Deep Learning-Based Detection of Intracranial Hemorrhages in Postmortem Computed Tomography: Comparative Study of 15 Transfer-Learned Models
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Postmortem Imaging Dataset
2.2. Non-Postmortem Imaging Dataset
2.3. Software and Hardware Environment
2.4. DL Models
2.5. Image Processing Steps for Each Case
2.6. Inference on Postmortem Images
2.7. Evaluation Methods
2.8. Statistical Analysis
2.9. Reader Study by a Radiology Resident
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| AUC | Area under the curve |
| CT | Computed tomography |
| DL | Deep learning |
| MRI | Magnetic resonance imaging |
| ROC | Receiver operating characteristic |
| ROC AUC | Area under the ROC curve |
| RSNA | Radiological Society of North America |
References
- Bolliger, S.A.; Thali, M.J. Imaging and virtual autopsy: Looking back and forward. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140253. [Google Scholar] [CrossRef]
- Roberts, I.S.D.; Benamore, R.E.; Benbow, E.W.; Lee, S.H.; Harris, J.N.; Jackson, A.; Mallett, S.; Patankar, T.; Peebles, C.; Roobottom, C.; et al. Post-mortem imaging as an alternative to autopsy in the diagnosis of adult deaths: A validation study. Lancet 2012, 379, 136–142. [Google Scholar] [CrossRef]
- Zhou, L.Q.; Wang, J.Y.; Yu, S.Y.; Wu, G.G.; Wei, Q.; Deng, Y.B.; Wu, X.L.; Cui, X.W.; Dietrich, C.F. Artificial intelligence in medical imaging of the liver. World J. Gastroenterol. 2019, 25, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Dey, D.; Slomka, P.J.; Leeson, P.; Comaniciu, D.; Shrestha, S.; Sengupta, P.P.; Marwick, T.H. Artificial intelligence in cardiovascular imaging: JACC state-of-the-art review. J. Am. Coll. Cardiol. 2019, 73, 1317–1335. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhu, J.; Duan, Z.; Liao, Z.; Wang, S.; Liu, W. Artificial intelligence in spinal imaging: Current status and future directions. Int. J. Environ. Res. Public Health 2022, 19, 11708. [Google Scholar] [CrossRef]
- Matsuo, H.; Kitajima, K.; Kono, A.K.; Kuribayashi, K.; Kijima, T.; Hashimoto, M.; Hasegawa, S.; Yamakado, K.; Murakami, T. Prognosis prediction of patients with malignant pleural mesothelioma using conditional variational autoencoder on 3D PET images and clinical data. Med. Phys. 2023, 50, 7548–7557. [Google Scholar] [CrossRef]
- Matsuo, H.; Nishio, M.; Kanda, T.; Kojita, Y.; Kono, A.K.; Hori, M.; Teshima, M.; Otsuki, N.; Nibu, K.I.; Murakami, T. Diagnostic accuracy of deep-learning with anomaly detection for a small amount of imbalanced data: Discriminating malignant parotid tumors in MRI. Sci. Rep. 2020, 10, 19388. [Google Scholar] [CrossRef]
- Nishio, M.; Noguchi, S.; Matsuo, H.; Murakami, T. Automatic classification between COVID-19 pneumonia, non-COVID-19 pneumonia, and the healthy on chest X-ray image: Combination of data augmentation methods. Sci. Rep. 2020, 10, 17532. [Google Scholar] [CrossRef]
- Kumari, R.; Nikki, S.; Beg, R.; Ranjan, S.; Gope, S.K.; Mallick, R.R.; Dutta, A. A review of image detection, recognition and classification with the help of machine learning and artificial intelligence. SSRN J. 2020. [Google Scholar] [CrossRef]
- Kim, H.E.; Cosa-Linan, A.; Santhanam, N.; Jannesari, M.; Maros, M.E.; Ganslandt, T. Transfer learning for medical image classification: A literature review. BMC Med. Imaging 2022, 22, 69. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Al-Amidie, M.; Al-Asadi, A.; Humaidi, A.J.; Al-Shamma, O.; Fadhel, M.A.; Zhang, J.; Santamaría, J.; Duan, Y. Novel Transfer Learning Approach for Medical Imaging with Limited Labeled Data. Cancers 2021, 13, 1590. [Google Scholar] [CrossRef]
- Li, M.; Jiang, Y.; Zhang, Y.; Zhu, H. Medical image analysis using deep learning algorithms. Front. Public Health 2023, 11, 1273253. [Google Scholar] [CrossRef]
- Thali, M.J.; Yen, K.; Schweitzer, W.; Vock, P.; Boesch, C.; Ozdoba, C.; Schroth, G.; Ith, M.; Sonnenschein, M.; Doernhoefer, T.; et al. Virtopsy, a new imaging horizon in forensic pathology: Virtual autopsy by postmortem multislice computed tomography (MSCT) and magnetic resonance imaging (MRI)—A feasibility study. J. Forensic Sci. 2003, 48, 386–403. [Google Scholar] [CrossRef]
- Arbabshirani, M.R.; Fornwalt, B.K.; Mongelluzzo, G.J.; Suever, J.D.; Geise, B.D.; Patel, A.A.; Moore, G.J. Advanced machine learning in action: Identification of intracranial hemorrhage on computed tomography scans of the head with clinical workflow integration. npj Digit. Med. 2018, 1, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shen, T.; Yang, S.; Lan, J.; Xu, Y.; Wang, M.; Zhang, J.; Han, X. A deep learning algorithm for automatic detection and classification of acute intracranial hemorrhages in head CT scans. NeuroImage Clin. 2021, 32, 102785. [Google Scholar] [CrossRef] [PubMed]
- Flanders, A.E.; Prevedello, L.M.; Shih, G.; Halabi, S.S.; Kalpathy-Cramer, J.; Ball, R.; Mongan, J.T.; Stein, A.; Kitamura, F.C.; Lungren, M.P.; et al. RSNA-ASNR 2019 Brain Hemorrhage CT Annotators, Construction of a machine learning dataset through collaboration: The RSNA 2019 brain CT hemorrhage challenge. Radiol. Artif. Intell. 2020, 2, e190211. [Google Scholar] [CrossRef]
- PyTorch-Image-Models: The Largest Collection of PyTorch Image Encoders/Backbones. Including Train, Eval, Inference, Export Scripts, and Pretrained Weights—ResNet, ResNeXT, EfficientNet, NFNet, Vision Transformer (ViT), MobileNetV4, MobileNet-V3 & V2, RegNet, DPN, CSPNet, Swin Transformer, MaxViT, CoAtNet, ConvNeXt, and More. GitHub. Available online: https://github.com/huggingface/pytorch-image-models (accessed on 26 January 2025).
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. arXiv 2016, arXiv:1608.06993. Available online: http://arxiv.org/abs/1608.06993 (accessed on 26 January 2025).
- Brock, A.; De, S.; Smith, S.L.; Simonyan, K. High-performance large-scale image recognition without normalization. arXiv 2021, arXiv:2102.06171. Available online: http://arxiv.org/abs/2102.06171 (accessed on 26 January 2025).
- Tan, M.; Le, Q.V. EfficientNet: Rethinking model scaling for convolutional Neural Networks. arXiv 2019, arXiv:1905.11946. Available online: http://arxiv.org/abs/1905.11946 (accessed on 26 January 2025).
- Tan, M.; Le, Q.V. EfficientNetV2: Smaller models and faster training. arXiv 2021, arXiv:2104.00298. Available online: http://arxiv.org/abs/2104.00298 (accessed on 26 January 2025).
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.C.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. Searching for MobileNetV3. arXiv 2019, arXiv:1905.02244. Available online: http://arxiv.org/abs/1905.02244 (accessed on 26 January 2025).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. arXiv 2016, arXiv:1603.05027. Available online: http://arxiv.org/abs/1603.05027 (accessed on 26 January 2025).
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. arXiv 2016, arXiv:1611.05431. Available online: http://arxiv.org/abs/1611.05431 (accessed on 26 January 2025).
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical vision Transformer using shifted windows. arXiv 2021, arXiv:2103.14030. Available online: http://arxiv.org/abs/2103.14030 (accessed on 26 January 2025).
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. Available online: http://arxiv.org/abs/1409.1556 (accessed on 26 January 2025).
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterhiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. Available online: http://arxiv.org/abs/2010.11929 (accessed on 26 January 2025).
- Cubuk, E.D.; Zoph, B.; Shlens, J.; Le, Q.V. RandAugment: Practical automated data augmentation with a reduced search space. arXiv 2019, arXiv:1909.13719. Available online: http://arxiv.org/abs/1909.13719 (accessed on 26 January 2025). [CrossRef]
- Nishio, M.; Koyasu, S.; Noguchi, S.; Kiguchi, T.; Nakatsu, K.; Akasaka, T.; Yamada, H.; Itoh, K. Automatic detection of acute ischemic stroke using non-contrast computed tomography and two-stage deep learning model. Comput. Methods Programs Biomed. 2020, 196, 105711. [Google Scholar] [CrossRef] [PubMed]


| Model | Parameter | Date |
|---|---|---|
| densenet201 [18] | 1.81 × 107 | 25 August 2016 |
| dm_nfnet_f0 [19] | 6.84 × 107 | 11 February 2021 |
| efficientnet_b2 [20] | 7.71 × 106 | 28 May 2019 |
| efficientnetv2_rw_m [21] | 5.11 × 107 | 01 April 2021 |
| efficientnetv2_rw_s [21] | 2.22 × 107 | 01 April 2021 |
| mobilenetv3_rw [22] | 4.21 × 106 | 06 May 2019 |
| resnetv2_101×1_bitm [23] | 4.25 × 107 | 16 March 2016 |
| resnetv2_101×1_bitm_in21k [23] | 4.25 × 107 | 16 March 2016 |
| resnext101_32×8d [24] | 8.68 × 107 | 16 November 2016 |
| resnext101_64×4d [24] | 8.14 × 107 | 16 November 2016 |
| swin_large_patch4_window7_224 [25] | 6.23 × 106 | 25 May 2021 |
| vgg16 [26] | 1.34 × 108 | 04 September 2014 |
| vgg16_bn [26] | 1.34 × 108 | 04 September 2014 |
| vit_base_patch32_224 [27] | 8.74 × 107 | 22 October 2020 |
| vit_large_patch16_224 [27] | 3.03 × 108 | 20 October 2020 |
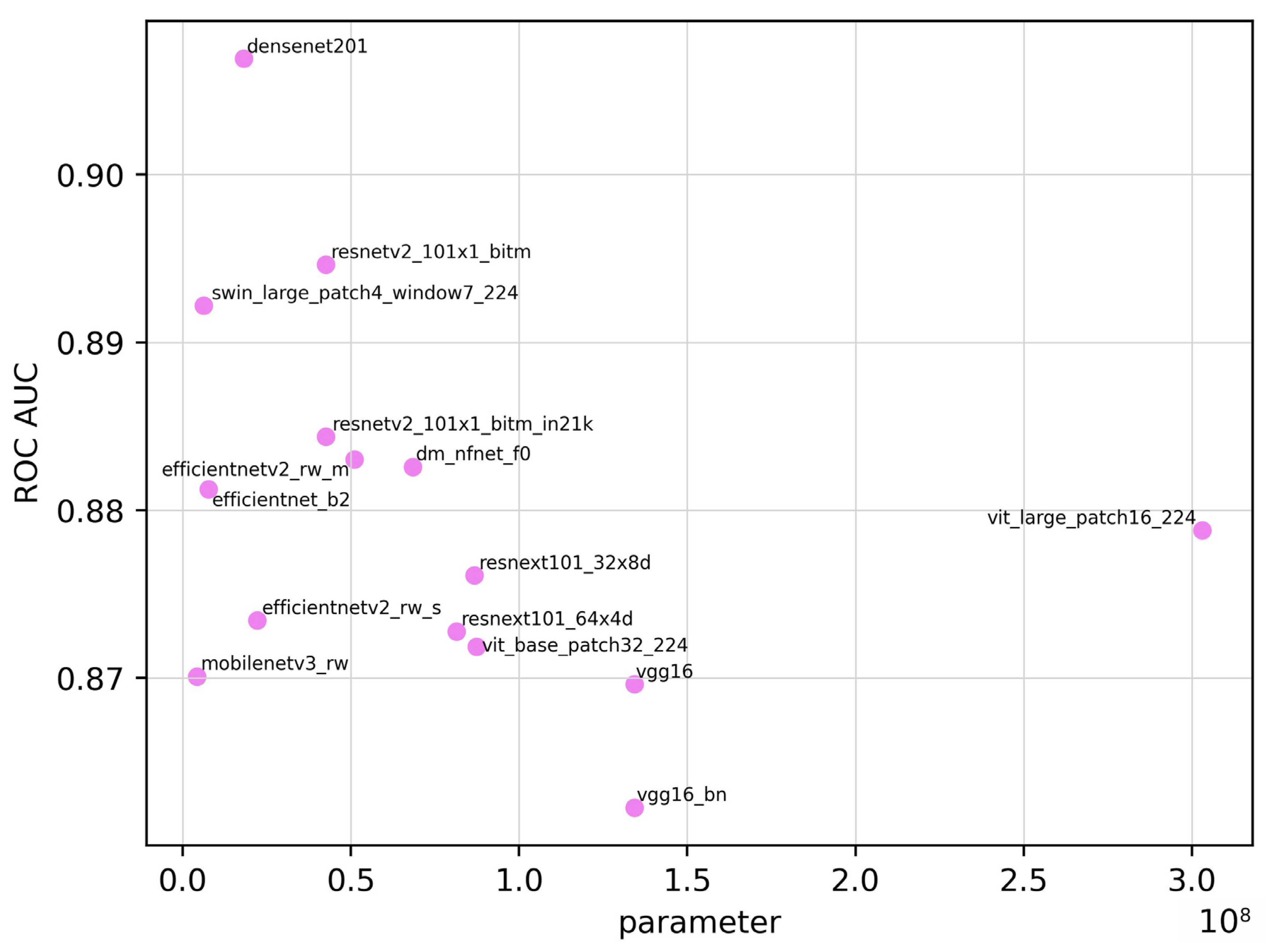
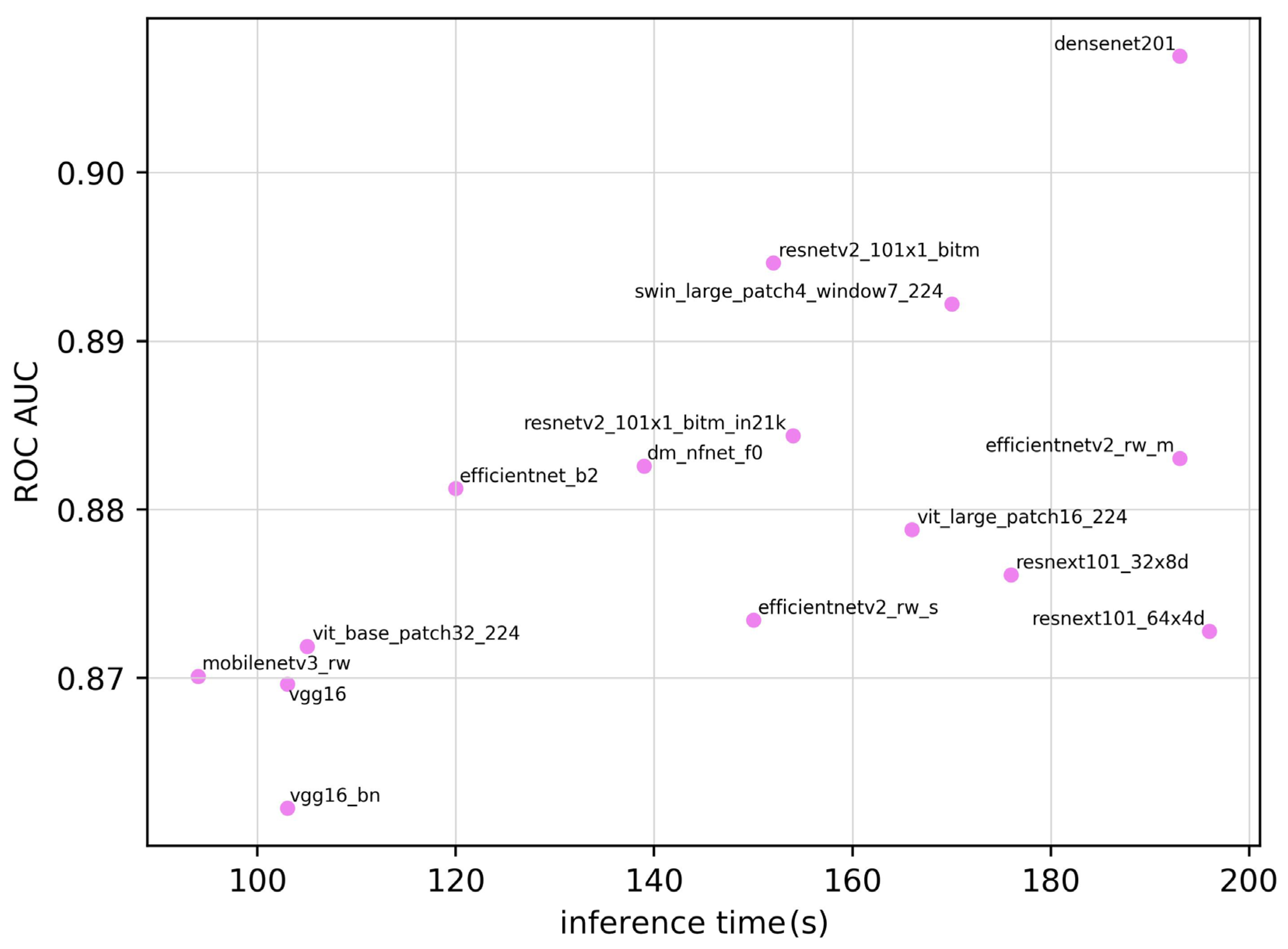
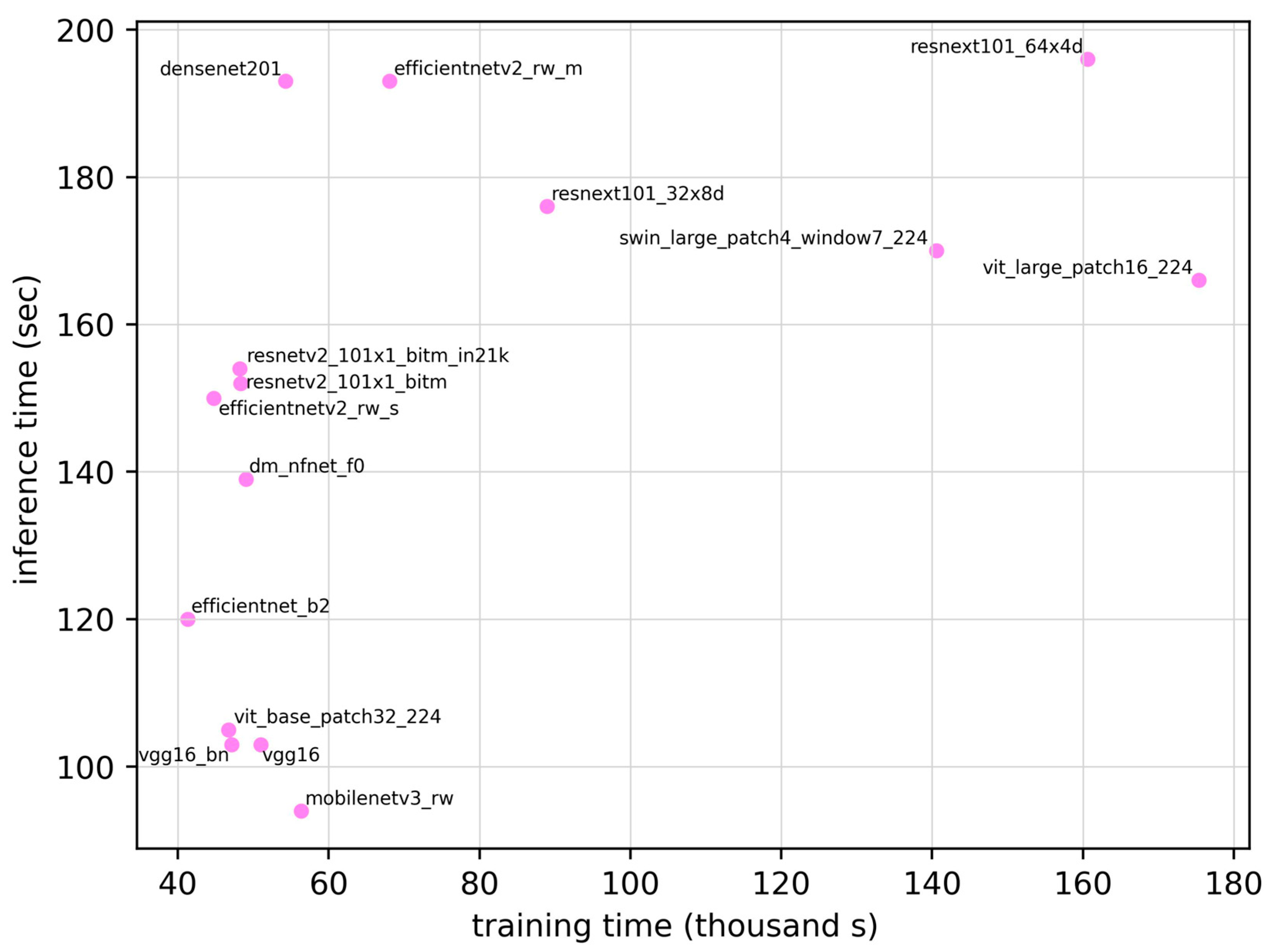
| Model | ROC AUC | ROC AUC 95% CI | Training Time (seconds) | Inference Time (seconds) | Parameter | Sensitivity | Specificity | Accuracy | F1 Score |
|---|---|---|---|---|---|---|---|---|---|
| densenet201 | 0.907 | 0.854–0.960 | 5.43 × 104 | 193 | 1.81 × 107 | 0.828 | 0.871 | 0.850 | 0.841 |
| dm_nfnet_f0 | 0.883 | 0.824–0.942 | 4.90 × 104 | 139 | 6.84 × 107 | 0.828 | 0.814 | 0.821 | 0.815 |
| efficientnet_b2 | 0.881 | 0.821–0.941 | 4.13 × 104 | 120 | 7.71 × 106 | 0.859 | 0.814 | 0.835 | 0.832 |
| efficientnetv2_rw_m | 0.883 | 0.824–0.942 | 6.80 × 104 | 194 | 5.11 × 107 | 0.703 | 0.957 | 0.836 | 0.803 |
| efficientnetv2_rw_s | 0.873 | 0.811–0.935 | 4.48 × 104 | 151 | 2.22 × 107 | 0.672 | 0.986 | 0.836 | 0.797 |
| mobilenetv3_rw | 0.870 | 0.807–0.933 | 5.64 × 104 | 95 | 4.21 × 106 | 0.656 | 0.957 | 0.813 | 0.770 |
| resnetv2_101×1_bitm | 0.895 | 0.839–0.951 | 4.83 × 104 | 153 | 4.25 × 107 | 0.734 | 0.957 | 0.850 | 0.824 |
| resnetv2_101×1_bitm_in21k | 0.884 | 0.825–0.943 | 4.82 × 104 | 154 | 4.25 × 107 | 0.781 | 0.886 | 0.836 | 0.820 |
| resnext101_32×8d | 0.876 | 0.815–0.937 | 8.89 × 104 | 176 | 8.68 × 107 | 0.750 | 0.886 | 0.821 | 0.800 |
| resnext101_64×4d | 0.873 | 0.811–0.935 | 1.61 × 105 | 197 | 8.14 × 107 | 0.734 | 0.843 | 0.791 | 0.770 |
| swin_large_patch4_window7_224 | 0.892 | 0.835–0.949 | 1.41 × 105 | 170 | 6.23 × 106 | 0.906 | 0.700 | 0.798 | 0.811 |
| vgg16 | 0.870 | 0.807–0.933 | 5.10 × 104 | 103 | 1.34 × 108 | 0.750 | 0.900 | 0.828 | 0.807 |
| vgg16_bn | 0.862 | 0.798–0.926 | 4.72 × 104 | 104 | 1.34 × 108 | 0.750 | 0.871 | 0.813 | 0.793 |
| vit_base_patch32_224 | 0.872 | 0.810–0.934 | 4.67 × 104 | 105 | 8.74 × 107 | 0.750 | 0.871 | 0.813 | 0.793 |
| vit_base_patch16_224 | 0.879 | 0.819–0.939 | 1.75 × 105 | 167 | 3.03 × 108 | 0.672 | 0.929 | 0.806 | 0.768 |
| Radiology Resident | 0.810 | 0.736–0.884 | 0.600 | 1.000 | 0.809 | 0.750 |
| Confusion Matrix | Predicted | ||
|---|---|---|---|
| Positive | Negative | ||
| Actual | Positive | 53 | 11 |
| Negative | 9 | 61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsumoto, R.; Matsuo, H.; Sugimoto, M.; Matsunaga, T.; Nishio, M.; Kono, A.K.; Yamasaki, G.; Takahashi, M.; Kondo, T.; Ueno, Y.; et al. Deep Learning-Based Detection of Intracranial Hemorrhages in Postmortem Computed Tomography: Comparative Study of 15 Transfer-Learned Models. Appl. Sci. 2025, 15, 10513. https://doi.org/10.3390/app151910513
Matsumoto R, Matsuo H, Sugimoto M, Matsunaga T, Nishio M, Kono AK, Yamasaki G, Takahashi M, Kondo T, Ueno Y, et al. Deep Learning-Based Detection of Intracranial Hemorrhages in Postmortem Computed Tomography: Comparative Study of 15 Transfer-Learned Models. Applied Sciences. 2025; 15(19):10513. https://doi.org/10.3390/app151910513
Chicago/Turabian StyleMatsumoto, Rentaro, Hidetoshi Matsuo, Marie Sugimoto, Takaaki Matsunaga, Mizuho Nishio, Atsushi K. Kono, Gentaro Yamasaki, Motonori Takahashi, Takeshi Kondo, Yasuhiro Ueno, and et al. 2025. "Deep Learning-Based Detection of Intracranial Hemorrhages in Postmortem Computed Tomography: Comparative Study of 15 Transfer-Learned Models" Applied Sciences 15, no. 19: 10513. https://doi.org/10.3390/app151910513
APA StyleMatsumoto, R., Matsuo, H., Sugimoto, M., Matsunaga, T., Nishio, M., Kono, A. K., Yamasaki, G., Takahashi, M., Kondo, T., Ueno, Y., Katada, R., & Murakami, T. (2025). Deep Learning-Based Detection of Intracranial Hemorrhages in Postmortem Computed Tomography: Comparative Study of 15 Transfer-Learned Models. Applied Sciences, 15(19), 10513. https://doi.org/10.3390/app151910513







