Automated Segmentation and Quantification of Histology Fragments for Enhanced Macroscopic Reporting
Abstract
1. Introduction
1.1. Gross Examination Technologies
1.2. Instance Segmentation Overview
1.3. Contributions
- The design and implementation of a compact, 3D-printed imaging platform specifically engineered for tissue fragment documentation.
- The creation of a comprehensive dataset of biopsy and resection fragments, meticulously annotated using the Segment Anything Model (SAM).
- The integration of a YOLO-based instance segmentation approach for the detection and segmentation of tissue fragments.
- The automatic generation of visual reports summarizing the number and size of detected fragments to support quality control in pathology workflows.
2. Materials and Methods
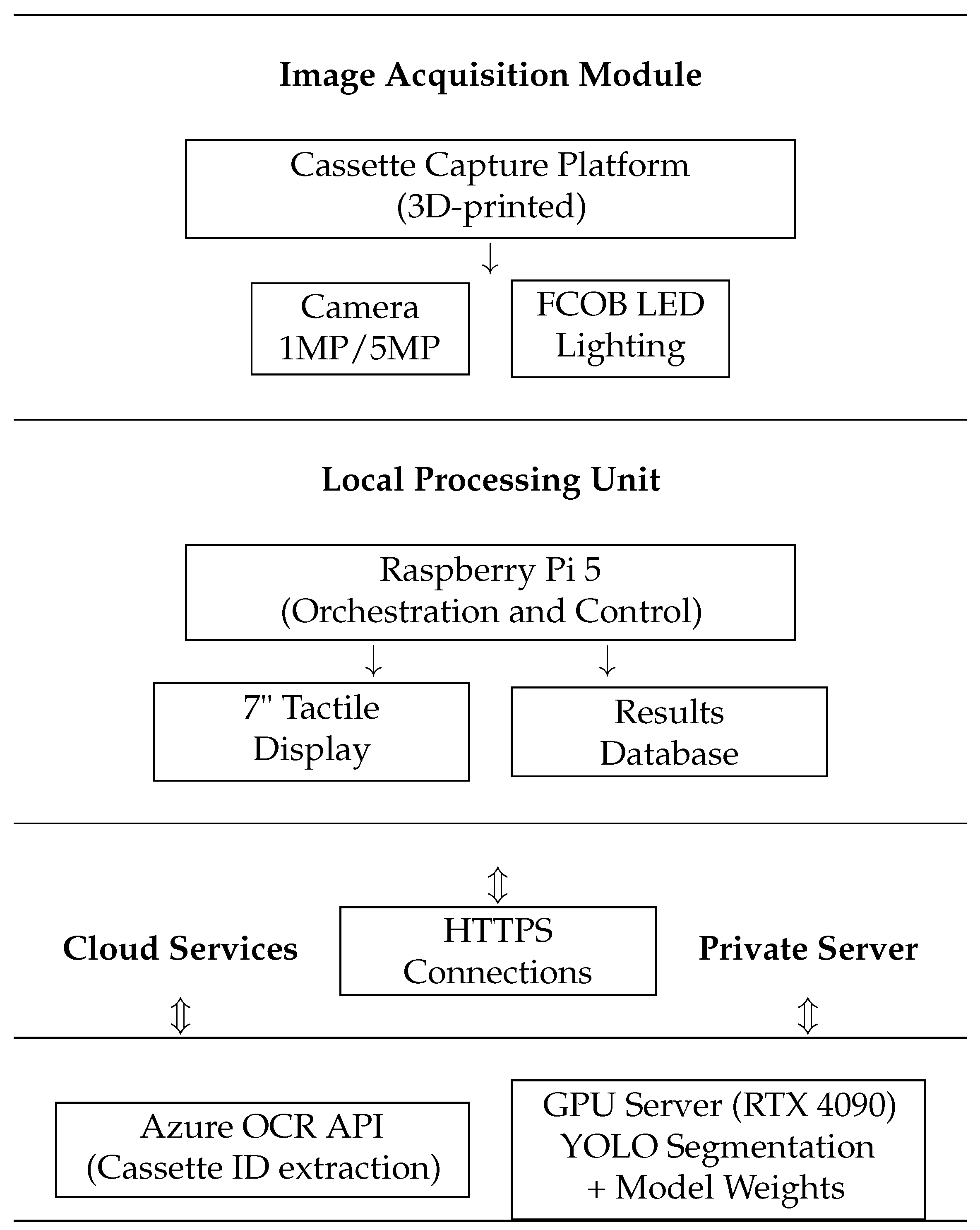
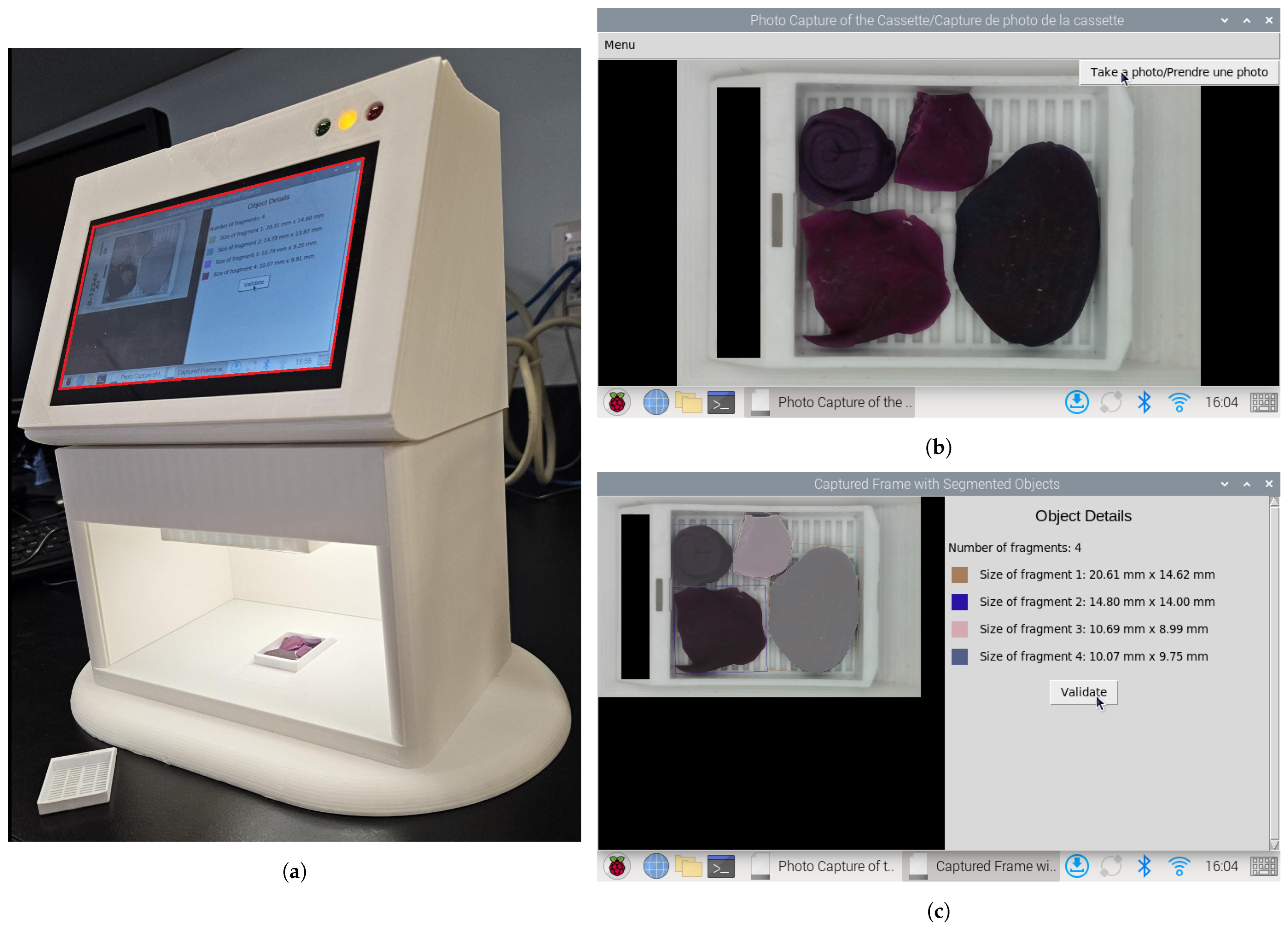
2.1. Cassette Image Capture Platform
2.2. Patient Identifier Processing and Data Security
2.3. Data Annotation
2.4. Fragment Segmentation
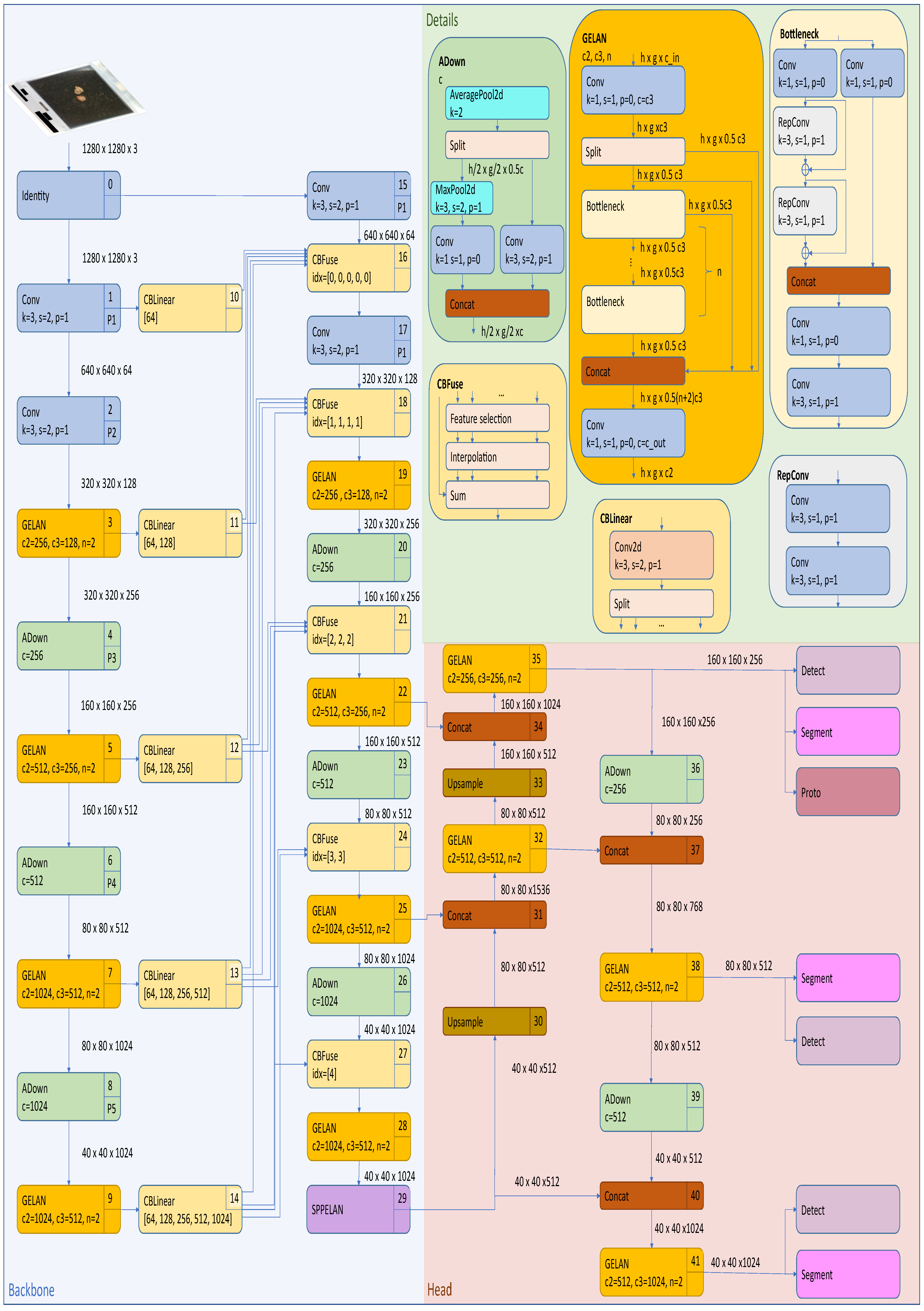
2.4.1. Yolov8 Instance Segmentation Architecture
- Cross-Stage Partial Darknet (CSPDarknet) backboneThe backbone employs a modified CSPDarknet architecture that integrates CSP modules to reduce computational redundancy while enhancing gradient flow. The CSP design in YOLOv8 splits the feature map of each stage into two parts, with one part undergoing a dense block of convolutions and the other being directly concatenated with the dense block’s output, reducing computational complexity while preserving accuracy. The backbone consists of multiple CSP blocks, each comprising a split operation, a dense block, a transition layer, and a concatenation operation. YOLOv8 also utilizes Sigmoid Linear Unit (SiLU) for non-linear feature mapping, which enhances gradient flow and feature expressiveness. These design elements reduce computational complexity, improve gradient flow, enhance feature reuse, and maintain high accuracy while reducing model size. The backbone generates feature maps at three scales (P3, P4, and P5) with strides of 8, 16, and 32 pixels, respectively, thus capturing hierarchical spatial information from fine-grained details to high-level semantics.
- Path Aggregation Network (PANet) neckThe PANet neck in YOLOv8 enhances information flow and feature fusion across different network layers by building on the Feature Pyramid Network (FPN) design. It includes a bottom-up path for feature extraction, a top-down path for semantic feature propagation, and an additional bottom-up path for further feature hierarchy enhancement. At each level, features from corresponding bottom-up and top-down paths are fused through element-wise addition or concatenation, while adaptive feature pooling is introduced to enhance multi-scale feature fusion by pooling features from all levels for each region of interest. This design improves the network’s ability to detect objects at various scales, boosts performance relating to small object detection, and enhances information flow between different feature levels, making it crucial for edge applications where objects may appear at different scales and distances.
- Detection and segmentation headsThe detection–segmentation head operates through two parallel branches. The detection branch uses an anchor-free mechanism to predict center points via a 1 × 1 convolution with 4 + C4 + C outputs (box coordinates, objectness, and class probabilities). Its loss function combines Distribution Focal Loss (DFL) for classification and CIoU for regression. The segmentation branch includes a mask prototype network that generates 32 prototype masks per image through a 3 × 3 convolution layer. Mask coefficients are predicted alongside detection outputs and are combined through matrix multiplication. The segmentation branch’s loss function uses binary cross-entropy.
2.4.2. Yolov9 Instance Segmentation Architecture
- Generalized Efficient Layer Aggregation Network (GELAN) backboneThe backbone in YOLOv9-seg is designed to extract multi-scale features from the input image. It leverages the GELAN, which combines the strengths of CSPNet and the Efficient Layer Aggregation Network (ELAN). GELAN incorporates various computational blocks such as CSPblocks, Resblocks, and Darkblocks, ensuring efficient feature extraction while preserving key hierarchical features across the network’s layers.
- Programmable Gradient Information (PGI) neckThe neck component in YOLOv9-seg enhances the feature fusion process using PGI; this introduces an auxiliary reversible branch that ensures reliable gradient flow across the network, addressing the problem of information loss during training. This reversible architecture ensures that no crucial data are lost during the forward and backward passes, leading to more reliable predictions.
- Detection and segmentation headsThe head in YOLOv9-seg utilizes an anchor-free bounding box prediction method, similar to that of YOLOv8, but also benefits from the reversible functions provided by PGI. The head is divided into two parts—the main branch and the multi-level auxiliary branch. The auxiliary branch focuses on capturing and retaining gradient information during training, supporting the main branch by preserving essential gradient information.
2.5. Fragment Dimension Measurement
2.6. Evaluation Metrics
2.6.1. Intersection over Union (IoU)
- A: Prediction mask;
- B: Ground truth mask;
- : Intersection area between the prediction and the ground truth;
- : Union area between the prediction and the ground truth.
2.6.2. Dice Coefficient
- : Number of pixels in the prediction mask;
- : Number of pixels in the ground truth mask.
2.6.3. Mean Average Precision (mAP)
- is the Average Precision for masks, calculated at an IoU threshold t;
- t ranges from 0.50 to 0.95 in steps of 0.05, representing a total of 10 thresholds.
- is the precision as a function of Recall r;
- The integral is calculated over the Recall interval from 0 to 1.
2.6.4. Spatial Accuracy
- (True Positive): Number of predicted masks that match ground truth masks with IoU ;
- (False Positive): Number of predicted masks that do not match any ground truth mask (also evaluated independently as a standalone metric);
- (False Negative): Number of ground truth masks that are not matched by any prediction;
- : IoU threshold (set to 0.5 in our experiments).
2.7. Dataset
2.8. Training and Validation Setup
- freeze3: Initial layers preceding the three-scale feature map generation are frozen (modules: input convolution, initial convolution, C2f block).
- freeze5: Layers generating the first-scale feature map and earlier blocks are frozen.
- freeze10: All backbone layers are frozen, including the SPPF module (covers feature extraction and spatial pyramid pooling).
- freeze16: Backbone and top-down path neck layers are frozen.
- freeze22: All layers except prediction heads are frozen, enabling the fine-tuning of final detection outputs only.
2.9. Computational Environment and Reproducibility
3. Results
3.1. Impact of Model Size
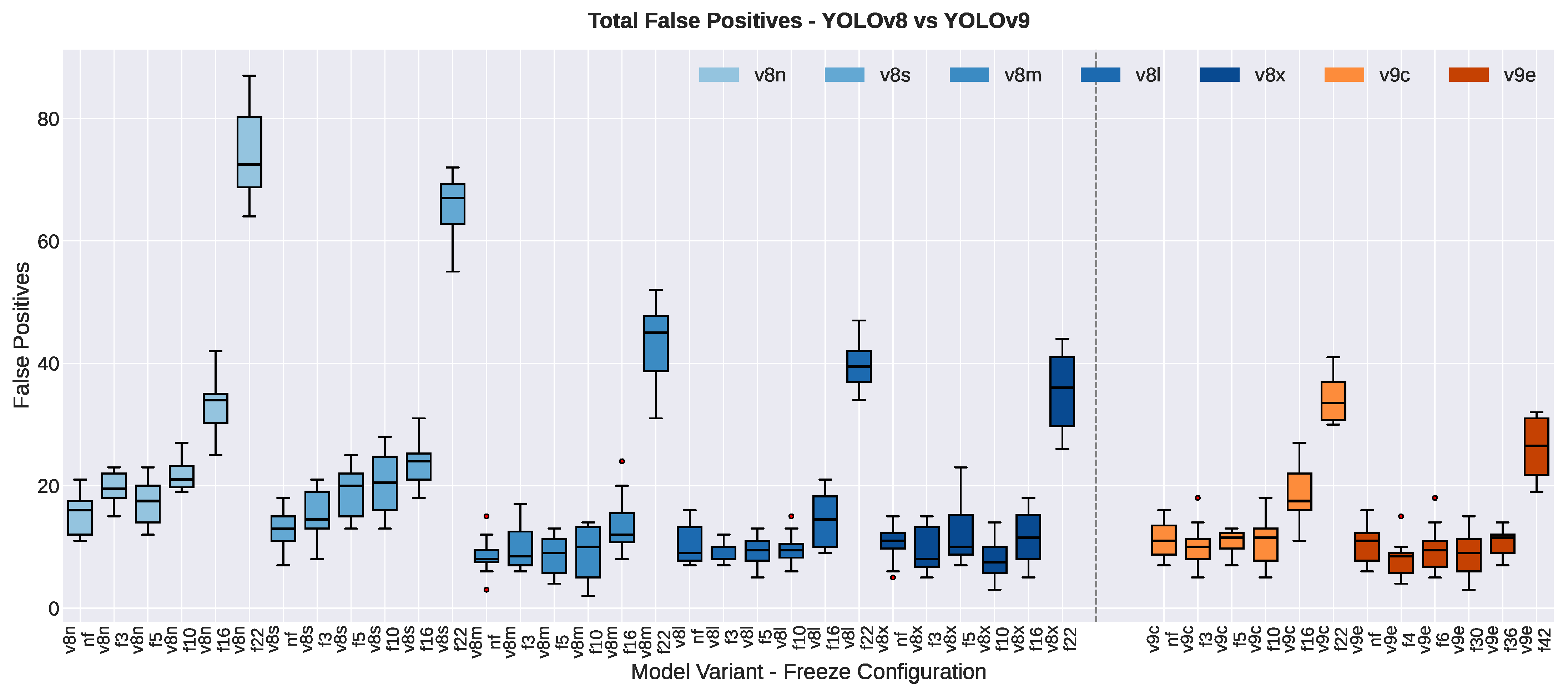
3.1.1. False Positive Count
3.1.2. Spatial Accuracy Improvements
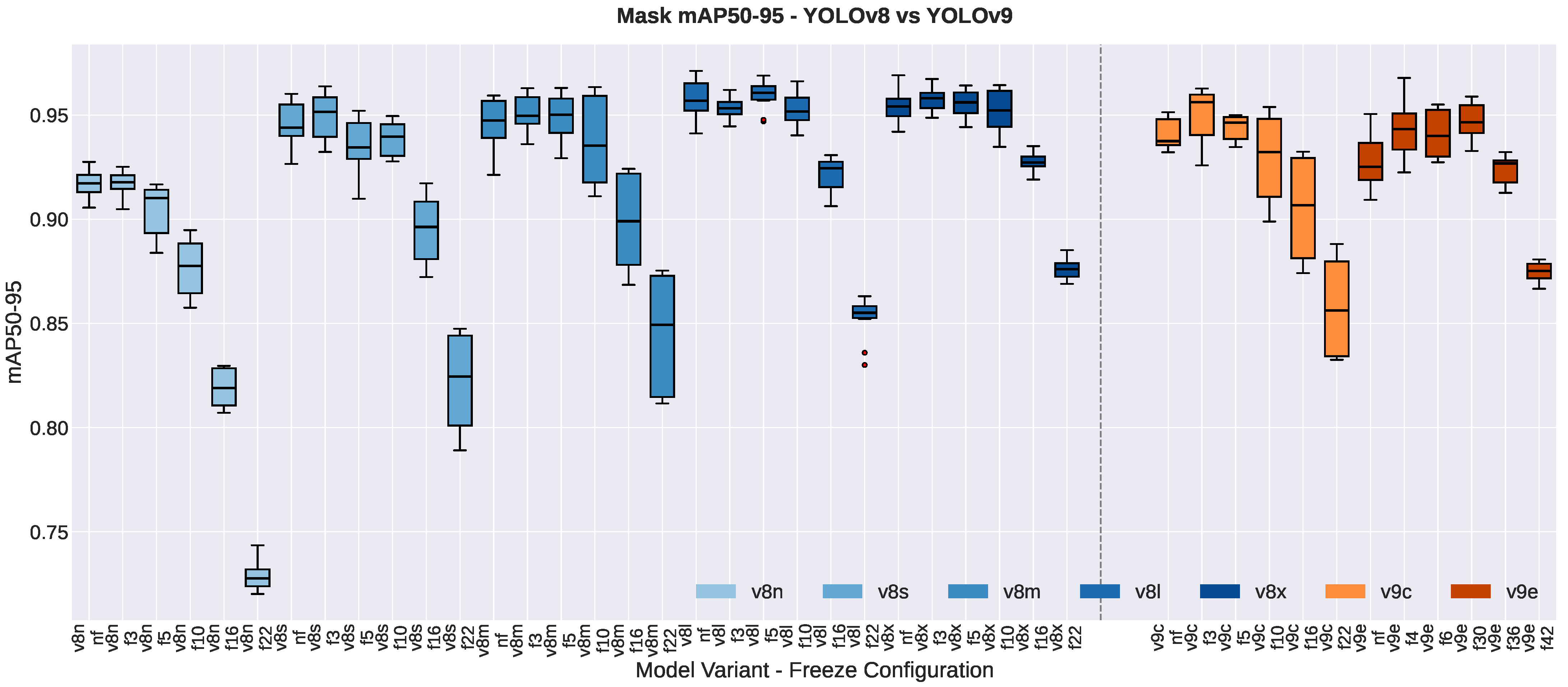
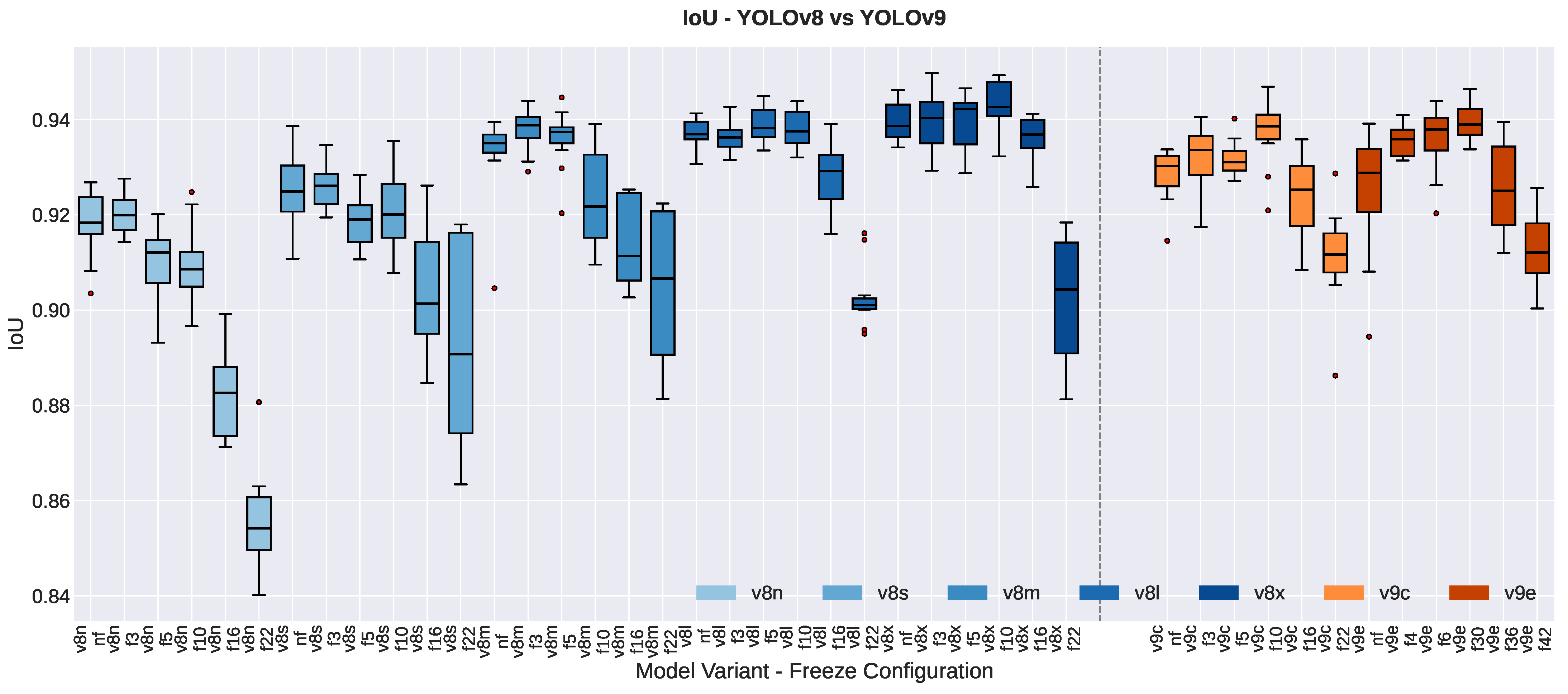
3.1.3. Segmentation Quality Metrics
3.1.4. Resolution Effects
3.2. Impact of Freezing Strategies
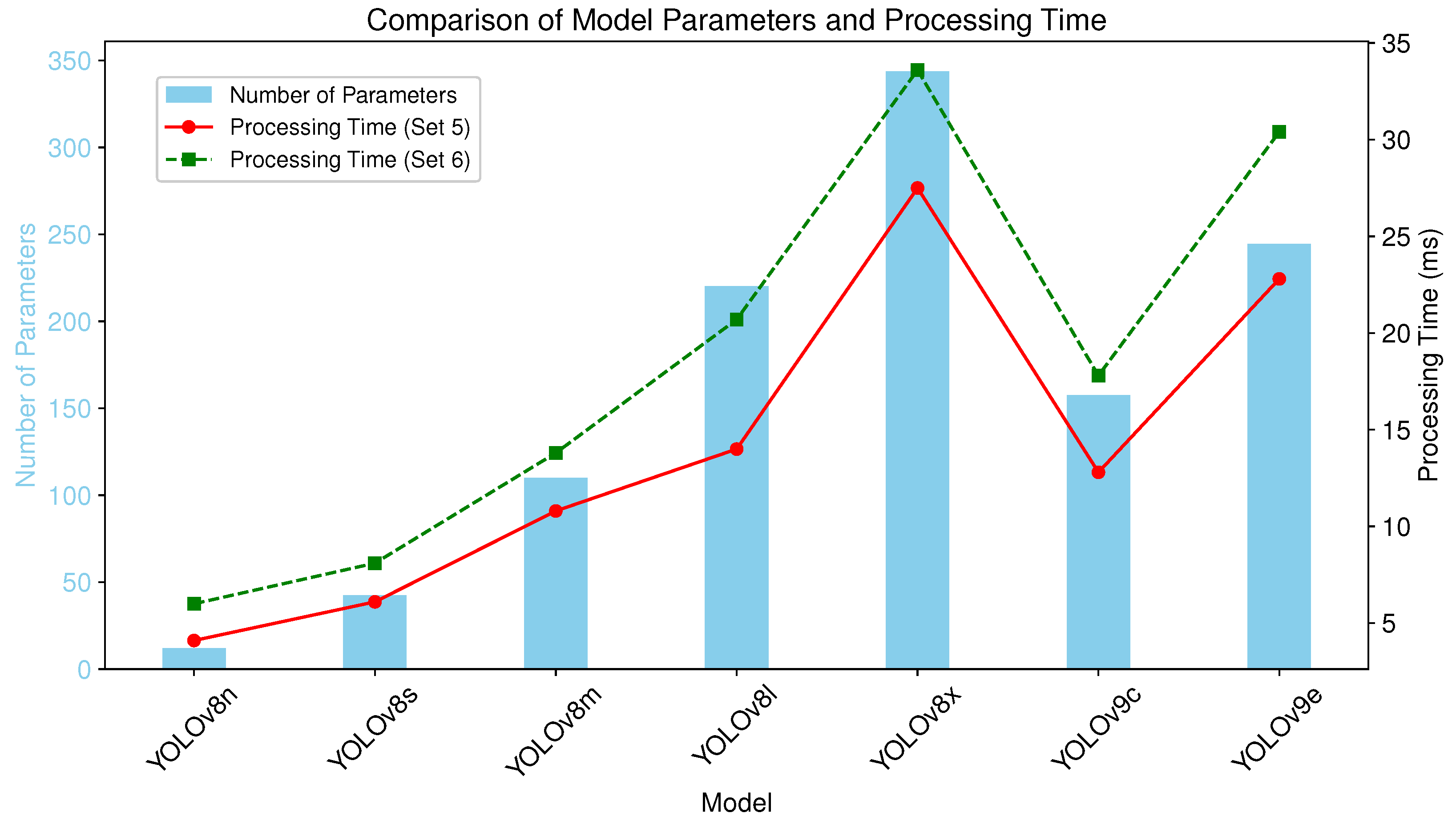
3.3. Computational Configuration and Efficiency
3.4. Performance–Efficiency Trade-Offs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AP | Average Precision |
| C2f | Cross Stage Partial with Two Fusion |
| CIoU | Complete Intersection over Union |
| CSPDarknet | Cross-Stage Partial Darknet |
| DFL | Distribution Focal Loss |
| E-ELAN | Extended-Efficient Layer Aggregation Network |
| ELAN | Efficient Layer Aggregation Network |
| FCOB LED | Flexible Circuit Board Light-Emitting Diode |
| FPN | Feature Pyramid Network |
| GELAN | Generalized Efficient Layer Aggregation Network |
| HD-YOLO | Histology-based Detection using YOLO |
| hsv | Hue Saturation Value |
| IoU | Intersection over Union |
| lr | Learning Rate |
| mAP | mean Average Precision |
| MRI | Magnetic Resonance Imaging |
| OCR | Optical Character Recognition |
| PANet | Path Aggregation Network |
| PGI | Programmable Gradient Information |
| PLA | Polylactic Acid |
| R-ELAN | Residual Efficient Layer Aggregation Network |
| SAM | Segment Anything Model |
| SiLU | Sigmoid Linear Unit |
| SIoU | SCYLLA Intersection over Union |
| SPPF | Spatial Pyramid Pooling Fast |
| YOLO | You Only Look Once |
| Yolo-seg | YOLO-based segmentation model |
| YOLOv8l | YOLO version 8 Large |
| YOLOv8m | YOLO version 8 Medium |
| YOLOv8n | YOLO version 8 Nano |
| YOLOv8s | YOLO version 8 Small |
| YOLOv8x | YOLO version 8 Extra Large |
| YOLOv9c | YOLO version 9 Compact |
| YOLOv9e | YOLO version 9 Extended |
References
- Varma, M.; Collins, L.C.; Chetty, R.; Karamchandani, D.M.; Talia, K.; Dormer, J.; Vyas, M.; Conn, B.; Guzmán-Arocho, Y.D.; Jones, A.V.; et al. Macroscopic examination of pathology specimens: A critical reappraisal. J. Clin. Pathol. 2024, 77, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Cleary, A.S.; Lester, S.C. The Critical Role of Breast Specimen Gross Evaluation for Optimal Personalized Cancer Care. Surg. Pathol. Clin. 2022, 15, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Bell, W.C.; Young, E.S.; Billings, P.E.; Grizzle, W.E. The efficient operation of the surgical pathology gross room. Biotech. Histochem. 2008, 83, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Diepeveen, A. A Dilemma in Pathology: Overwork, Depression, and Burnout. Available online: https://lumeadigital.com/pathologists-are-overworked/ (accessed on 28 March 2025).
- Vistapath. Vistapath Is Building the Next Generation of Pathology Labs: Optimize Your Grossing Workflows with Sentinel. Available online: https://www.vistapath.ai/sentinel/ (accessed on 28 March 2025).
- Druffel, E.; Bridgeman, A.; Brandegee, K.; Bodell, A.; McClintock, D.; Garcia, J. Current State of Intra-/Interobserver Accuracy and Reproducibility in Tissue Biopsy Grossing and Comparison to an Automated Vision System. In Proceedings of the United States and Canada Association of Pathologists (USCAP) Annual Meeting, Baltimore, MD, USA, 23–28 March 2024. Poster #198. [Google Scholar]
- Tissue-Tek AutoTEC® a120: An Automated Tissue Embedding System. Available online: https://www.sakuraus.com/Products/Embedding/AutoTEC-a120.html (accessed on 28 March 2025).
- Greenlee, J.; Webster, S.; Gray, H.; von Bueren, E. Reducing Common Embedding Errors Through Automation: Manual Embedding Versus Automated Embedding Using the Tissue-Tek AutoTEC® a120 Automated Embedding System and the Tissue-Tek® Paraform® Sectionable Cassette System. In Proceedings of the National Society for Histotechnology (NSH) Annual Symposium, New Orleans, LA, USA, 20–25 September 2019; Sakura Finetek USA, Inc.: Torrance, CA, USA, 2019. Available online: https://www.sakuraus.com/getattachment/Products/Embedding/AutoTEC-a120/MPUB0012-Poster-NSH-2019-with-title-Reducing-common-embedding-errors-t.pdf?lang=en-US (accessed on 28 March 2025).
- deBram Hart, M.; von Bueren, E.; Wander, B.; Fussner, M.; Scancich, C.; Reed, C.; Cockerell, C.J. Continuous Specimen Flow Changes Night Shifts to Day Shifts While Reducing Turn-Around-Time (TAT). In Proceedings of the National Society for Histotechnology (NSH) Annual Symposium, Washington, DC, USA, 28 August–2 September 2015; Cockerell Dermatopathology. Sakura Finetek USA, Inc.: Torrance, CA, USA, 2015. Available online: https://www.sakuraus.com/SakuraWebsite/media/Sakura-Document/Poster_1_-_AutoTEC_a120_-_NSH_2015.pdf (accessed on 28 March 2025).
- Milestone Medical. eGROSS pro-x: Ergonomic Mobile Grossing Station. Product Brochure. 2019. Available online: https://www.milestonemedsrl.com/products/grossing-and-macro-digital/egross/ (accessed on 28 March 2025).
- Roß, T.; Reinke, A.; Full, P.M.; Wagner, M.; Kenngott, H.; Apitz, M.; Hempe, H.; Mindroc-Filimon, D.; Scholz, P.; Tran, T.N.; et al. Comparative validation of multi-instance instrument segmentation in endoscopy: Results of the ROBUST-MIS 2019 challenge. Med. Image Anal. 2021, 70, 101920. [Google Scholar] [CrossRef] [PubMed]
- Maier-Hein, L.; Wagner, M.; Ross, T.; Reinke, A.; Bodenstedt, S.; Full, P.M.; Hempe, H.; Mindroc-Filimon, D.; Scholz, P.; Tran, T.N.; et al. Heidelberg colorectal data set for surgical data science in the sensor operating room. Sci. Data 2021, 8, 101. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Fu, B.; Ye, J.; Wang, G.; Li, T.; Wang, H.; Li, R.; Yao, H.; Cheng, J.; Li, J.; et al. Interactive medical image segmentation: A benchmark dataset and baseline. In Proceedings of the Computer Vision and Pattern Recognition Conference, Nashville, TN, USA, 10–17 June 2025; pp. 20841–20851. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLO9000: Better, Faster, Stronger. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 6517–6525. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar] [CrossRef]
- Bochkovskiy, A.; Wang, C.; Liao, H.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar] [CrossRef]
- Jocher, G. Ultralytics YOLOv5; Version 7.0; Ultralytics: Frederick, MD, USA, 2020; Available online: https://github.com/ultralytics/yolov5 (accessed on 28 March 2025).
- Li, C.; Li, L.; Jiang, H.; Weng, K.; Geng, Y.; Li, L.; Ke, Z.; Li, Q.; Cheng, M.; Nie, W.; et al. YOLOv6: A Single-Stage Object Detection Framework for Industrial Applications. arXiv 2022, arXiv:2209.02976. [Google Scholar] [CrossRef]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. YOLOv7: Trainable Bag-of-Freebies Sets New State-of-the-Art for Real-Time Object Detectors. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 7464–7475. [Google Scholar] [CrossRef]
- Jocher, G.; Chaurasia, A.; Qiu, J. Ultralytics YOLOv8; Version 8.0.0; Ultralytics: Frederick, MD, USA, 2023; Available online: https://docs.ultralytics.com/models/yolov8/ (accessed on 28 March 2025).
- Wang, C.Y.; Yeh, I.H.; Mark Liao, H.Y. YOLOv9: Learning What You Want to Learn Using Programmable Gradient Information. In Proceedings of the Computer Vision—ECCV 2024, Milan, Italy, 29 September–4 October 2024; Leonardis, A., Ricci, E., Roth, S., Russakovsky, O., Sattler, T., Varol, G., Eds.; Springer Nature: Cham, Switzerland, 2025; pp. 1–21. [Google Scholar]
- Wang, A.; Chen, H.; Liu, L.; CHEN, K.; Lin, Z.; Han, J.; Ding, G. YOLOv10: Real-Time End-to-End Object Detection. In Proceedings of the International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 10–15 December 2024. [Google Scholar]
- Jocher, G.; Qiu, J. Ultralytics YOLO11; Version 11.0.0; Ultralytics: Frederick, MD, USA, 2024; Available online: https://github.com/ultralytics/ultralytics (accessed on 28 March 2025).
- Tian, Y.; Ye, Q.; Doermann, D. YOLOv12: Attention-Centric Real-Time Object Detectors. arXiv 2025, arXiv:2502.12524. [Google Scholar]
- Ragab, M.G.; Abdulkadir, S.J.; Muneer, A.; Alqushaibi, A.; Sumiea, E.H.; Qureshi, R.; Al-Selwi, S.M.; Alhussian, H. A Comprehensive Systematic Review of YOLO for Medical Object Detection (2018 to 2023). IEEE Access 2024, 12, 57815–57836. [Google Scholar] [CrossRef]
- Kumar, K.S.; Juliet, A.V. Microfluidic droplet detection for bio medical application using YOLO with COA based segmentation. Evol. Syst. 2024, 16, 21. [Google Scholar] [CrossRef]
- AlSadhan, N.A.; Alamri, S.A.; Ben Ismail, M.M.; Bchir, O. Skin Cancer Recognition Using Unified Deep Convolutional Neural Networks. Cancers 2024, 16, 1246. [Google Scholar] [CrossRef] [PubMed]
- Rong, R.; Sheng, H.; Jin, K.W.; Wu, F.; Luo, D.; Wen, Z.; Tang, C.; Yang, D.M.; Jia, L.; Amgad, M.; et al. A Deep Learning Approach for Histology-Based Nucleus Segmentation and Tumor Microenvironment Characterization. Mod. Pathol. 2023, 36. [Google Scholar] [CrossRef] [PubMed]
- Zade, A.A.T.; Aziz, M.J.; Majedi, H.; Mirbagheri, A.; Ahmadian, A. Spatiotemporal analysis of speckle dynamics to track invisible needle in ultrasound sequences using convolutional neural networks: A phantom study. Int. J. Comput. Assist. Radiol. Surg. 2023, 18, 1373–1382. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Huangfu, T.; Wu, L.; Chen, W. Comparison of RetinaNet, SSD, and YOLO v3 for real-time pill identification. Bmc Med. Inform. Decis. Mak. 2021, 21, 324. [Google Scholar] [CrossRef] [PubMed]
- Iriawan, N.; Pravitasari, A.A.; Nuraini, U.S.; Nirmalasari, N.I.; Azmi, T.; Nasrudin, M.; Fandisyah, A.F.; Fithriasari, K.; Purnami, S.W.; Irhamah; et al. YOLO-UNet Architecture for Detecting and Segmenting the Localized MRI Brain Tumor Image. Appl. Comput. Intell. Soft Comput. 2024, 2024, 3819801. [Google Scholar] [CrossRef]
- Hossain, A.; Islam, M.T.; Almutairi, A.F. A deep learning model to classify and detect brain abnormalities in portable microwave based imaging system. Sci. Rep. 2022, 12, 6319. [Google Scholar] [CrossRef] [PubMed]
- Almufareh, M.F.; Imran, M.; Khan, A.; Humayun, M.; Asim, M. Automated Brain Tumor Segmentation and Classification in MRI Using YOLO-Based Deep Learning. IEEE Access 2024, 12, 16189–16207. [Google Scholar] [CrossRef]
- George, J.; Hemanth, T.S.; Raju, J.; Mattapallil, J.G.; Naveen, N. Dental Radiography Analysis and Diagnosis using YOLOv8. In Proceedings of the International Conference on Smart Computing and Communications (ICSCC), Kochi, India, 17–19 August 2023; pp. 102–107. [Google Scholar] [CrossRef]
- Microsoft Corporation. OCR—Optical Character Recognition—Azure AI Services. Available online: https://learn.microsoft.com/fr-fr/azure/ai-services/computer-vision/overview-ocr (accessed on 28 April 2025).
- Baraneedharan, P.; Kalaivani, S.; Vaishnavi, S.; Somasundaram, K. Revolutionizing healthcare: A review on cutting-edge innovations in Raspberry Pi-powered health monitoring sensors. Comput. Biol. Med. 2025, 190, 110109. [Google Scholar] [CrossRef] [PubMed]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y.; et al. Segment Anything. In Proceedings of the International Conference on Computer Vision, Paris, France, 2–6 October 2023; pp. 4015–4026. [Google Scholar]
- Ultralytics. Brief Summary of YOLOv8 Model Structure. 2023. Available online: https://github.com/ultralytics/ultralytics/issues/189 (accessed on 24 July 2025).
- Lin, T.Y.; Maire, M.; Belongie, S.; Bourdev, L.; Girshick, R.; Hays, J.; Perona, P.; Ramanan, D.; Zitnick, C.L.; Dollár, P. Microsoft COCO: Common Objects in Context. arXiv 2015, arXiv:1405.0312. [Google Scholar] [CrossRef]







| System | Functionality | Advantages | Limitations |
|---|---|---|---|
| VistaPath Sentinel [5] | AI-powered augmentation system for grossing | Reduces grossing time by 93%; improves labeling accuracy by 43% | High cost; requires extra space for device and display in grossing station |
| FormaPath nToto [6] | Automated transfer and documentation of small biopsies using computer vision and robotics | Hands-free documentation; suitable for biopsies < 1 cm2; tested in clinical settings | Limited to small biopsies; not suitable for large or complex specimens |
| eGROSS [10] | Digital macro documentation with integrated high-resolution imaging, audio, and traceability tools | Enhances traceability with high-resolution images and voice notes; useful for education and audits | Limited portability; not easily adaptable to compact or space-constrained grossing areas |
| Application Area | YOLO Version | Imaging Modality | Key Metrics/Highlights |
|---|---|---|---|
| Microfluidic droplet detection for biomedical research [27] | YOLOv7 | Microscopic images of microfluidic droplets | Achieves 96% accuracy and 93% F1-score. Uses Cheetah Optimization Algorithm for optimal selection of anchor boxes. |
| Skin cancer recognition [28] | YOLOv3, YOLOv4, YOLOv5, YOLOv7 | Dermoscopy images | Experimental results show that the YOLOv7 model achieves the best performance—IoU: 86.3%, mAP: 75.4%, and F1: 77.9%. |
| Nucleus detection, segmentation, and classification [29] | HD-YOLO | WSI | Achieved an F1-score of 0.7409 and a mean Intersection over Union (mIoU) of 0.8423 in lung cancer detection. |
| Needle tracking in ultrasound interventions (biopsies, epidural injections) [30] | Modified YOLOv3 (nYolo) | Ultrasound | Uses spatiotemporal features of speckle dynamics to track needles, especially when they are invisible. Achieved angle and tip localization errors of and mm, respectively, in challenging scenarios. |
| Real-time pill identification [31] | YOLOv3 | RGB pill images | Addresses the issue of pharmacists struggling to distinguish pills due to the lack of imprint codes. Compares YOLOv3 to RetinaNet and SSD to reduce medication errors and the waste of medical resources. YOLOv3 achieved the best mAP of 93.4%. |
| Brain tumor detection and segmentation [32] | YOLO + FCN-UNet | MRI | Achieved a correct classification rate of about 97% using YOLOv3-UNet for segmenting original and Gaussian noisy images. YOLOv4-UNet performed less satisfactorily. |
| Brain abnormality detection and classification [33] | YOLOv5 | Reconstructed microwave brain images | The YOLOv5l model performed better than YOLOv5s and YOLOv5m, acheiving an 85.65% Area Under the Curve (AUC) for benign tumor classification and a 91.36% AUC for malignant tumor classification. |
| Brain tumor segmentation and classification [34] | YOLOv5, YOLOv7 | MRI | The models were trained and validated on the Figshare brain tumor dataset. YOLOv5 achieved a box mAP of 0.947 and a mask mAP of 0.947, while YOLOv7 achieved a box mAP of 0.940 and a mask mAP of 0.941. |
| Dental radiography analysis [35] | YOLOv8 | Panoramic dental radiography | Precision of 82.36% in detecting and classifying dental diseases (cavities, periodontal disease, oral cancers). |
| Component | Justification | Alternatives Considered | Reason for Exclusion |
|---|---|---|---|
| Raspberry Pi 5 | Compact; low power consumption; Linux support; sufficient for image capture and display control | NVIDIA Jetson Nano, Arduino | Jetson Nano not tested due to higher cost; Arduino lacks support for complex vision tasks |
| HBV-W202012HD Camera (1 MP) | Minimal optical distortion; stable USB interface; cost-effective; no calibration needed | 5 MP Raspberry Pi Camera | Minimal accuracy improvement; increased processing time and storage requirements |
| Custom Light Box with FCOB LED Strip | Uniform illumination achieved through custom-built diffusion box integrated into 3D-printed platform | Ring light | Ring light difficult to integrate into compact design |
| Touchscreen Interface | Direct interaction for technicians; immediate result validation; compact integration | External monitor; mobile app | Monitor requires peripherals and larger footprint; mobile app needs network setup and device management |
| 3D-printed PLA Platform | Cost-effective; easily customizable; rapid prototyping | Nylon (3D printing) | Nylon is difficult to print and is prone to warping |
| Azure OCR API | High accuracy on cassette text; handles various orientations | Tesseract OCR; EasyOCR; PaddleOCR | Lower accuracy on cassette IDs; Tesseract required extensive preprocessing; PaddleOCR and EasyOCR showed inconsistent character recognition |
| Set Number | Camera Resolution (MP) | Number of Images | Number of Instances | Image Size (Pixels) | Purpose | Notes |
|---|---|---|---|---|---|---|
| Set 1 | 1 MP | 32 | 65 | 1280 × 800 | Training | Some cassettes overlap with Set 2 to enhance training consistency across different resolutions. |
| Set 2 | 5 MP | 32 | 68 | 2592 × 1944 | Training | High-resolution images; overlaps with Set 1 for consistency. |
| Set 3 | 1 MP | 42 | 95 | 1280 × 800 | Validation | Used for model validation and hyperparameter tuning. |
| Set 4 | 1 MP | 139 | 328 | 800 × 640 | Training | Lower resolution to diversify training scenarios. |
| Set 5 | 1 MP | 100 | 237 | 1280 × 800 | Testing | Images of the same cassettes as those in Set 6; used for robust model testing across different resolutions. |
| Set 6 | 5 MP | 100 | 237 | 2592 × 1944 | Testing | Same cassettes as Set 5; captured with a 5 MP camera to evaluate model performance across varied image qualities. |
| Parameter | Value |
|---|---|
| Initial Learning Rate (lr0) | 0.00129 |
| Final Learning Rate (lrf) | 0.00881 |
| Optimizer | SGD |
| Momentum | 0.937 |
| Weight decay | 0.0005 |
| Warmup epochs | 3.0 |
| Warmup momentum | 0.8 |
| Warmup bias LR | 0.1 |
| Auto Augment | randaugment |
| Parameter | Value |
|---|---|
| Hue Adjustment (hsv_h) | 0.00761 |
| Saturation Adjustment (hsv_s) | 0.60615 |
| Value Adjustment (hsv_v) | 0.22315 |
| Translation (translate) | 0.11023 |
| Scaling (scale) | 0.34352 |
| Horizontal Flip (fliplr) | 0.20548 |
| Mosaic (mosaic) | 1.0 |
| Random Erasing (erasing) | 0.4 |
| Crop Fraction (crop_fraction) | 1.0 |
| Model | Number of Parameters |
|---|---|
| YOLOv8n | 3,258,259 |
| YOLOv8s | 11,779,987 |
| YOLOv8m | 27,222,963 |
| YOLOv8l | 45,912,659 |
| YOLOv8x | 71,721,619 |
| YOLOv9c | 27,625,299 |
| YOLOv9e | 59,682,451 |
| Model | Metric | Val5 (1 MP) | Val6 (5 MP) | Difference | t-Stat | p-Value | Cohen’s d |
|---|---|---|---|---|---|---|---|
| Spatial Accuracy (%) | |||||||
| YOLOv8n | 93.39 ± 0.62 | 93.13 ± 1.07 | −0.26 | 0.82 | 0.448 | −0.34 | |
| YOLOv8s | 94.00 ± 1.15 | 93.75 ± 1.50 | −0.25 | 0.43 | 0.687 | −0.17 | |
| YOLOv8m | 94.78 ± 0.81 | 93.83 ± 2.76 | −0.95 | 0.85 | 0.435 | −0.35 | |
| YOLOv8l | 94.88 ± 0.76 | 94.81 ± 0.88 | −0.06 | 0.14 | 0.897 | −0.06 | |
| YOLOv8x | 94.44 ± 0.95 | 95.33 ± 0.94 | +0.89 | −1.21 | 0.279 | 0.50 | |
| YOLOv9c | 93.63 ± 1.16 | 94.12 ± 0.77 | +0.49 | −0.82 | 0.449 | 0.34 | |
| YOLOv9e | 91.86 ± 3.10 | 94.31 ± 1.79 | +2.45 | −1.59 | 0.173 | 0.65 | |
| False Positive Count | |||||||
| YOLOv8n | 18 ± 3 | 13 ± 2 | −4.50 | 4.26 | 0.008 ** | −1.74 | |
| YOLOv8s | 13 ± 3 | 13 ± 3 | −0.17 | 0.18 | 0.867 | −0.07 | |
| YOLOv8m | 10 ± 3 | 7 ± 2 | −3.33 | 2.29 | 0.070 | −0.94 | |
| YOLOv8l | 11 ± 4 | 10 ± 3 | −1.83 | 1.03 | 0.350 | −0.42 | |
| YOLOv8x | 13 ± 2 | 9 ± 3 | −4.00 | 2.78 | 0.039 * | −1.14 | |
| YOLOv9c | 12 ± 4 | 11 ± 3 | −0.83 | 0.64 | 0.550 | −0.26 | |
| YOLOv9e | 12 ± 3 | 9 ± 3 | −3.00 | 1.91 | 0.114 | −0.78 | |
| IoU | |||||||
| YOLOv8n | 0.9205 ± 0.0052 | 0.9161 ± 0.0085 | −0.0044 | 1.97 | 0.105 | −0.81 | |
| YOLOv8s | 0.9259 ± 0.0100 | 0.9238 ± 0.0064 | −0.0022 | 0.64 | 0.548 | −0.26 | |
| YOLOv8m | 0.9367 ± 0.0030 | 0.9290 ± 0.0121 | −0.0076 | 1.96 | 0.108 | −0.80 | |
| YOLOv8l | 0.9387 ± 0.0030 | 0.9354 ± 0.0024 | −0.0033 | 6.19 | 0.002 ** | −2.53 | |
| YOLOv8x | 0.9423 ± 0.0029 | 0.9361 ± 0.0021 | −0.0062 | 3.60 | 0.016 * | −1.47 | |
| YOLOv9c | 0.9302 ± 0.0029 | 0.9265 ± 0.0072 | −0.0037 | 1.03 | 0.349 | −0.42 | |
| YOLOv9e | 0.9210 ± 0.0172 | 0.9289 ± 0.0066 | +0.0079 | −1.22 | 0.275 | 0.50 | |
| Dice Coefficient | |||||||
| YOLOv8n | 0.9479 ± 0.0063 | 0.9475 ± 0.0084 | −0.0004 | 0.20 | 0.850 | −0.08 | |
| YOLOv8s | 0.9507 ± 0.0107 | 0.9517 ± 0.0057 | +0.0010 | −0.31 | 0.769 | 0.13 | |
| YOLOv8m | 0.9611 ± 0.0033 | 0.9554 ± 0.0121 | −0.0057 | 1.55 | 0.182 | −0.63 | |
| YOLOv8l | 0.9629 ± 0.0018 | 0.9612 ± 0.0014 | −0.0018 | 5.35 | 0.003 ** | −2.18 | |
| YOLOv8x | 0.9663 ± 0.0034 | 0.9623 ± 0.0027 | −0.0040 | 1.77 | 0.137 | −0.72 | |
| YOLOv9c | 0.9557 ± 0.0039 | 0.9537 ± 0.0076 | −0.0021 | 0.53 | 0.622 | −0.21 | |
| YOLOv9e | 0.9469 ± 0.0178 | 0.9580 ± 0.0066 | +0.0111 | −1.55 | 0.181 | 0.63 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaiani, M.; Selouani, S.A.; Mailhot, S. Automated Segmentation and Quantification of Histology Fragments for Enhanced Macroscopic Reporting. Appl. Sci. 2025, 15, 9276. https://doi.org/10.3390/app15179276
Chaiani M, Selouani SA, Mailhot S. Automated Segmentation and Quantification of Histology Fragments for Enhanced Macroscopic Reporting. Applied Sciences. 2025; 15(17):9276. https://doi.org/10.3390/app15179276
Chicago/Turabian StyleChaiani, Mounira, Sid Ahmed Selouani, and Sylvain Mailhot. 2025. "Automated Segmentation and Quantification of Histology Fragments for Enhanced Macroscopic Reporting" Applied Sciences 15, no. 17: 9276. https://doi.org/10.3390/app15179276
APA StyleChaiani, M., Selouani, S. A., & Mailhot, S. (2025). Automated Segmentation and Quantification of Histology Fragments for Enhanced Macroscopic Reporting. Applied Sciences, 15(17), 9276. https://doi.org/10.3390/app15179276







