This section presents the results obtained from the two main classification approaches explored in this study: the Fuzzy ARTMAP classifier, evaluated under various feature selection strategies, and the ANFIS model, analyzed under both automatic and manual configurations. Model performance was assessed using confusion matrices, ROC curves, and standard evaluation metrics, including sens, spe, pre, F1-score, and overall acc. This comparative analysis highlights how feature selection and membership function design influence each classifier’s ability to generalize and discriminate among multiple classes
5.2. Automatic ANFIS Analysis
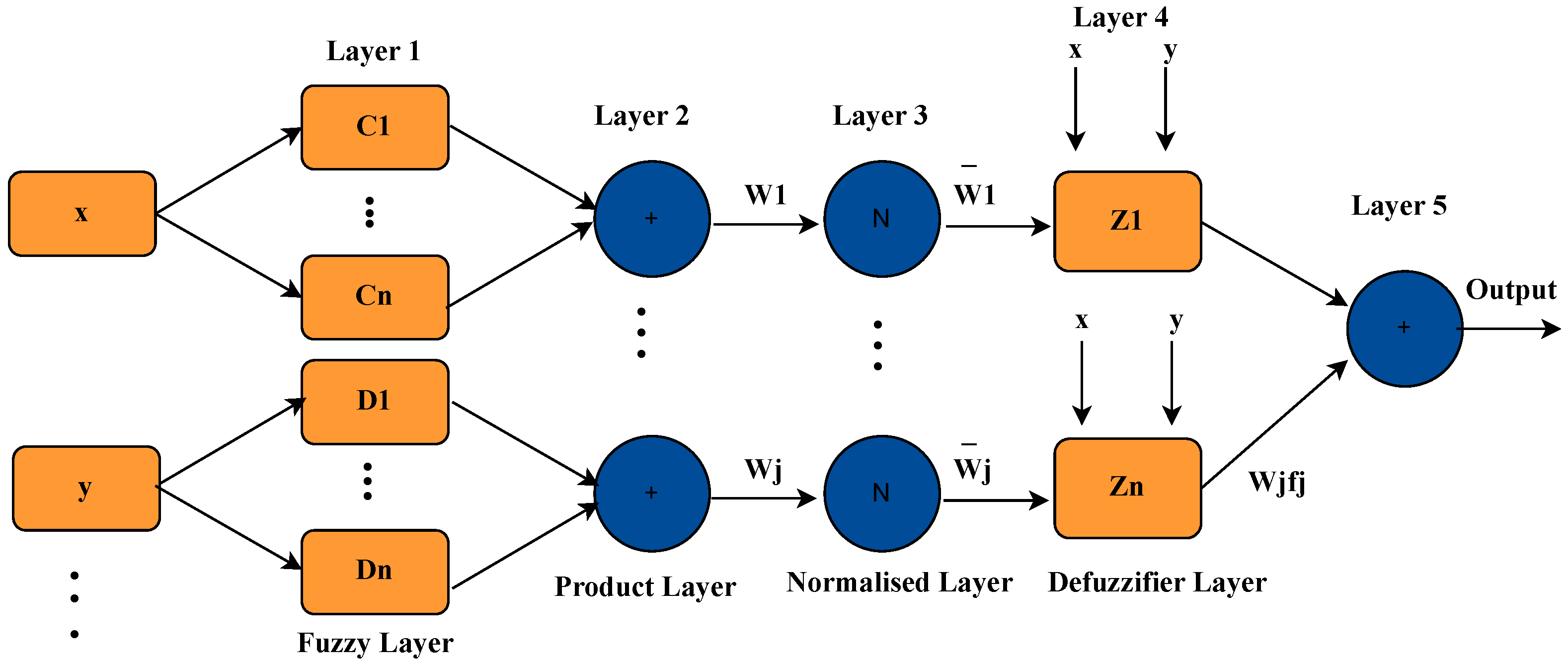
The ANFIS system was trained using 64 parameters previously selected from the CNN, which represent relevant and discriminative characteristics extracted from the ECG signals. These parameters were used as input for the adaptive fuzzy system with the goal of generating a reduced and efficient set of inference rules.
During the supervised training process, ANFIS generated a total of 10 fuzzy rules, which were constructed from the combination of multiple input parameters and their respective class labels (Normal, VF, VT, etc.). It is important to note that these rules are not individually assigned to each parameter; instead, each grouping of several attributes defines specific decision regions in the multidimensional feature space. Each rule represents a typical pattern associated with a class and defines a particular combination of input conditions that lead to a determined output. In this way, the system can capture complex relationships between physiological features without requiring a large number of rules, which favors interpretability and computational efficiency.
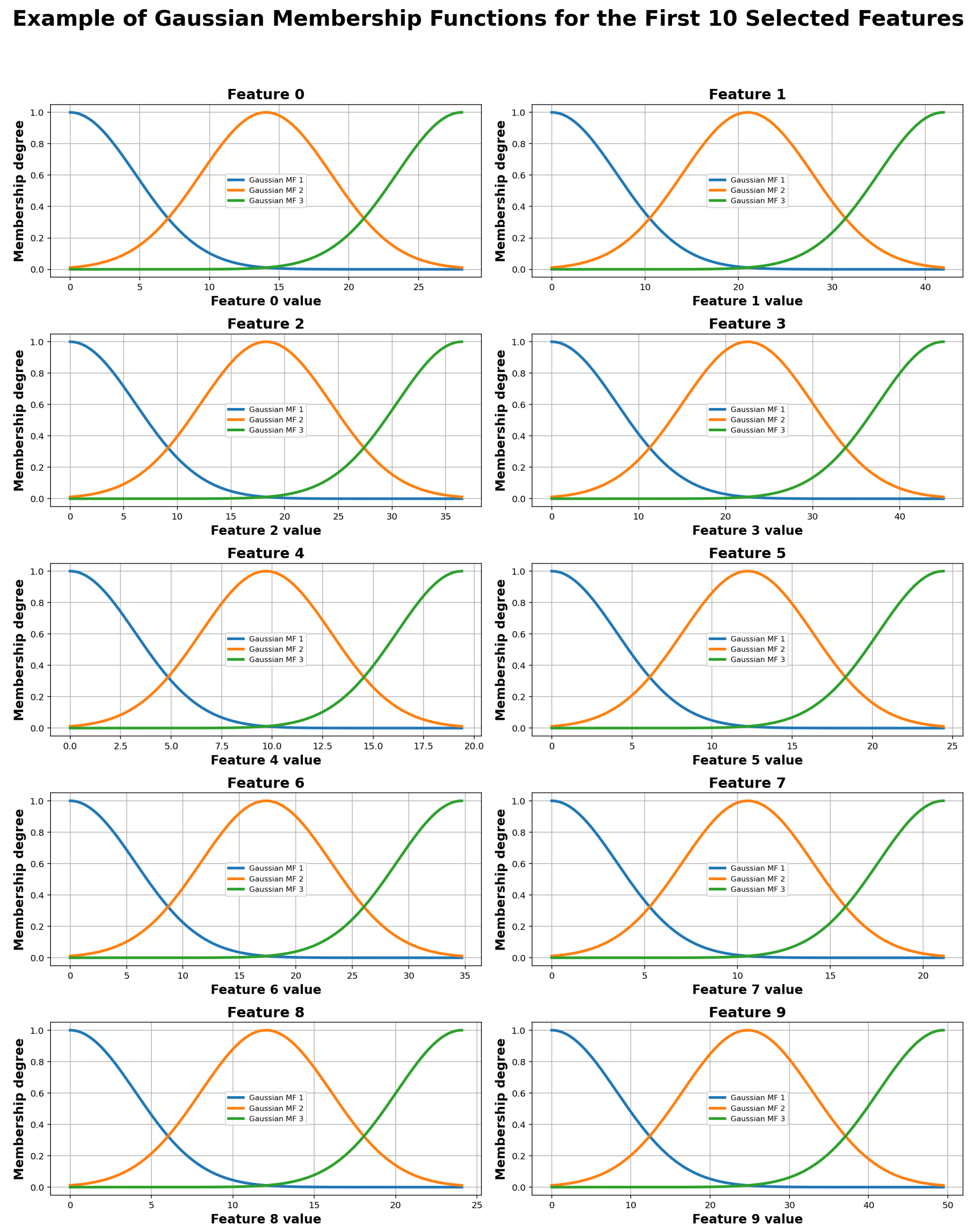
In this approach, training not only focuses on adjusting membership functions over the parameters but also centers on the generation and refinement of rules that encapsulate the knowledge learned from labeled data. This set of rules forms the basis for decision making in the ANFIS model and constitutes a key component of its generalization capability. An example of the trapezoidal membership functions generated for four selected parameters (0, 1, 6, and 7) is shown in
Figure 7, where each parameter generates 10 membership functions.
5.3. Analysis of the ARTMAP Classifier
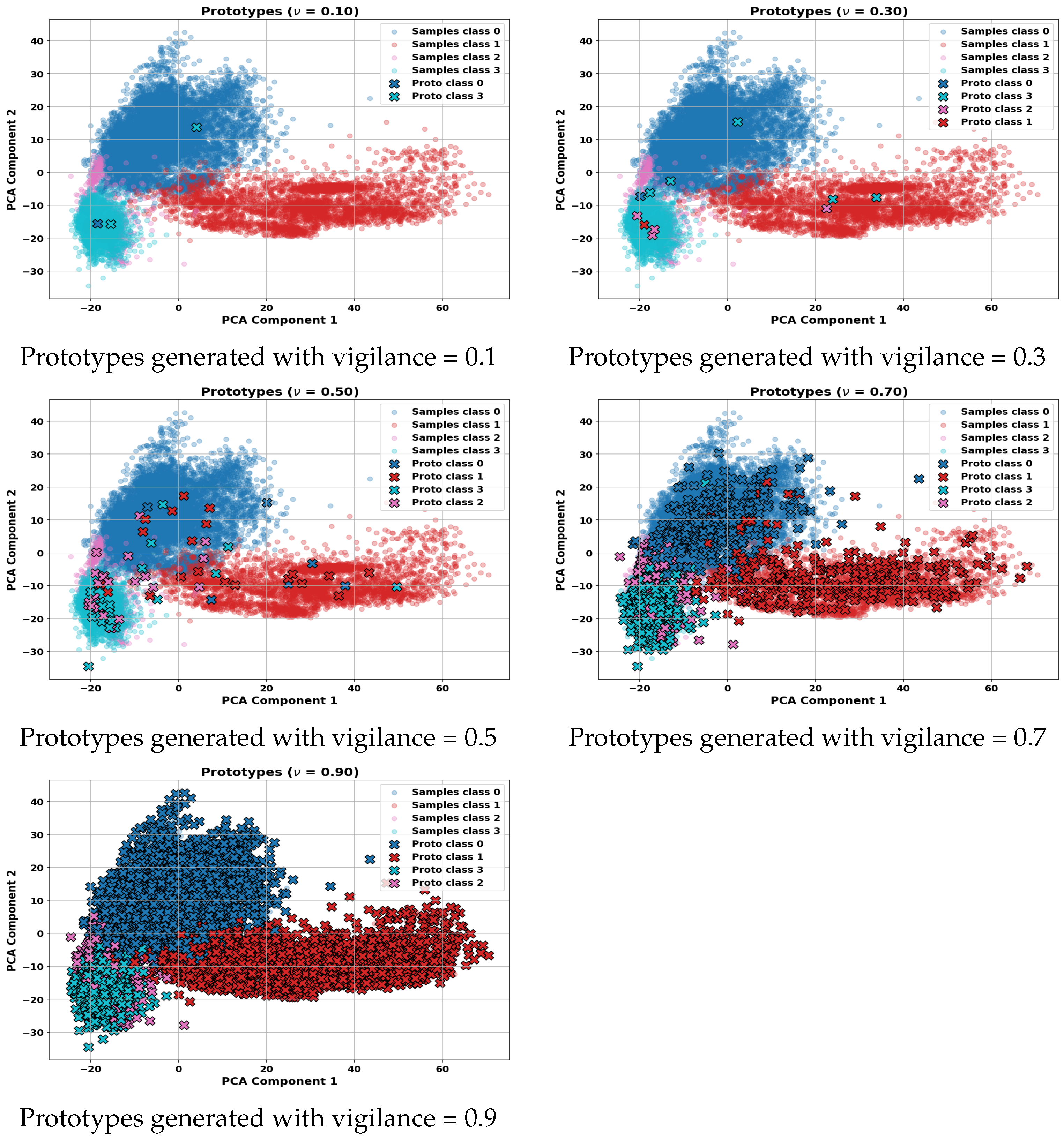
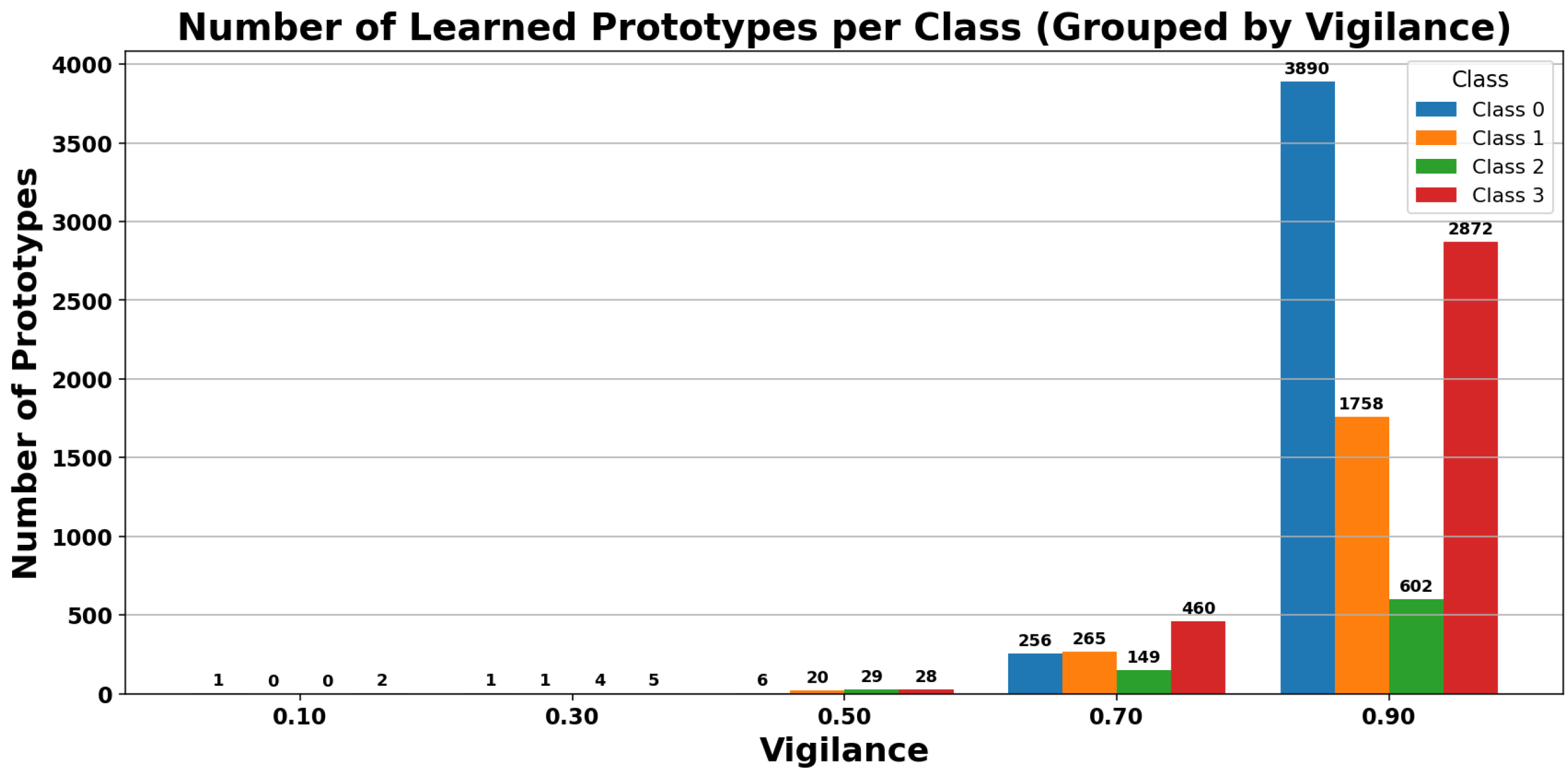
Figure 8 shows visual representations of the prototypes generated by the ARTMAP classifier at different levels of vigilance parameters, projected onto the plane of the first two principal components (PC1 and PC2). As the vigilance value increases, the number of prototypes generated also increases, reflecting greater specialization and segmentation of the feature space. This behavior allows for better class discrimination, but also increases model complexity and the risk of overfitting if not properly regulated.
Figure 9,
Figure 10 and
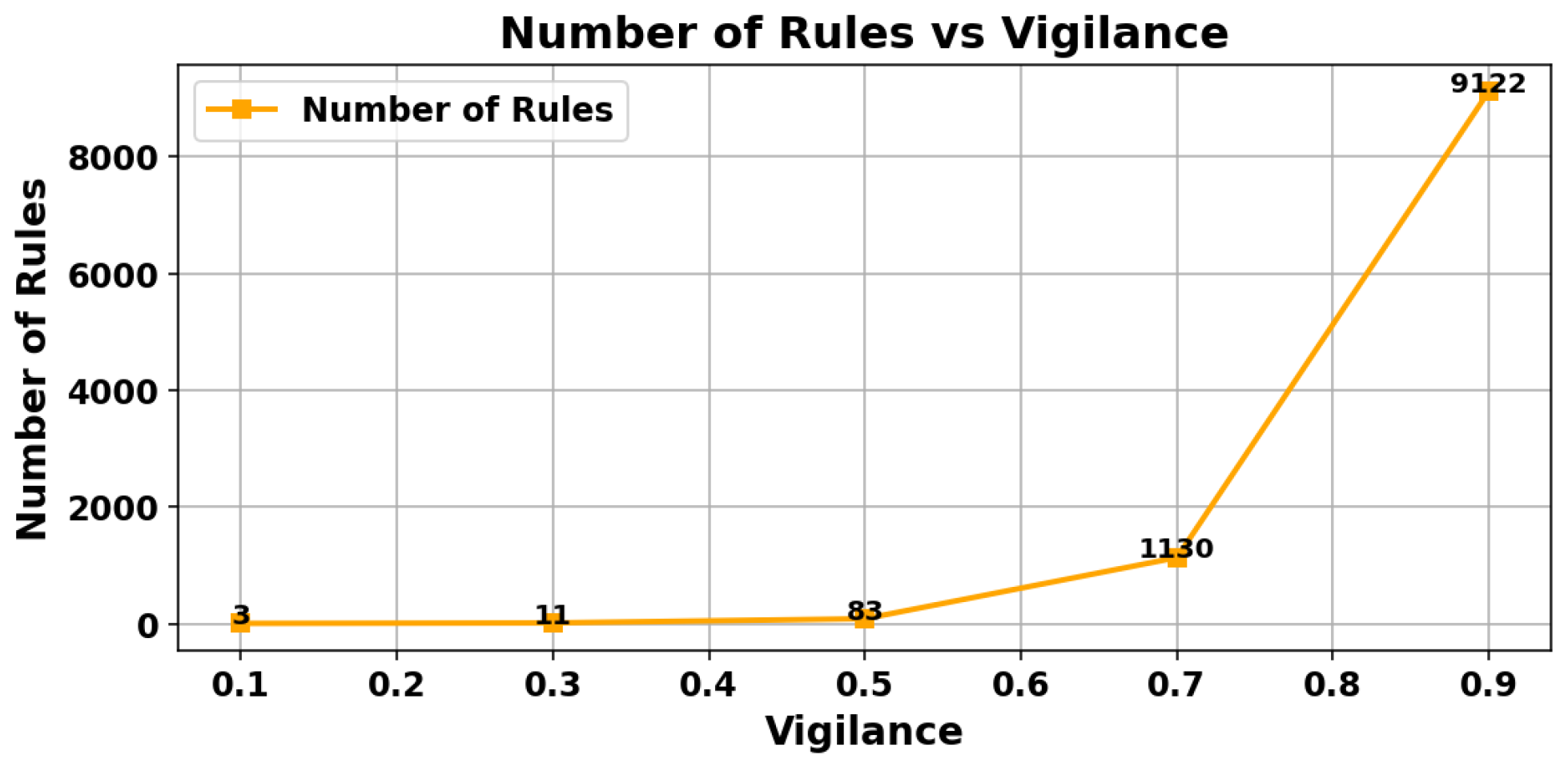
Figure 11 present a quantitative analysis of the effect of the vigilance parameter on the performance of the ARTMAP classifier.
Figure 9 shows that the total number of prototypes increases as the vigilance parameter increases, confirming a greater segmentation of the feature space.
Figure 10 presents the number of rules generated per class, allowing analysis of the complexity associated with each class and its contribution to the overall model behavior. Finally,
Figure 11 illustrates the variation in classifier accuracy. Although initially an increase in vigilance improves performance, a saturation point is reached (around vigilance = 0.7), after which the accuracy stabilizes or even decreases, indicating potential overfitting of the model.
Selection of the Vigilance Parameter
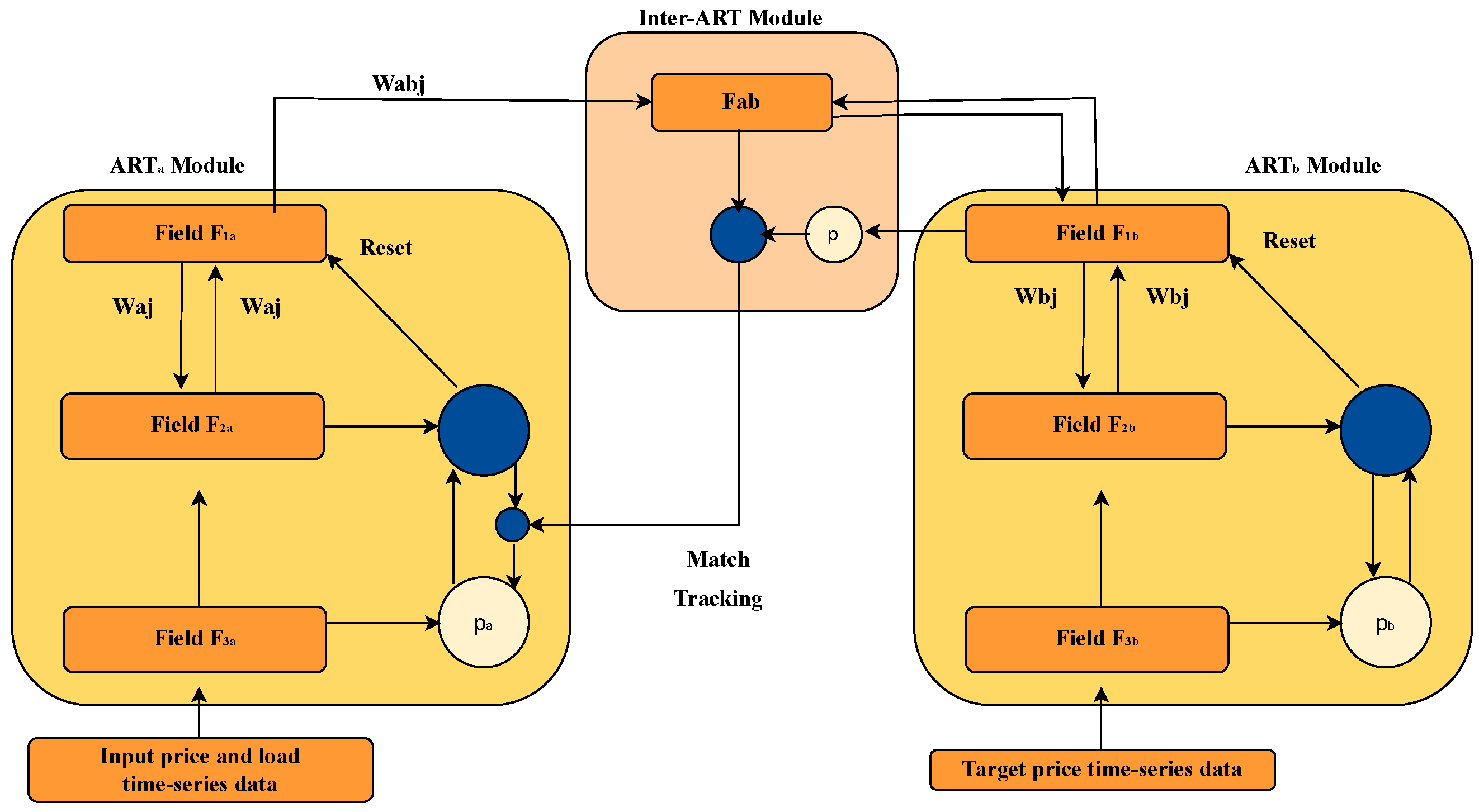
In theory, the ART-B algorithm allows dynamic adjustment of the vigilance parameter through a mechanism known as match tracking. However, in this study, the standard ARTMAP version with manual vigilance tuning was employed, which enabled controlled analysis of the model’s behavior across different values of this parameter.
Vigilance values ranging from 0.1 to 0.9 were evaluated. The analysis focused solely on the accuracy metric, observing how it varied as a function of the parameter. Although values such as 0.7 offered an acceptable balance, the value of 0.9 achieved the highest classification accuracy. For this reason, vigilance = 0.9 was selected as the final configuration for the subsequent phases of this study, including its use in comparison with other classifiers such as ANFIS. This decision is based exclusively on performance criteria (accuracy), without considering complexity or the number of rules.
5.4. Execution Times and Classifier Performance
First, the performance of the FuzzyARTMAP classifier was evaluated using different feature selection methods, obtaining processing, selection, and prediction times for each case (see
Table 3). Among all methods, Random Forest (RF) provided the best balance between accuracy and speed, with a total execution time of approximately 0.470–0.480 s, far below other methods such as RFE (∼97 s), LDA (∼196 s), or PCA (∼295 s), and relatively close to the times obtained by ANFIS. Due to this result, FuzzyARTMAP with RF was selected as a reference for comparative tests with ANFIS, as it offered high performance with low computational cost.
Subsequently, the execution time of the ANFIS classifier was analyzed, both in manual and automatic configurations, using different membership functions (triangular, Gaussian, trapezoidal, and combinations), as shown in
Table 4. In all cases, ANFIS showed very low total execution times, ranging from 0.230 to 0.331 s, with a slight advantage for manual configurations in the prediction stage (≈0.9 ms versus 2.8–5.1 ms in automatic mode). Differences between membership types were minimal, showing that ANFIS, regardless of configuration, offers significantly faster execution than FuzzyARTMAP with heavier feature selection methods, and only slightly faster execution than FuzzyARTMAP with RF.
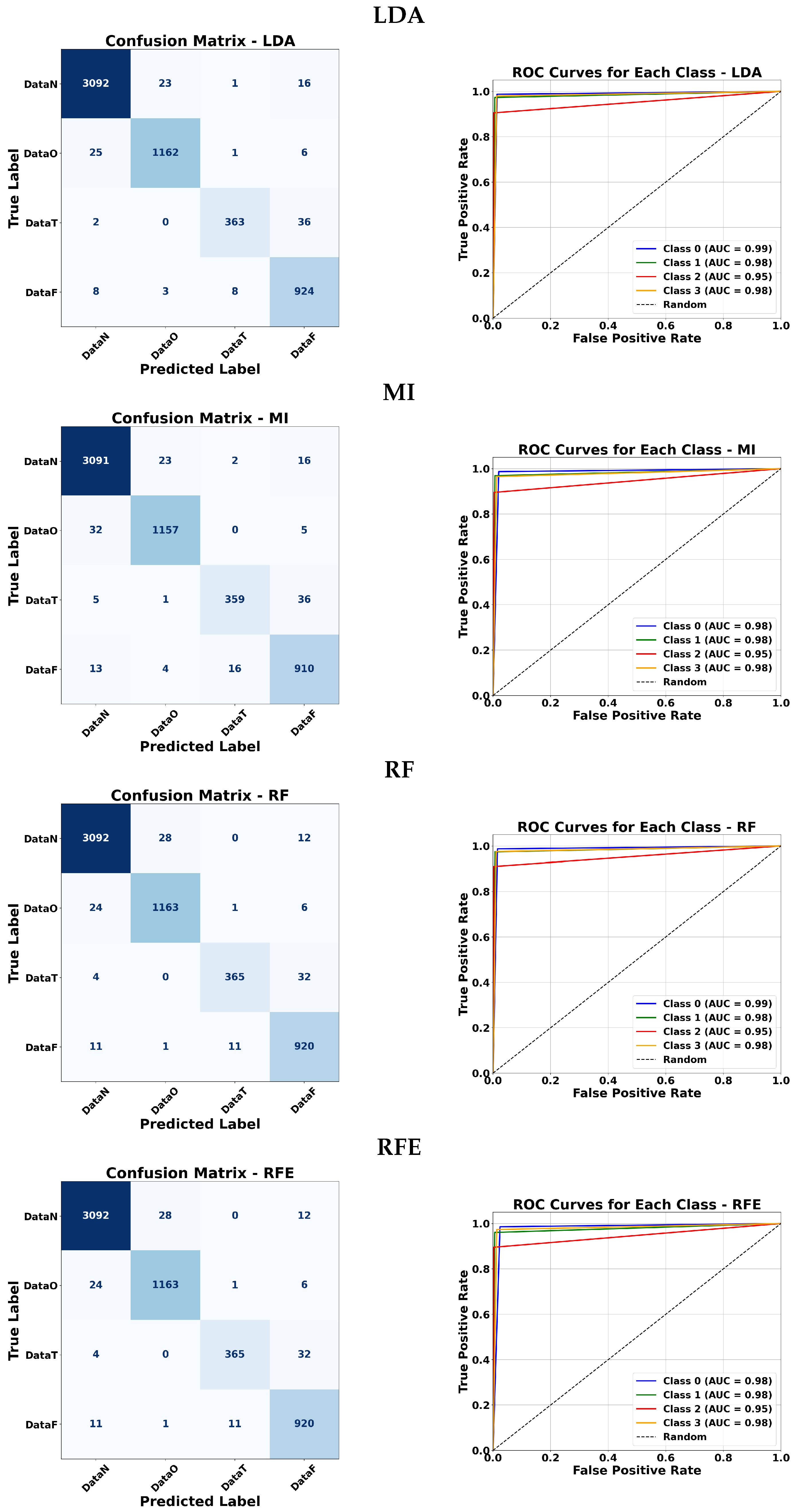
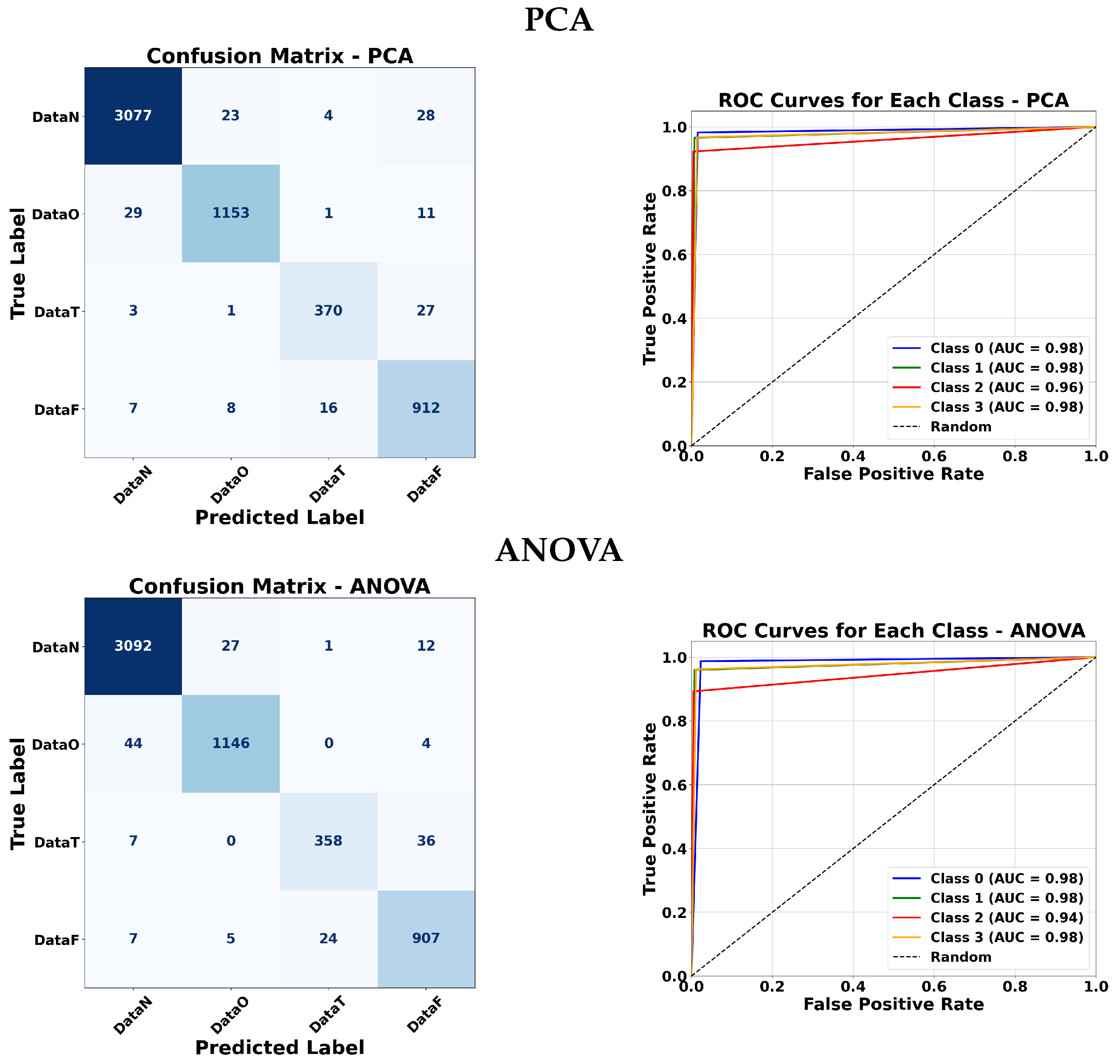
5.5. Confusion Matrices and ROC Curve Analysis
To further evaluate the effectiveness of each classifier, confusion matrices and ROC curves are presented.
Figure 12,
Figure 13,
Figure 14 and
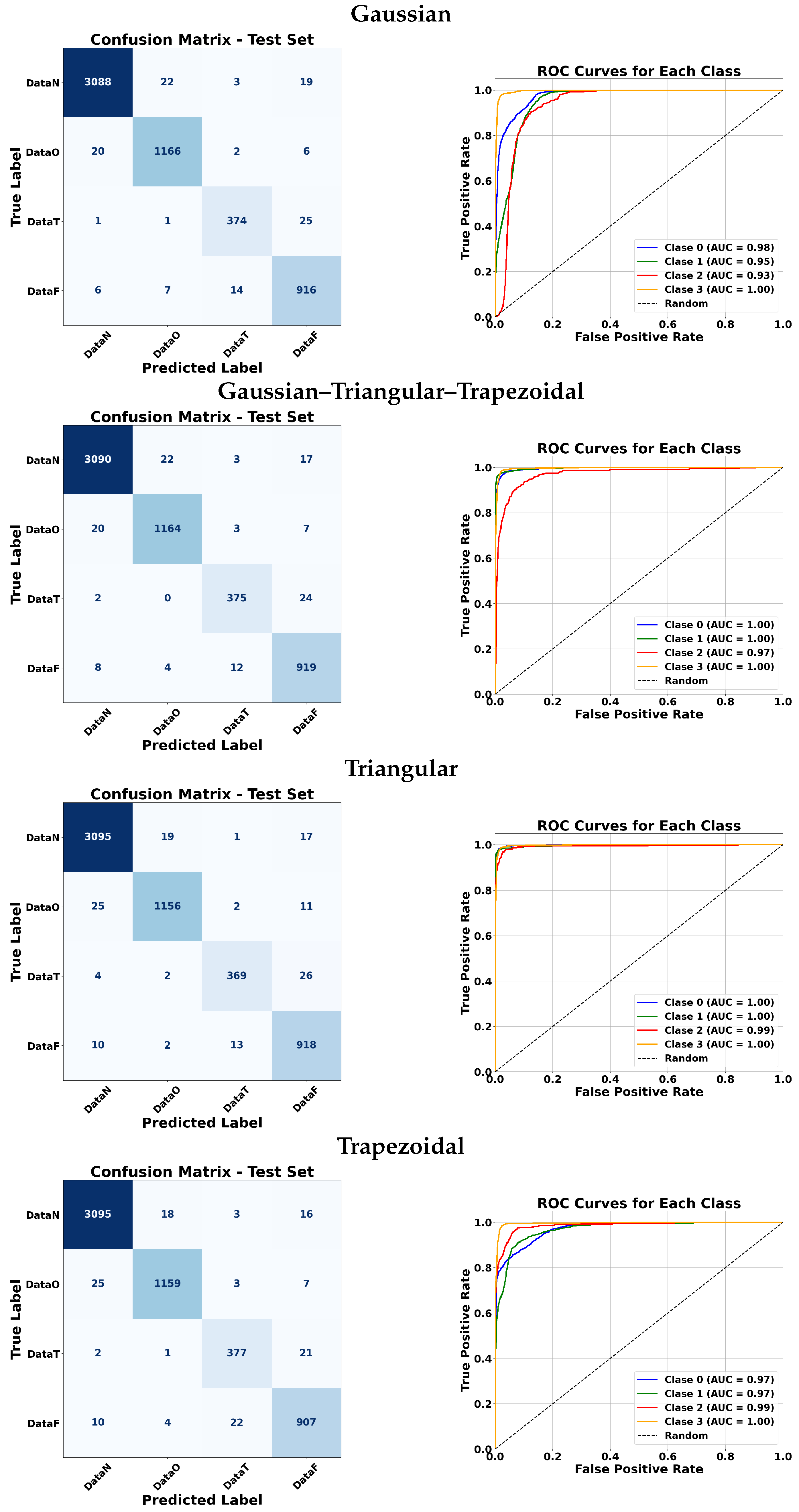
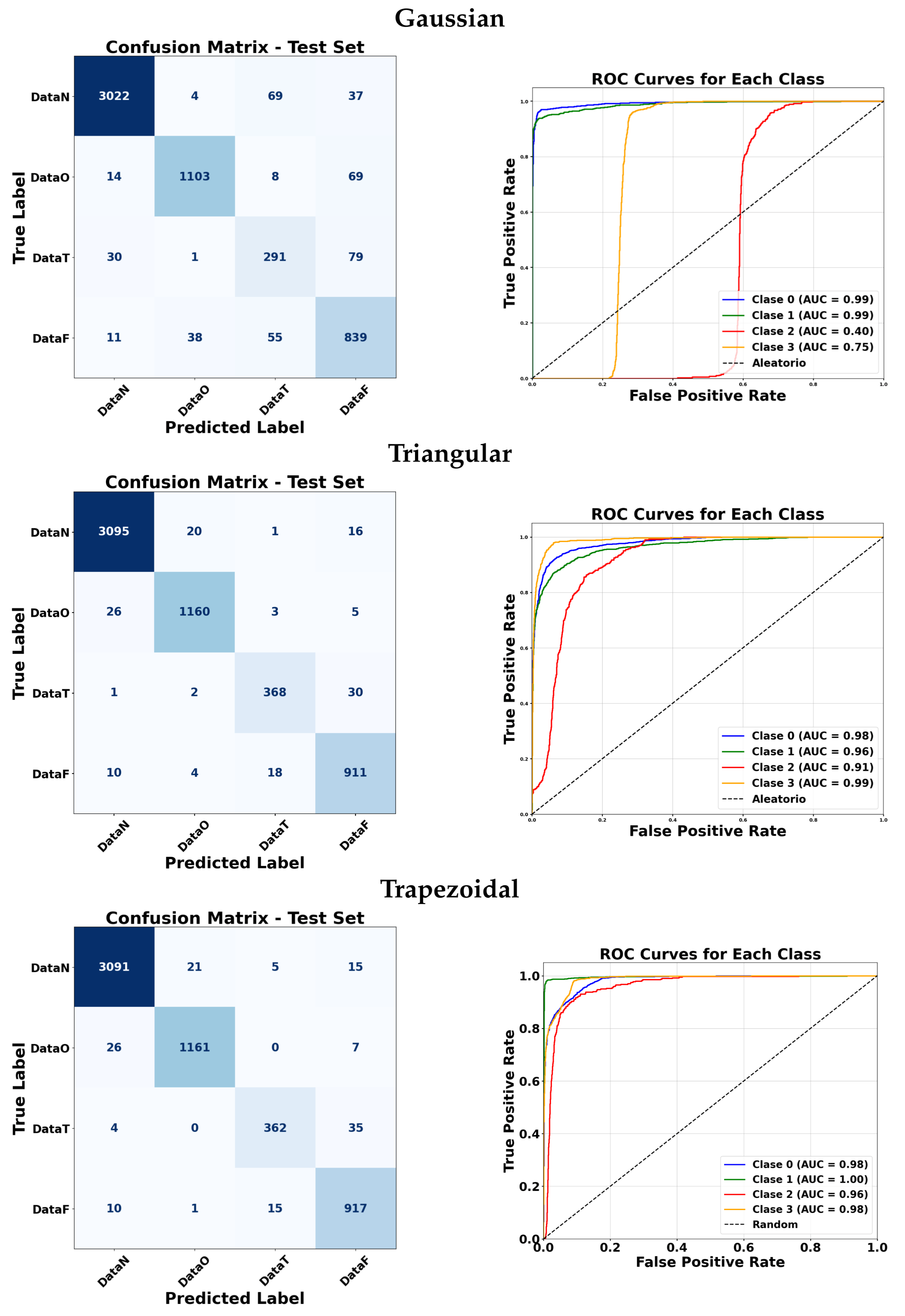
Figure 15 illustrate the classification performance of ANFIS under various membership function configurations and of the Fuzzy ARTMAP model under different feature selection strategies. These visualizations allow a more detailed comparison in terms of sensitivity and specificity across the different approaches. In the confusion matrices, the background color indicates the magnitude of the values: darker triangles correspond to higher detection values, while lighter or white cells indicate lower values. Similarly, for the triangular membership functions, more intense colors represent higher values.
5.5.1. ANFIS Analysis
ANFIS classifiers were evaluated using four membership function types: Gaussian, triangular, trapezoidal, and a combination of the three. For the manual approach, Gaussian membership functions achieved very high AUC values for most classes, with slightly lower performance for class 3. Triangular and trapezoidal functions generally produced comparable results, with some cases reaching perfect AUC for classes 0, 1, and 3. The combination of all three membership functions maintained excellent performance across almost all classes.
For the automatic approach, Gaussian membership functions yielded a high AUC for classes 0 and 1, but a lower AUC for class 2. Triangular and trapezoidal functions generally improved the performance for class 2 while maintaining a high AUC for other classes. Overall, most membership functions achieved strong classification results, with minor variations between classes.
5.5.2. Fuzzy ARTMAP Analysis
Fuzzy ARTMAP classifiers were tested with various feature selection methods: LDA, MI, RF, RFE, PCA, and ANOVA. Across all methods, classes 0 and 1 consistently achieved very high AUC values (close to 0.98–0.99). Class 2 tended to have slightly lower values around 0.95–0.96, except for ANOVA which dropped slightly for class 1. Class 3 maintained high performance for all methods. Overall, LDA, MI, RF, and RFE produced nearly identical results, while PCA and ANOVA showed only minor variations. This indicates that the Fuzzy ARTMAP classifier is robust to different feature selection strategies and performs reliably across most classes.
5.6. Comparative Results: FuzzyARTMAPClassifier, ANFIS Manual, and ANFIS Automatic
The results presented in
Table 5,
Table 6,
Table 7 and
Table 8 highlight the main performance indicators—precision, sensitivity, specificity, F1-score, and overall accuracy—for each class and configuration. These tables allow a direct comparison between models and feature selection methods, showing which combinations achieve superior performance.
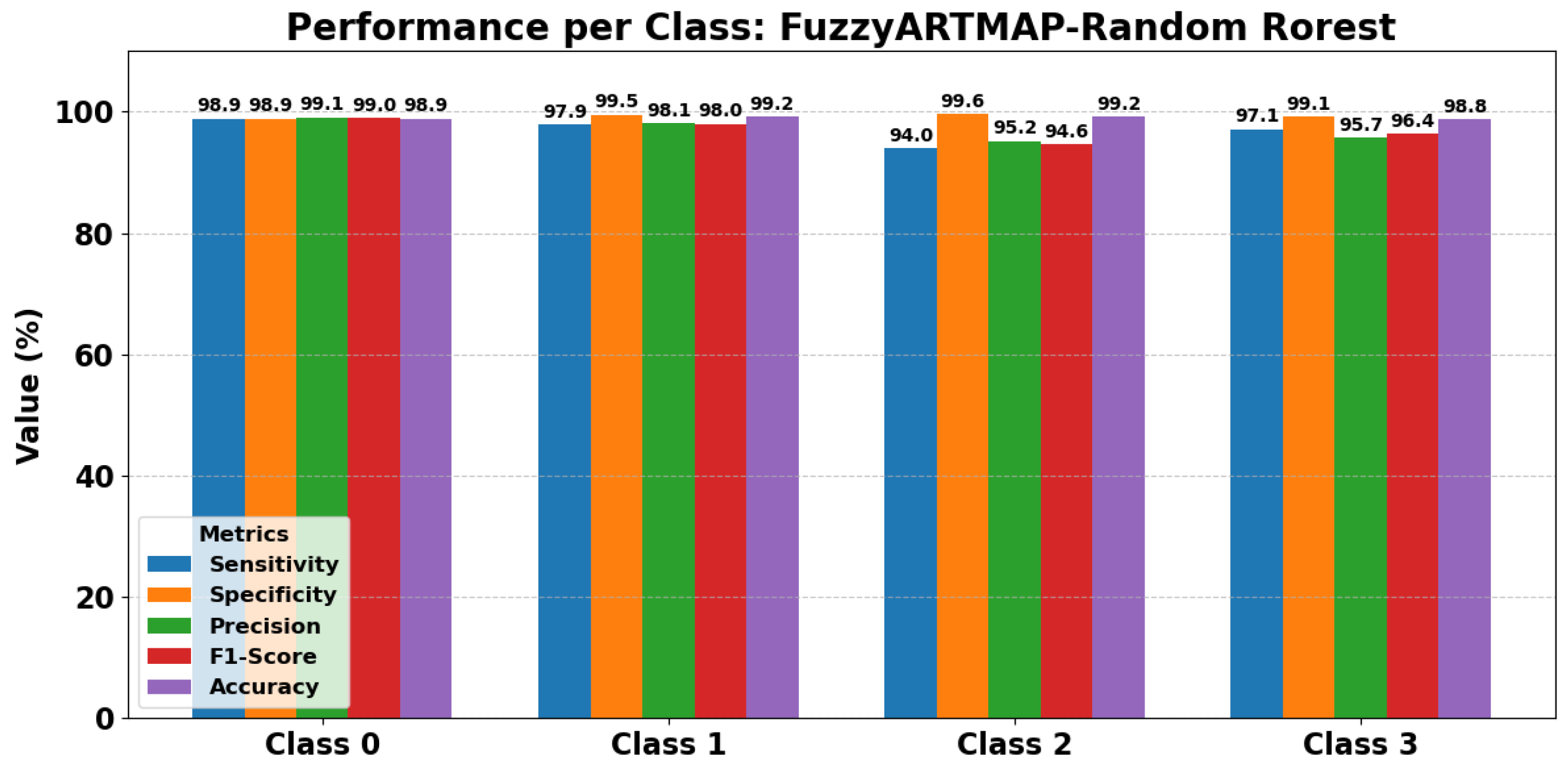
In particular, the FuzzyARTMAP model combined with Random Forest consistently achieves the highest performance across all four classes (Normal, Others, VT, and VF). Most metrics approach 99%, confirming that this combination outperforms other feature selection techniques such as LDA or PCA in terms of both accuracy and computational efficiency.
Figure 16 complements the tabular results by providing a visual overview of class-wise precision, sensitivity, specificity, F1-score, and accuracy. It clearly illustrates the robustness and consistently high-level performance of FuzzyARTMAP with Random Forest, reinforcing the numerical findings.
Overall, the combination of tabular data and graphical visualization provides a comprehensive understanding of model performance, demonstrating the superiority of FuzzyARTMAP with Random Forest over alternative configurations.
These results provide a strong baseline for comparing the performance of our FuzzyARTMAP configurations with other approaches reported in the literature. In the following section, we discuss how the achieved metrics—precision, sensitivity, specificity, F1-score, and overall accuracy—stand relative to other state-of-the-art methods, highlighting the improvements offered by the combination of FuzzyARTMAP with Random Forest.
As shown in
Table 9, the proposed FuzzyARTMAP classifiers, combined with Random Forest, were evaluated against several existing approaches to classify ventricular arrhythmias, including VT and VF. Importantly, our approach relies on a minimal set of input parameters extracted from the ECG signals, rather than using the entire signal as input, as is common in many other methods. This reduction in dimensionality results in significantly faster execution times while maintaining high classification performance.
The FuzzyARTMAP models achieve highly competitive results, with most performance metrics exceeding 98–99% across all classes. For example, sensitivity and specificity for VT and VF are comparable to or surpass those reported for deep learning models such as
Ht_TFR_CNN [
43],
InceptionV3, and
AlexNet. Other techniques in the literature, including ANNC, SSVR, and TDA [
40,
44], also report strong performance, but generally rely on more complex feature extraction or processing of the entire time–frequency representation of the signals.
These comparative results underscore the robustness and efficiency of our FuzzyARTMAP-based approach, demonstrating that highly accurate classification can be achieved with fewer input parameters and lower computational cost. The findings confirm the practical advantages of integrating FuzzyARTMAP with optimized feature selection methods for real-time detection of shockable arrhythmias.
Overall, the proposed FuzzyARTMAP + Random Forest approach consistently delivers superior performance across all classes, highlighting its robustness and suitability for clinical applications in arrhythmia detection.
Table 10 presents a comparative analysis of different techniques aimed at distinguishing shockable (i.e., VT and VF) from non-shockable rhythms. This classification task is crucial for real-time applications such as AEDs and ICDs, where rapid and reliable detection of treatable arrhythmias is essential. Several methods in the literature have achieved strong performance in this task. For instance, Mjahad et al. [
44] employed Topological Data Analysis (TDA), achieving a sensitivity of 99.03%, specificity of 99.67%, and an accuracy of 99.51%. In the same study, a Phase Distribution Index (PDI) approach yielded lower results with an accuracy of 95.12%. Acharya et al. [
45] proposed an eleven-layer CNN, reaching 95.32% sensitivity, 91.04% specificity, and 93.20% accuracy. Similarly, Buscema et al. [
46] obtained a high accuracy of 99.72% using a Recurrent Neural Network (RNN), while Kumar et al. [
47] reached 98.8% accuracy using a hybrid CNN and Improved Ensemble Neural Network (IENN). Alonso-Atienza et al. [
48] achieved 98.6% accuracy, 95.0% sensitivity, and 99.0% specificity using feature selection combined with SVM. Cheng and Dong [
49] used a personalized feature extraction method and SVM, obtaining 95.5% accuracy and 95.6% specificity. Li et al. [
50] combined genetic algorithm-based feature selection with SVM classification and achieved 98.1% accuracy, 98.4% sensitivity, and 98.0% specificity. Finally, Xu et al. [
51] reported 98.29% accuracy using adaptive variational techniques and boosted classification and regression trees (CART), along with high sensitivity (97.32%) and specificity (98.95%).
The results of the FuzzyARTMAP model proposed in this work demonstrate strong performance in distinguishing shockable rhythms, achieving a sensitivity of 99.56%, specificity of 98.81%, and an overall accuracy of 99.40% for VT and VF detection. These results highlight the effectiveness of the FuzzyARTMAP approach in identifying critical arrhythmias, providing a reliable and robust decision-support mechanism for real-time clinical applications. Compared with other methods in the literature, the proposed approach shows competitive or superior performance, particularly in sensitivity, which is crucial for avoiding missed detections of shockable rhythms.
Overall, these findings indicate that FuzzyARTMAP is well suited for deployment in real-time devices such as AEDs and ICDs, offering rapid and accurate discrimination between shockable and non-shockable arrhythmias. The method’s high sensitivity and specificity ensure reliable performance in critical care scenarios, supporting timely and appropriate interventions.