Abstract
Pulmonary neuroendocrine tumors (PULMONARY NETs) are heterogeneous tumors ranging from well-differentiated to highly aggressive neoplasms. The aim of this study is to prospectively test pre-operative circulating free DNA (cfDNA) in PULMONARY NET patients undergoing surgery and evaluate its relationship to clinicopathological features. From February to December 2024, 136 patients with suspected primary lung cancer underwent pre-operative blood sampling, of whom 21 were diagnosed with PULMONARY NETs. Total cell-free nucleic acid extraction was performed using the Genexus Purification System (Thermofisher). cfDNA was quantified using a fluorometric assay with the Qubit dsDNA HS Assay kit (Thermofisher) and a capillary electrophoresis-based assay (cell-free DNA ScreenTape kit) on the Tape Station 4200 systems (Agilent). A cfDNA quality assessment was also obtained (cfDNA sizing and % cfDNA). Most patients had Stage I (18/21.85.7%) typical carcinoids (16/21.76.2%). Nodal involvement was detected in one patient (0.5%). Six months after surgery, all patients were alive without recurrence. Larger tumors presented higher levels of cfDNA. The mean tumor size in patients with cfDNA > 40 ng was 266 mm (±16.7 mm), compared to 13.2 mm (±7.3 mm) for cfDNA < 40 ng (p-value = 0.018). Higher levels of cfDNA were observed in patients with pStages greater than IA (p-value = 0.007). Although limited by a small sample group and biases of a surgical series, we observed that larger/advanced PULMONARY NETs presented higher cfDNA levels pre-operatively.
1. Introduction
Neuroendocrine pulmonary tumors (PULMONARY NETs) are a group of heterogeneous tumors ranging from well-differentiated, low-grade neuroendocrine tumors—like typical and atypical carcinoids—to high-grade, poorly differentiated aggressive malignancies, such as large cell neuroendocrine carcinoma (LCNEC) and small cell lung cancer (SCLC) [1]. On the basis of the degree of differentiation, proliferative index, mitotic rate, and necrosis status, PULMONARY NETs are divided into low-, intermediate-, and high-grade tumors [2]. Low- to intermediate-grade tumors (carcinoids/atypical carcinoids) are quite rare as they represent around 1–2% of all lung neoplasms, typically indolent in behavior compared to more aggressive high-grade PULMONARY NETs [3].
Lung carcinoids exhibit different clinicopathologic characteristics when compared to other types of lung cancer. Patients with lung PULMONARY NETs are typically diagnosed at a younger age, show a slight female predominance, often express somatostatin receptors (SSTs), and may be associated with genetic or functional syndromes [4,5]. Usually, pre-operative workup includes CT scans and octreotide, 68-gallium-dotatate PET/CT, and 18F-FDG-PET/CT scans. Indeed, somatostatin-targeting peptides (e.g., [68Ga]Ga- DOTA-TOC and -TATE) and [18F] DOPA are valuable in neuroendocrine tumors because of their peculiar receptor expression [6]. Even though these approaches may help in predicting the suspicion of a pulmonary neuroendocrine tumor vs. other histologies, due to a different radionuclide uptake, it is not possible to predict the grade of the eventual neuroendocrine tumor. Surgical resection is the treatment of choice for localized diseases. This approach is supported by a retrospective study by the European Society of Thoracic Surgeons—NETs of the Lung Working Group, which, furthermore, indicated that anatomical resection is superior to wedge resection for typical carcinoids [7].
To increase the accuracy of PULMONARY NET pre-op diagnoses, a breakthrough and mostly unexplored tool is the analysis of the circulating cell-free DNA (cfDNA) and circulating tumor DNA (ctDNA). The role of cfDNA as a prognostic marker has already been documented in many kinds of tumors such as breast, prostate, lung, and colorectal cancers [8,9,10]. In 2022, the European Society of Medical Oncology (ESMO) published a recommendation manuscript focused on ctDNA detected in plasma as a liquid biopsy analyte and highlighted that ctDNA has sufficiently strong evidence to be used routinely in clinical practice to genotype cancers and direct molecularly targeted therapies [11]. Boons and colleagues underlined the role of cfDNA in the diagnosis, prognosis, and follow-up of metastatic PULMONARY NETs [12]. Another experience from Zakka and coworkers confirmed the feasibility of ctDNA testing in patients with neuroendocrine tumors and characterized common alterations in the genomic landscape [13]. These, however, were general analyses focused on different kinds of NETs, not just pulmonary ones. Concerning cfDNA, few studies in the literature [12,14,15,16,17,18,19] analyze its implications in pancreatic and prostatic neuroendocrine tumors. To date, there is only one paper available in the literature that analyzes cfDNA in lung neuroendocrine tumors.
Zhou and coworkers [20] explored the possibility of genomic subtyping from cfDNA to improve prognostication and therapeutic decision-making in large cell neuroendocrine lung cancer patients.
As still reported by Zhou et al. [20], cfDNA analysis may be a reliable alternative for genomic profiling of LCNEC.
To the best of our knowledge, predicting the relationship between cfDNA levels and clinicopathological PULMONARY NET features has not yet been explored. This could help in pre-operatory diagnosis, avoiding pre operatory biopsy, and in differential diagnosis with lung nodules looking like carcinoids and radiological imaging (i.e., amartomas).
In the framework of the LANTERN project, [21] pre-operative blood samples were collected in patients with suspected lung cancer; we herein focused our attention in those patients who underwent surgery and had a final pathological diagnosis of PULMONARY NETs. The aim of the present study is to explore in this subset of patients the relationship between pre-operative circulating cfDNA and clinicopathological features on surgical specimens.
2. Methods
This is a prospective observational study performed in our institution between February 2024 and December 2024.
This study was conducted after the Ethical Committee approval (no. of approval 5420, 19 January 2023) for research use of prospectively collected data (observational) stemming from standard clinical practice. This study was therefore performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
Inclusion criteria were as follows:
- The collection of a blood sample available for the analysis, as reported below in detail;
- The pathological confirmation of PULMONARY NETs after curative surgery.;
- Written patients’ signed consent to enter the study.
Among 136 patients initially enrolled for whom pre-operative blood sampling was taken, a total of 21 met the inclusion criteria and represented the study population of the present analysis.
2.1. Preo-Perative Workup, Surgical Management, and Pathological Evaluation
The pre-operative workup included an evaluation of cardio-pulmonary function, a computed tomography (CT) brain–thorax–abdomen scan with contrast, and 18-FDG-PET. EBUS with selective biopsy was performed in patients with suspected mediastinal nodal involvement (lymph node with short axis > 1 cm or with uptake at 18-FDG-PET). A multidisciplinary team, consisting of oncologists and surgeons, managed pre- and post-operative treatments, planned follow-up schedules, and determined eventual adjuvant treatments based on the final pathological reports, histology, and nodal status.
In the event of poor pulmonary function, if surgically feasible (i.e., peripheral lung nodule smaller than 2 cm), sublobar resection (wedge resection or segmentectomy) was performed. Lobectomy was indicated in all cases with central nodules greater than 2 cm and with good pulmonary function. All 21 patients were operated on using the uniportal-VATS approach.
A final pathological diagnosis on the surgical specimen was performed by a dedicated pathologist. Criteria to distinguish typical from atypical specimens were based on the number of mitoses per field, according to the WHO classification of lung tumors. The Ki-67 index is useful in separating carcinoids from high-grade tumors, including small cell lung cancer and large cell neuroendocrine tumors. Nevertheless, an official role for Ki-67 in the diagnosis is not given due to the absence of a validated score for it in lung tumors. Ki-67 values are generally below 5% for typical carcinoids and range from 9% to 18% for atypical ones. A Ki-67 index value exceeding 30% is usually indicative of high-grade small cell lung cancer and large cell neuroendocrine tumors [22].
2.2. Sample Collection and cfDNA Processing
Blood samples were collected for each enrolled patient using two ethylenediaminetetraacetic (EDTA-K2) tubes with a total of 18 mL (2 mL × 9 mL) of blood. Whole-blood samples were processed within 2 h of collection to isolate plasma via centrifugation. Samples were centrifuged according to a two-step protocol: first, it was spun for 20 min at 2000× g (4 °C); then supernatants were again centrifuged for 10 min at 13,000× g (4 °C). Plasma samples were stored at −80 °C until needed. Cell-free total nucleic acid (cfTNA) extraction was performed within two days to limit the degradation of nucleic acids. cfTNA isolation was performed using the GenexusTM Purification System (© Copyright 2006–2025 Thermo Fisher Scientific Inc., Waltham, MA, USA) according to the manufacturer’s instructions. The automated method used for cfTNA extraction is as follows:
MagMAX® technology with a magnetic bead-based purification: Extracted cfDNA was quantified using a fluorometric assay with the Qubit dsDNA HS Assay kit (Thermofisher) on the Qubit 2.0 Fluorometer (data described as total ng). cfDNA was also evaluated for quantity and quality (cfDNA subcomponents) assessments using the cell-free DNA ScreenTape assay on the capillary electrophoresis (CE) Tape Station 4200 system (Agilent, Santa Clara, CA, USA). We evaluated the quality of the extracted cfDNA from the electropherogram profile that contains a prominent peak of the mononucleosome (at approximately 170 bp) component and other smaller peaks (nucleosome multimers). The CE analysis included quality checking for the presence of high-molecular-weight (HMW) DNA (>700 bp). CE output results were related to the cfDNA mean sizing in base pair (bp) and the percentage of cfDNA compared to the total DNA (cfDNA%) for the regions 50–700 bp and 75–300 bp.
All experimental analyses were carried out in the Genomics Research Core Facility of the Gemelli Science and Technology Park (GSTeP). The centralization of the study for both tissue and liquid biopsy evaluations minimized variability and ensured reproducibility and accuracy of the results.
2.3. Statistics
From the quantitation of cfDNA obtained using a fluorometric assay, cfDNA values were categorized based on the statistical distribution of the studied condition (a mean value of cfDNBA = 25 ng) and subdivided into quartiles, with the patients falling within the following groups: <20 ng, 20–40 ng, and >40 ng. From the capillary electrophoresis analyses, we collected the following dataset: a mean bp value of cfDNA; cfDNA% at the 75–300 bp region and the total ng of cfDNA at the 75–300 bp region.
Exploratory descriptive analysis was performed to describe the conditions under study using mean/SD and percentage frequencies. A subsequent investigation through association tests was performed using Fisher’s exact test, and conditions with a p-value ≤0.05 were considered significant.
3. Results
Data collected related to gender, age, signs and symptoms, lab tests, tumor location and stage, surgeons’ notes, pathological features, and post-operative therapy are reported (see Table 1). The majority of patients were women (61.9%), non-smokers (90%), and not affected by COPD (76%). PET uptake (considered positive with a SUVmax > 2.5, as discussed with nuclear medicine) was detected in 66.6% of patients. As expected in a surgical series, the majority of patients were diagnosed with Stage I (18/21, 85.7%) typical carcinoid (16/21, 76.2%) cancer. All surgeries were performed via the Uniportal VATS approach. Nodal involvement was detected in one patient (4.8%). The mean tumor dimension was 16.4 ± 10.11 mm. Follow-up was performed 6 months after surgery; no recurrence was detected, and all patients, both the ones affected with typical carcinoids and the ones affected with atypical carcinoid/LCNEC, were alive. From the quantitative and qualitative evaluation of the extracted cfDNA, we obtained the following: total cfDNA weights (median [IQR[Q1-Q3]-]) of 25.5 [18.2–38.2] (FLUO) and 13.5 ng [9–24.8] (CE at 75–300 bp region); a cfDNA bp value (median [IQR[Q1-Q3]-) of 279 [263–289], calculated at the 50–700 bp region; and cfDNA% (median [IQR[Q1-Q3]-]) of 85.4 [81.6–90.2] (at the 50–700 bp region) of 60 [56–67] (at the 75–300 bp region). The comparable results of the cfDNA% in the two regions of 50–700 bp and 75–300 bp indicated the high quality of the extracted cfDNA, enriched in the mononucleosome fraction instead of other high-molecular-weight cfDNA subcomponents.

Table 1.
Clinical, surgical, and pathological characteristics of the population and cfDNA distribution. The concentration of cfDNA was reported according to the fluorometric assay.
cfDNA levels detected by the fluorometric assay and capillary electrophoresis are reported in Figure 1. During statistical analysis, we observed that larger tumors presented with higher levels of cfDNA (fluorometric assay method). The mean tumor size in patients with cfDNA > 40 ng was 26.6 mm (±16.7 mm) compared to 13.2 mm (±7.3 mm) in patients reporting cfDNA values < 40 ng (p-value = 0.018). Consistently, higher levels of cfDNA were observed in patients with pStages greater than IA (p-value = 0.007). The same outcome was obtained with the capillary assay, with a higher value of cfDNA identified for larger tumors but without a statistically significant difference.
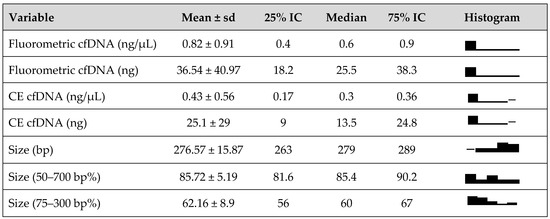
Figure 1.
cfDNA size distribution and levels detected by fluorometric and capillary electrophoresis (CE) assays.
No statistically significant correlations were found between the cfDNA levels and any histological or pathological features (Table 1).
4. Discussion
Currently, the diagnosis of neuroendocrine lung cancer is largely dependent on histology and immunohistochemistry assessments, which may not always accurately reflect true tumor biology.
Although relatively rare, their incidence appears to be increasing over time, highlighting the importance of better understanding how they are managed [22]. The WHO criteria [23] for differentiating typical versus atypical lung carcinoids are currently based on the mitotic rate. In our cohort, the mitotic rate data was reported as > or <2/2 mm2, with a higher value associated with atypical histology, in line with what was reported by the WHO criteria.
Another important parameter that we considered was Ki67, which is not present in the WHO criteria but is described in results from different studies [24,25]. They report emerging evidence for a more routine role of Ki67 as a good prognostic marker in lung carcinoids. In particular, in our series, the Ki67 data was always present in the final pathological report as a percentage value (> or <5%), with a higher value often associated with atypical histology.
Concerning pre-operative diagnosis, in our center, a PET-CT scan with 18-FDG is routinely performed if neuroendocrine lung cancer is suspected. Concerning 68-gallium-dotatate, it was not routinely used in our series of patients. In our study, PET uptake with 18-FDG had a SUV max >2.5 in 66.6% of the entire cohort.
The management of lung PULMONARY NETs depends on several factors, including the disease stage and proliferative rate. Surgery is the treatment of choice for patients who have no evidence of distant metastases [26]. The surgical approaches used in the management of PULMONARY NETs include lobectomy and sublobar resection, with no difference in disease-specific survival and overall survival, as reported in the study by Fox and coworkers. The likelihood of cure through radical surgery is considered high for both typical and atypical carcinoids [27].
For many cancer types, liquid biopsies are emerging as minimally invasive biomarkers, and the analysis of cfDNA is especially gaining a lot of interest. In patients with cancer, part of the cfDNA is tumor-derived and is known as ctDNA, and it can be used for the molecular characterization of a tumor without requiring an invasive tissue biopsy [28]. Currently, knowledge and research on liquid biopsies in NENs (neuroendocrine neoplasms) remain limited to a few studies. Our team has recently developed a multi-omics platform of research (LANTERN project) [21] that stands as a beacon of innovation, contributing valuable insights that align seamlessly with the evolving paradigm of personalized and preventive healthcare. One of the fields explored in the application of this platform is the prediction of the complexity of lung cancer from liquid biopsies.
In this framework, the aim of our prospective analysis was to test the pre-operative circulating cfDNA in PULMONARY NET patients and evaluate the relationship between it and clinicopathological features on surgical specimens. The clinical utility of cfDNA for neuroendocrine tumors has been well described for pancreatic and prostatic cancers. For example, in the analysis by Cowzer and colleagues [15], concerning metastatic pancreatic NETs, cfDNA-based NGS identified tumor-associated mutations in 66% of plasma samples, with a high level of plasma–tissue agreement in PanNEN-associated genes. The group by Franceschini and colleagues [16] observed extensive DNA methylation changes associated with prostate cancer neuroendocrine tumors, and such changes can be detected in cfDNA.
To date, the only analysis addressing neuroendocrine lung cancer was made by Zhuo and colleagues [20], but they only focused on large cell histology and thus are not representative of all PULMONARY NET entities.
In the present analysis, cfDNA was quantified with fluorometric assays and capillary electrophoresis, the latter also giving a qualitative analysis with cfDNA sizing and % cfDNA. For both approaches, no statistically significant correlation was found between cfDNA levels and histology (typical vs. atypical or large cell carcinoids), the presence of necrosis, %Ki67, or the number of mitoses. We have hypothesized several reasons to explain this result. Firstly, we may consider the small sample size analyzed, particularly the small number of atypical/large cell carcinoids compared to the typical ones (5 vs. 16, respectively). Indeed, the analysis by Zhou and his group [20] included a greater cohort of 45 LCNEC tumors (a more aggressive disease), and they could observe a 90% correspondence between cfDNA and tissue findings.
Nevertheless, in our analysis, the cfDNA levels detected with the fluorometric assay method showed a statistically significant correlation with some clinical features. In particular, a higher level of cfDNA correlated with a greater tumor dimension and consequently to a higher pStage.
This result deserves attention because it signifies a correlation between the tumor extension and its presence in the blood stream, opening future investigation into its possible application as baseline biomarkers for early diagnosis, together with the study of radiological imagines or post-treatment biomarkers in predicting recurrence and managing adjuvant treatments. Our findings are especially interesting if they can be inserted into artificial intelligence programs, such as the LANTERN study 21], in which this data can be combined with other clinical (i.e., radiomics) and pathological features to predict diagnosis and plan surgery or the best medical treatment. Once a correlation between cfDNA and tumor dimensions for PULMONARY NETs is demonstrated, an important analysis would be to analyze circulating tumor DNA and perform a sequencing analysis in this rare tumor type. This could be instrumental in predicting prognoses and risk stratification or surveillance strategies or in analyzing the multi-omics framework, which can be performed in the LANTERN study.
Finally, no correlation was found with clinical variables such as being affected with COPD or being a smoker, thus showing that cfDNA detection can be used independently of smoking status or COPD presence without the risk that the cfDNA levels found could be biased by pre-operative features.
Limitations and Points of Strength
Our study has some limitations: the small number of our sample size and the inherent bias due to the fact that our cohort was composed only of patients eligible for surgery.
Concerning the first point, it is important to consider that PULMONARY NETs are very uncommon, accounting for about 1–2% of all lung cancers [29]. Moreover, pre-operative diagnosis is not always feasible, especially for small peripheral tumors [30], limiting pre-operative enrollment even in prospective trials. Indeed, one of our key strengths lies in the fact that we prospectively collected data and blood samples of all the suspected primary lung cancer, not suspected PULMONARY NETs only. Thus, all the cases found to have PULMONARY NETs after surgery were prospectively enrolled. Otherwise, performing a focused prospective study on PULMONARY NETs with a clear histology before treatment would likely result in the enrollment of only 3–4 cases per year in our center.
We also deliberated the possible logistical difficulties of this kind of study when extending the project to a multicentric setting (to increase the sample size). Confounding biases related to lab analysis and clinical management performed in different centers would have been considerable. Indeed, the Genomics Core Research Facility in our center for liquid biopsy sample processing ensures that blood sample analyses are accurately performed. Even if sample processing was centralized, logistical delays in the blood analysis could present further issues. Thus, while we are conscious that these are preliminary results based on a small sample size, our approach provides a methodologically more accurate evaluation of this rare patient population.
Efforts in the early diagnosis and identification of small cell lung cancer subtypes using a noninvasive liquid biopsy approach have been made [31]. The experimental data described here were obtained using two common and feasible methods available in most leading molecular laboratories. These approaches provide quantitative and qualitative evaluations of the cfDNA. We also need to take into account that since the cut-off values of the cfDNA were based on the median cfDNA value (25 ng) of the present cohort of patients, these should be validated in a prospective clinical cohort to be applicable in daily clinical practice.
Nevertheless, our results are an encouraging starting point in the pre-operative prediction of PULMONARY NET features, especially considering the emerging role of multiomics-based predictive approaches in diagnosing and managing lung cancer, as reported in the aims of the LANTERN study.
While our data may suggest that cfDNA detection is a feasible test that physicians could take into account when evaluating a pulmonary NET, data are not so robust to state that this marker may be of help to estimate the feasibility of surgery or of other treatments. A validation study is going to be planned with our experimental cohort, another validation cohort from an external comprehensive high-volume cancer center (which can guarantee high-quality sample analysis), and a final control group of healthy people (20 cases) to overcome the bias due to the small sample size.
5. Conclusions
Despite being limited by a small sample group and biases of a surgical series, we observed that larger/advanced PULMONARY NETs presented with higher cfDNA levels before surgery. On the contrary, cfDNA dosage seems to not be useful in distinguishing histology or pathological features in PULMONARY NETs. Multicenter studies or collaborative datasets that overcome the biases due to a small sample size are needed to be able to validate our preliminary results.
Author Contributions
Conceptualization, F.L. and C.S.; methodology, C.S.; validation, F.L., J.E. and C.S.; formal analysis, A.C. and J.E., investigation, C.S., E.D.P., J.E. and A.M.; resources, C.S.; data curation, G.S.; writing—original draft preparation, C.S., E.D.P., J.E. and A.M.; writing—review and editing, C.S. and F.L.; visualization, A.U.; supervision, F.L., S.M. and A.U.; project administration, S.M., F.L., A.M. and A.U. All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported by the Ministry of Health under the frame of ERA PerMed JTC2022.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Fondazione Policlinico Universitario Agostino Gemelli IRCCS (protocol code 5420 and 19 January 2023).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data are available upon request from the author.
Acknowledgments
Thanks to Akshaya Balamurugan for her contribute to the paper.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sen, T.; Dotsu, Y.; Corbett, V.; Puri, S.; Sen, U.; A Boyle, T.; Mack, P.; Hirsch, F.; Aljumaily, R.; Naqash, A.R.; et al. Pulmonary neuroendocrine neoplasms: The molecular landscape, therapeutic challenges, and diagnosis and management strategies. Lancet Oncol. 2025, 26, e13–e33. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.; Alrehaili, M.; Marginean, H.; Goodwin, R.; Wheatley-Price, P. Low to intermediate grade lung neuroendocrine tumours. A single centre real world experience. Cancer Treat. Res. Commun. 2024, 41, 100846. [Google Scholar] [CrossRef] [PubMed]
- Al-Toubah, T.; Morse, B.; Strosberg, J. Capecitabine and temozolomide in advanced lung neuroendocrine neoplasms. Oncologist 2020, 25, e48–e52. [Google Scholar] [CrossRef] [PubMed]
- Araujo-Castro, M.; Pascual-Corrales, E.; Molina-Cerrillo, J.; Mata, N.M.; Alonso-Gordoa, T. Bronchial carcinoids: From molecular background to treatment approach. Cancers 2022, 14, 520. [Google Scholar] [CrossRef] [PubMed]
- Rekhtman, N. Lung neuroendocrine neoplasms: Recent progress and persistent challenges. Mod. Pathol. 2022, 35 (Suppl. S1), 36–50. [Google Scholar] [CrossRef]
- Jiang, Y.; Hou, G.; Cheng, W. The utility of 18F-FDG and 68Ga-DOTA-Peptide PET/CT in the evaluation of primary pulmonary carcinoid: A systematic review and meta-analysis. Medicine 2019, 98, e14769. [Google Scholar] [CrossRef]
- Filosso, P.L.; Guerrera, F.; Falco, N.R.; Thomas, P.; Yuste, M.G.; Rocco, G.; Welter, S.; Casado, P.M.; Rendina, E.A.; Venuta, F.; et al. Anatomical resections are superior to wedge resections for overall survival in patients with stage 1 typical carcinoids. Eur. J. Cardiothorac. Surg. 2019, 55, 273–279. [Google Scholar] [CrossRef]
- Scher, H.I.; Jia, X.; de Bono, J.S.; Fleisher, M.; Pienta, K.J.; Raghavan, D.; Heller, G. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: A reanalysis of IMMC38 trial data. Lancet Oncol. 2009, 10, 233–239. [Google Scholar] [CrossRef]
- Krebs, M.G.; Sloane, R.; Priest, L.; Lancashire, L.; Hou, J.M.; Greystoke, A.; Ward, T.H.; Ferraldeschi, R.; Hughes, A.; Clack, G.; et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 2011, 29, 1556–1563. [Google Scholar] [CrossRef]
- Aggarwal, C.; Meropol, N.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.A.; Mitchell, E.; et al. Relationship among circulating tumor cells, CEA and overall survival in patients with metastatic colorectal cancer. Ann. Oncol. 2013, 24, 420–428. [Google Scholar] [CrossRef]
- Pascual, J.; Attard, G.; Bidard, F.C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.D.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef] [PubMed]
- Boons, G.; Vandamme, T.; Mariën, L.; Lybaert, W.; Roeyen, G.; Rondou, T.; Papadimitriou, K.; Janssens, K.; Op de Beeck, B.; Simoens, M.; et al. Longitudinal Copy-Number Alteration Analysis in Plasma Cell-Free DNA of Neuroendocrine Neoplasms is a Novel Specific Biomarker for Diagnosis, Prognosis, and Follow-up. Clin. Cancer Res. 2022, 28, 338–349. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zakka, K.; Nagy, R.; Drusbosky, L.; Akce, M.; Wu, C.; Alese, O.B.; El-Rayes, B.F.; Kasi, P.M.; Mody, K.; Starr, J.; et al. Blood-based next-generation sequencing analysis of neuroendocrine neoplasms. Oncotarget 2020, 11, 1749–1757. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sipola, J.; Munzur, A.D.; Kwan, E.M.; Seo, C.C.; Hauk, B.J.; Parekh, K.; Liao, Y.J.R.; Bernales, C.Q.; Donnellan, G.; Bloise, I.; et al. Plasma Cell–Free DNA Chromatin Immunoprecipitation Profiling Depicts Phenotypic and Clinical Heterogeneity in Advanced Prostate Cancer. Cancer Res. 2025, 85, 791–807. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cowzer, D.; Shah, R.H.; Chou, J.F.; Kundra, R.; Punn, S.; Fiedler, L.; DeMore, A.; Capanu, M.; Berger, M.F.; Reidy-Lagunes, D.; et al. Clinical utility of plasma cell-free DNA in pancreatic neuroendocrine neoplasms. Endocr. Relat. Cancer 2024, 31, e230292. [Google Scholar] [CrossRef]
- Franceschini, G.M.; Quaini, O.; Mizuno, K.; Orlando, F.; Ciani, Y.; Ku, S.Y.; Sigouros, M.; Rothmann, E.; Alonso, A.; Benelli, M.; et al. Noninvasive Detection of Neuroendocrine Prostate Cancer through Targeted Cell-free DNA Methylation. Cancer Discov. 2024, 14, 424–445. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shin, H.J.; Hua, J.T.; Li, H. Recent advances in understanding DNA methylation of prostate cancer. Front. Oncol. 2023, 13, 1182727. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Szkukalek, J.; Dóczi, R.; Dirner, A.; Boldizsár, Á.; Varga, Á.; Déri, J.; Lakatos, D.; Tihanyi, D.; Vodicska, B.; Schwáb, R.; et al. Personalized First-Line Treatment of Metastatic Pancreatic Neuroendocrine Carcinoma Facilitated by Liquid Biopsy and Computational Decision Support. Diagnostics 2021, 11, 1850. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Oversoe, S.K.; Sorensen, B.S.; Tabaksblat, E.M.; Gronbaek, H.; Kelsen, J. Cell-Free DNA and Clinical Characteristics in Patients with Small Intestinal or Pancreatic Neuroendocrine Tumors. Neuroendocrinology 2022, 112, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, M.; Guan, Y.; Yang, X.; Hong, L.; Wang, Y.; Li, Z.; Chen, R.; Abbas, H.A.; Chang, L.; Gong, Y.; et al. The Prognostic and Therapeutic Role of Genomic Subtyping by Sequencing Tumor or Cell-Free DNA in Pulmonary Large-Cell Neuroendocrine Carcinoma. Clin. Cancer Res. 2020, 26, 892–901. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lococo, F.; Ghaly, G.; Flamini, S.; Campanella, A.; Chiappetta, M.; Bria, E.; Vita, E.; Tortora, G.; Evangelista, J.; Sassorossi, C.; et al. Artificial intelligence applications in personalizing lung cancer management: State of the art and future perspectives. J. Thorac. Dis. 2024, 16, 7096–7110. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Modlin, L.M.; Lye, K.D.; Kidd, M. A 5-decade analysis of 13,715 carcinoid tumors. Cancer 2003, 97, 934–959. [Google Scholar] [CrossRef] [PubMed]
- Brabander, T.; van der Zwan, W.A.; Teunissen, J.J.; Kam, B.L.; Feelders, R.A.; de Herder, W.W.; van Eijck, C.H.; Franssen, G.J.; Krenning, E.P.; Kwekkeboom, D.J. Long-term efficacy, survival, and safety of [177Lu-DOTA0,Tyr3]octreotate in patients with gastroenteropancreatic and bronchial neuroendocrine tumors. Clin. Cancer Res. 2017, 23, 4617–4624. [Google Scholar] [CrossRef] [PubMed]
- Dermawan, J.K.T.; Farver, C.F. The role of histologic grading and Ki-67 Index in predicting outcomes in pulmonary carcinoid tumors. Am. J. Surg. Pathol. 2020, 44, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Marchiò, C.; Gatti, G.; Massa, F.; Bertero, L.; Filosso, P.; Pelosi, G.; Cassoni, P.; Volante, M.; Papotti, M. Distinctive pathological and clinical features of lung carcinoids with high proliferation index. Virchows Arch. 2017, 471, 713–720. [Google Scholar] [CrossRef]
- Savu, C.; Melinte, A.; Diaconu, C.; Stiru, O.; Gherghiceanu, F.; Tudorica, Ș.D.O.; Dumitrașcu, O.C.; Bratu, A.; Balescu, I.; Bacalbasa, N. Lung neuroendocrine tumors: A systematic literature review (Review). Exp. Ther. Med. 2021, 23, 176. [Google Scholar] [CrossRef]
- Fox, M.; Van Berkel, V.; Bousamra, M.; Sloan, S.; Martin, R.C.G. Surgical management of pulmonary carcinoid tumors: Sublobar resection versus lobectomy. Am. J. Surg. 2013, 205, 200–208. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Lococo, F.; Boldrini, L.; Diepriye, C.D.; Evangelista, J.; Nero, C.; Flamini, S.; Minucci, A.; De Paolis, E.; Vita, E.; Cesario, A.; et al. Lung cancer multi-omics digital human avatars for integrating precision medicine into clinical practice: The LANTERN study. BMC Cancer 2023, 23, 1082, Erratum in BMC Cancer 2023, 23, 540. [Google Scholar]
- Lococo, F.; Rapicetta, C.; Mengoli, M.C.; Filice, A.; Paci, M.; Di Stefano, T.; Coruzzi, C.; Versari, A. Diagnostic performances of 68Ga-DOTATOC versus 18Fluorodeoxyglucose positron emission tomography in pulmonary carcinoid tumours and interrelationship with histological features. Interact. Cardiovasc. Thorac. Surg. 2019, 28, 957–960. [Google Scholar] [CrossRef] [PubMed]
- Heeke, S.; Gay, C.M.; Estecio, M.R.; Tran, H.; Morris, B.B.; Zhang, B.; Tang, X.; Raso, M.G.; Rocha, P.; Lai, S.; et al. Tumor- and circulating-free DNA methylation identifies clinically relevant small cell lung cancer subtypes. Cancer Cell 2024, 42, 225–237.e5. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).