-Zyoud, R.; Abu Elsoud, E.; Al-Hassan, M.; Abdallah, A.E. Deep Learning Framework for Advanced De-Identification of Protected Health Information. Future Internet 2025, 17, 47. [Google Scholar] [CrossRef]
- Shahid, A.; Bazargani, M.H.; Banahan, P.; Mac Namee, B.; Kechadi, T.; Treacy, C.; Regan, G.; MacMahon, P. A Two-Stage De-Identification Process for Privacy-Preserving Medical Image Analysis. Healthcare 2022, 10, 755. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, M.; Kreiner, K.; Wiesmüller, F.; Hayn, D.; Puelacher, C.; Schreier, G. Masketeer: An Ensemble-Based Pseudonymization Tool with Entity Recognition for German Unstructured Medical Free Text. Future Internet 2024, 16, 281. [Google Scholar] [CrossRef]
- Mosteiro, P.; Wang, R.; Scheepers, F.; Spruit, M. Investigating De-Identification Methodologies in Dutch Medical Texts: A Replication Study of Deduce and Deidentify. Electronics 2025, 14, 1636. [Google Scholar] [CrossRef]
- Sousa, S.; Jantscher, M.; Kröll, M.; Kern, R. Large Language Models for Electronic Health Record De-Identification in English and German. Information 2025, 16, 112. [Google Scholar] [CrossRef]
- Negash, B.; Katz, A.; Neilson, C.J.; Moni, M.; Nesca, M.; Singer, A.; Enns, J.E. De-identification of free text data containing personal health information: A scoping review of reviews. Int. J. Popul. Data Sci. 2023, 8, 2153. [Google Scholar] [CrossRef]
- Johnson, A.E.W.; Bulgarelli, L.; Pollard, T.J. Deidentification of free-text medical records using pre-trained bidirectional transformers. Proc. ACM Conf. Health Inference Learn. 2020, 2020, 214–221. [Google Scholar] [CrossRef]
- Radhakrishnan, L.; Schenk, G.; Muenzen, K.; Oskotsky, B.; Ashouri, C.H.; Plunkett, T.; Israni, S.; Butte, A.J. A certified de-identification system for all clinical text documents for information extraction at scale. JAMIA Open 2023, 6, ooad045. [Google Scholar] [CrossRef]
- Rempe, M.; Heine, L.; Seibold, C.; Hörst, F.; Kleesiek, J. De-identification of medical imaging data: A comprehensive tool for ensuring patient privacy. Eur. Radiol. 2025; in press. [Google Scholar] [CrossRef]
- Liang, B.; Qin, W.; Liao, Z. A Differential Evolutionary-Based XGBoost for Solving Classification of Physical Fitness Test Data of College Students. Mathematics 2025, 13, 1405. [Google Scholar] [CrossRef]
- Wen, H.-T.; Wu, H.-Y.; Liao, K.-C. Using XGBoost Regression to Analyze the Importance of Input Features Applied to an Artificial Intelligence Model for the Biomass Gasification System. Inventions 2022, 7, 126. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.; Li, C.; Zhou, J. Application of XGBoost Model Optimized by Multi-Algorithm Ensemble in Predicting FRP-Concrete Interfacial Bond Strength. Materials 2025, 18, 2868. [Google Scholar] [CrossRef]
- Hidayaturrohman, Q.A.; Hanada, E. Impact of Data Pre-Processing Techniques on XGBoost Model Performance for Predicting All-Cause Readmission and Mortality Among Patients with Heart Failure. BioMedInformatics 2024, 4, 2201–2212. [Google Scholar] [CrossRef]
- Dong, T.; Oronti, I.B.; Sinha, S.; Freitas, A.; Zhai, B.; Chan, J.; Fudulu, D.P.; Caputo, M.; Angelini, G.D. Enhancing Cardiovascular Risk Prediction: Development of an Advanced Xgboost Model with Hospital-Level Random Effects. Bioengineering 2024, 11, 1039. [Google Scholar] [CrossRef]
- Amjad, M.; Ahmad, I.; Ahmad, M.; Wróblewski, P.; Kamiński, P.; Amjad, U. Prediction of Pile Bearing Capacity Using XGBoost Algorithm: Modeling and Performance Evaluation. Appl. Sci. 2022, 12, 2126. [Google Scholar] [CrossRef]
- Sun, Z.; Huang, Y.; Zhang, A.; Li, C.; Jiang, L.; Liao, X.; Li, R.; Wan, J. Hybrid Uncertainty Metrics-Based Privacy-Preserving Alternating Multimodal Representation Learning. Appl. Sci. 2025, 15, 5229. [Google Scholar] [CrossRef]
- Riaz, S.; Ali, S.; Wang, G. Latif MA, Iqbal MZ. Membership inference attack on differentially private block coordinate descent. Peer J. Comput. Sci. 2023, 9, e1616. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Zhong, T. Analysis of Application Examples of Differential Privacy in Deep Learning. Comput. Intell. Neurosci. 2021, 2021, 4244040. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Liu, Y.; Jin, B.; Yu, Z.; Cai, X.; Li, H.; Zha, Z.; Hou, D.; Peng, K. Leveraging Multiple Adversarial Perturbation Distances for Enhanced Membership Inference Attack in Federated Learning. Symmetry 2024, 16, 1677. [Google Scholar] [CrossRef]
- Jonnagaddala, J.; Wong, Z.S. Privacy preserving strategies for electronic health records in the era of large language models. NPJ Digit. Med. 2025, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Rathee, G.; Singh, A.; Singal, G.; Tomar, A. A distributed classification and prediction model using federated learning in healthcare. Knowl. Inf. Syst. 2025; in press. [Google Scholar] [CrossRef]
- Pati, S.; Kumar, S.; Varma, A.; Edwards, B.; Lu, C.; Qu, L.; Wang, J.J.; Lakshminarayanan, A.; Wang, S.-H.; Sheller, M.J.; et al. Privacy preservation for federated learning in health care. Patterns 2024, 5, 100974. [Google Scholar] [CrossRef]
- Abadi, M.; Chu, A.; Goodfellow, I.; McMahan, H.B.; Mironov, I.; Talwar, K.; Zhang, L. Deep learning with differential privacy. In Proceedings of the 2016 ACM SIGSAC Conference on Computer and Communications Security (CCS), Vienna, Austria, 24–28 October 2016; pp. 308–318. [Google Scholar] [CrossRef]
- Shokri, R.; Stronati, M.; Song, C.; Shmatikov, V. Membership inference attacks against machine learning models. In Proceedings of the 2017 IEEE Symposium on Security and Privacy (S&P), San Jose, CA, USA, 22–26 May 2017; pp. 3–18. [Google Scholar] [CrossRef]
- Wunderlich, S.; Haase, A.; Merkert, S.; Jahn, K.; Deest, M.; Frieling, H.; Glage, S.; Korte, W.; Martens, A.; Kirschning, A.; et al. Targeted biallelic integration of an inducible Caspase 9 suicide gene in iPSCs for safer therapies. Mol. Ther. Methods Clin. Dev. 2022, 26, 84–94. [Google Scholar] [CrossRef]
- Shang, S.; Shi, Y.; Zhang, Y.; Liu, M.; Zhang, H.; Wang, P.; Zhuang, L. Artificial intelligence for brain disease diagnosis using electroencephalogram signals. J. Zhejiang Univ. Sci. B. 2024, 25, 914–940. [Google Scholar] [CrossRef]
- Geng, Q.; Viswanath, P. Optimal noise adding mechanisms for approximate differential privacy. IEEE Trans. Inf. Theory 2015, 62, 952–969. [Google Scholar] [CrossRef]
- Stacey, T.; Li, L.; Ercan, G.M.; Wang, W.; Yu, L. Effects of differential privacy and data skewness on membership inference vulnerability. arXiv 2019, arXiv:1911.09777. [Google Scholar] [CrossRef]

| Parameter | Description |
|---|---|
| Programming language | Python 3.10 |
| Core libraries | pandas 2.2.2; numpy 1.26.4; scikit-learn 1.5.1; statsmodels 0.14.2; XGBoost 2.0.3; Splink 3.10.0; recordlinkage 0.15 |
| Differential privacy | PyTorch 2.4.0 with Rényi DP accountant; opacus (DP-SGD) 1.5.2 |
| Parallelization | joblib (bootstrap resampling, repeated runs) |
| CPU | 24-core Intel Xeon (Intel Corporation, Santa Clara, CA, USA) |
| GPU | NVIDIA RTX 5090 (24 GB memory) (NVIDIA Corporation, Santa Clara, CA, USA) |
| RAM | 128 GB |
| Parameter | Value | Description |
|---|---|---|
| Batch size (B) | 256 | Mini-batch size per iteration |
| Learning rate (η) | 0.01 | Initial step size for optimization |
| Clipping norm (C) | 1.0 | ℓ2 norm bound for per-sample gradients |
| Noise multiplier (σ) | 1.1 | Scale of Gaussian noise added to clipped gradients |
| Number of epochs (T) | 150 | Total training epochs |
| Privacy accountant | RDP | Used to compute (ε, δ) privacy guarantees |
| δ | 1 × 10−5 | Fixed delta parameter |
| Final ε | 2.488 | Reported privacy budget after training |
| Interval | Baseline | Enhanced Generalization | Enhanced Generalization with Suppression | |||
|---|---|---|---|---|---|---|
| ECs (n/%) | Recs (n/%) | ECs (n/%) | Recs (n/%) | ECs (n/%) | Recs (n/%) | |
| k = 1 | 23/26.44% | 23/0.36% | 10/15.87% | 10/0.16% | 10/15.87% | 10/0.16% |
| k = 2–4 | 24/27.59% | 66/1.03% | 15/23.81% | 42/0.66% | 15/23.81% | 42/0.66% |
| k = 5–9 | 12/13.79% | 79/1.24% | 8/12.70% | 57/0.89% | 8/12.70% | 56/0.88% |
| k ≥ 10 | 28/32.18% | 6221/97.37% | 30/47.62% | 6280/98.29% | 30/47.62% | 6278/98.31% |
| suppressed | - | - | - | - | - | 171/0.17% |
| Metric | Mean |
|---|---|
| Attack classifier AUROC | 0.515 (95% CI: 0.514–0.516) |
| Threshold AUROC | 0.504 (95% CI: 0.503–0.505) |
| Attack Type | AUROC | Best Accuracy | Advantage |
|---|---|---|---|
| Confidence | 0.503 (95% CI: 0.472–0.533) | 0.527 (95% CI: 0.512–0.540) | 0.027 (95% CI: 0.010–0.045) |
| Entropy | 0.502 (95% CI: 0.472–0.533) | 0.526 (95% CI: 0.511–0.541) | 0.026 (95% CI: 0.009–0.044) |
| YeomLoss | 0.503 (95% CI: 0.474–0.534) | 0.529 (95% CI: 0.514–0.543) | 0.030 (95% CI: 0.012–0.048) |
| Metric | Victim Model | Student Model |
|---|---|---|
| Correlation (prob.) | – | 0.9999 (95% CI: 0.9998–1.0000) |
| MAE | – | 0.0008 (95% CI: 0.0006–0.0011) |
| AUROC | 0.975 (95% CI: 0.936–1.000) | 0.9850 (95% CI: 0.9620–1.0000) |
| Accuracy | 0.996 (95% CI: 0.991–0.999) | 0.9960 (95% CI: 0.9910–0.9990) |
| F1-score | 0.967 (95% CI: 0.945–0.981) | 0.9680 (95% CI: 0.9470–0.9820) |
| Brier score | 0.002 (95% CI: 0.002–0.003) | 0.0020 (95% CI: 0.0016–0.0028) |
| Attack Type | AUROC | Best Accuracy | Advantage |
|---|---|---|---|
| Confidence | 0.501 (95% CI: 0.480–0.523) | 0.530 (95% CI: 0.506–0.568) | 0.002 (95% CI: −0.000–0.012) |
| Entropy | 0.501 (95% CI: 0.480–0.523) | 0.530 (95% CI: 0.506–0.568) | 0.002 (95% CI: 0.000–0.012) |
| YeomLoss | 0.498 (95% CI: 0.477–0.520) | 0.530 (95% CI: 0.505–0.568) | 0.002 (95% CI: 0.000–0.010) |
| Metric | Victim Model | Student Model | Victim–Student Agreement |
|---|---|---|---|
| AUROC | 0.729 (95% CI: 0.654–0.785) | 0.486 (95% CI: 0.382–0.646) | r = −0.008 (95% CI: −0.076–0.036) |
| Accuracy | 0.950 (95% CI: 0.943–0.959) | 0.950 (95% CI: 0.943–0.959) | MAE = 0.002 (95% CI: 0.001–0.005) |
| Brier score | 0.050 (95% CI: 0.040–0.056) | 0.050 (95% CI: 0.041–0.056) | – |
| Category | Metric | Value | Interpretation |
|---|---|---|---|
| Privacy | δ | 1 × 10−5 (fixed) | Preset privacy parameter |
| ε | 2.488 (95% CI 2.322–2.721) | Privacy budget (RDP accountant) | |
| Utility | AUROC | 0.729 (95% CI 0.654–0.785) | Discrimination ability |
| Accuracy | 0.950 (95% CI 0.943–0.959) | Classification performance | |
| log loss | 0.293 (95% CI 0.228–0.347) | Calibration performance |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Lee, K.H. Balancing Privacy and Utility in Artificial Intelligence-Based Clinical Decision Support: Empirical Evaluation Using De-Identified Electronic Health Record Data. Appl. Sci. 2025, 15, 10857. https://doi.org/10.3390/app151910857
Lee J, Lee KH. Balancing Privacy and Utility in Artificial Intelligence-Based Clinical Decision Support: Empirical Evaluation Using De-Identified Electronic Health Record Data. Applied Sciences. 2025; 15(19):10857. https://doi.org/10.3390/app151910857
Chicago/Turabian StyleLee, Jungwoo, and Kyu Hee Lee. 2025. "Balancing Privacy and Utility in Artificial Intelligence-Based Clinical Decision Support: Empirical Evaluation Using De-Identified Electronic Health Record Data" Applied Sciences 15, no. 19: 10857. https://doi.org/10.3390/app151910857
APA StyleLee, J., & Lee, K. H. (2025). Balancing Privacy and Utility in Artificial Intelligence-Based Clinical Decision Support: Empirical Evaluation Using De-Identified Electronic Health Record Data. Applied Sciences, 15(19), 10857. https://doi.org/10.3390/app151910857






