1. Introduction
The general term for any algae formation, whether microscopic, e.g., phytoplankton and cyanobacteria, or macroscopic, e.g., seagrass, is an algal bloom. Harmful algal blooms (HABs), red tides, water blooms, and algal mats or scums are other terms also used instead of algal bloom [
1]. Algal blooms have three major categories, viz., cyanobacteria blooms, which are generally blue and green [
1,
2]; dinoflagellate blooms, also known as red tides, which are red, brown, and yellow [
3]; and diatom blooms, which are brown in color [
4]. There are several reasons for the rapid growth of algae in aquatic ecosystems [
5]. The most significant factors are nutrient overloading [
6], increased water temperature [
7], and changes in water stratification [
8]. Algal blooms, while appearing vibrant, can wreak havoc on aquatic ecosystems. Their rapid growth has consequences like oxygen depletion [
9,
10], food web disruption [
10,
11], and habitat alterations [
12,
13].
MODIS plays a crucial role in monitoring Earth activities. Since its temporal resolution is 1–2 days, it is useful for monitoring daily activities. MODIS is an essential instrument on board NASA’s Terra, which launched in 1999, whereas Aqua launched in 2002. It collects data on a global scale across 36 spectral bands, and at four spatial resolutions: 250 m, 500 m, 1 km, and 5.6 km [
14]. These features make it popular for Earth observation, including ocean monitoring [
15] and other applications like land cover and land change detection, fire detection and tracking, and natural disaster response [
16].
MODIS leverages the power of multispectral data to observe Earth from space, collecting information across a range of wavelengths from visible (0.4 μm) to infrared (14.4 μm); MODIS provides a richer picture of the planet compared to sensors that capture data in a single band [
17]. Due to this, discrimination of terrestrial features is easy and efficient in multispectral data, as the spectral signature varies with the variation in the electromagnetic spectrum [
18]. Combining spectral bands with arithmetical operators has good effects in delineating and enhancing a feature by suppressing others in the imagery, better known as spectral indices [
19]. With its 36 spectral bands, MODIS offers a rich platform for developing and applying various spectral indices.
Due to the sudden change in environmental factors (like temperature, salinity, wind speed, artificial activities like agriculture, and industrial and pathogenic waste), aquatic ecosystems can experience algal blooms. Algal blooms are a kind of aquatic pollutant that has several consequences for aquatic animals, ecosystems, the environment, and human beings, directly and indirectly. An early detection and alarm system is required to avoid and reduce their consequences. Since MODIS has daily global coverage, it has been utilized to fulfill the objective of researching the extraction of algal blooms and their daily monitoring.
This research article has significant novelties. First, the interaction with the Earthdata website is automated. Second, the data is downloaded automatically based on the central coordinates of the location and specific date range. Third, if the date range has N (N is greater than 0) days, the data will be downloaded for all days. Fourth, the study area will be automatically clipped based on the LLC and URC. Fifth, a novel index is proposed to enhance algal bloom features in the clipped imageries. From downloading data to algal bloom extraction, the output data will be saved automatically in the designated folders created while defining the test site attributes.
2. Materials and Methods
Specific platforms and resources are utilized in the automation process of algal bloom extraction and daily monitoring, as discussed in the sections below.
2.1. Web Resources and Software
The NASA Earth Observatory (NEO) studies incidents of algal bloom and reports them at
https://earthobservatory.nasa.gov/, accessed on 14 May 2025; the test sites for this study were selected based on the NEO’s reports. The longitude and latitude of the LLC and URC were fetched from Google Maps. The script for the algal bloom extraction was written in Python 3.12 and used the libraries numpy, matplotlib, pandas, geopandas, earthaccess, geometry, rioxarray, xarray, rasterio, earthpy, and osgeo. The Python script interacts with the Earthdata website
https://www.earthdata.nasa.gov/ (accessed on 14 May 2025) to deal with the MODIS images. The Earthdata website needs login credentials to download the images; this login credential is available in a file saved with the. netrc extension in the USER’S directory.
2.2. Data Set
MODIS releases data in different forms and formats. The temporal resolution also varies. The data that were considered in this study are MOD09GQ and MOD09GA. MOD09GQ—MODIS Surface Reflectance Daily Level 2 Global coverage and 250 m spatial resolution: This file contains two bands of spectral data centered at 645 nm (RED) and 858 nm (NIR). MOD09GA—MODIS Surface Reflectance Daily Level 2 Global coverage of 500 m spatial resolution: this file contains seven bands of spectral data, which are tabulated in
Table 1.
2.3. Test Sites
During 2003 and 2023, numerous algal bloom incidents were reported on the NEO website. Nine sites were selected to test the automation process in this article. The details of the test sites and their LLC and URC are given in
Table 2.
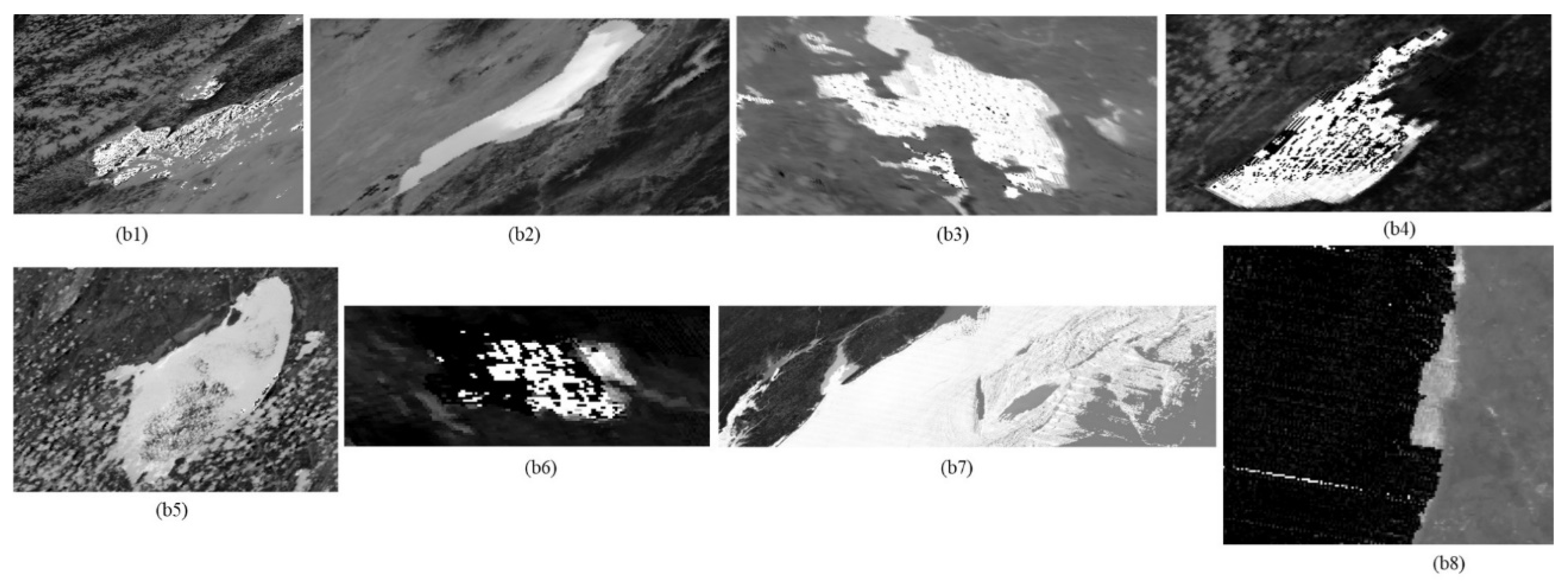
The images of the test sites and the highlighted regions of algal bloom incidents are reported on the NEO’s website [
20,
21,
22,
23,
24,
25,
26,
27]. A collage of images of the test site is shown in
Figure 1.
2.4. Proposed Method for Automatic Extraction
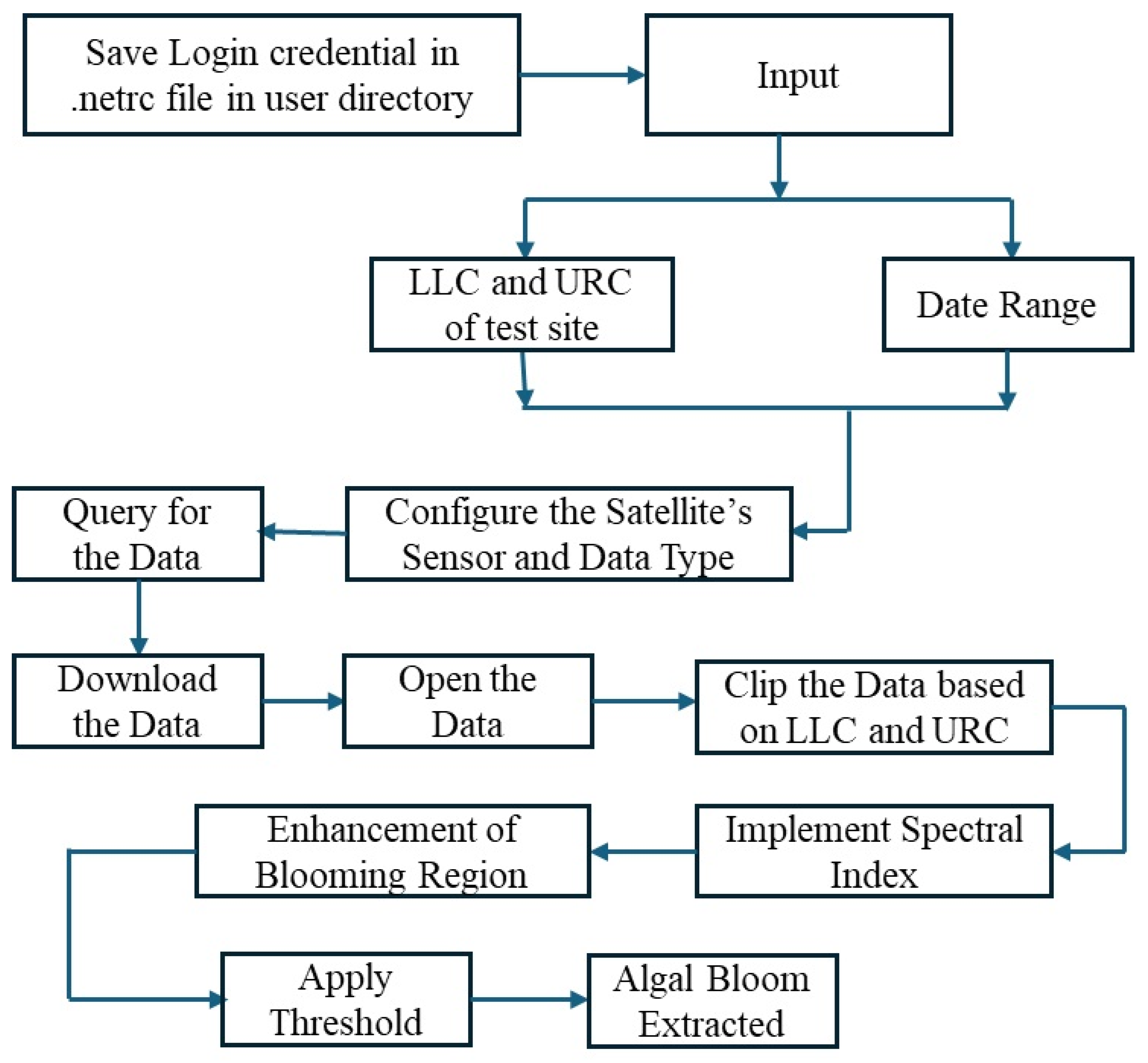
The automatic extraction of algal blooms and their daily monitoring using the Earthdata website were implemented in eight modules, depicted in
Figure 2.
Module 1—Study Area (test_site_dict): This module deals with the study area and the period of algal bloom monitoring by maintaining a dictionary of all test sites. There can be more than one study area, so N defines the number of test sites. The date range defines the dates considered for the study; it can be one day or more than a day; the dates are inputted in the YYYY-MM-DD format. Next, a location name is required to create a folder in which to save the processed data. Finally, the LLC and URC of the study area are input to fetch the study area in one of the granules available in the website’s database.
Module 2—Configuration of Satellite Sensor (config): This module configures the satellite sensor’s name and the required data type. Since MODIS provides data at three spatial resolutions (1 km, 500 m, and 250 m), fetching the correct data from the correct sensor is of the utmost importance.
Module 3—Query Data (query_data): This module utilizes the “earthaccess” library for Python. The search\textunderscore data function requires four parameters: the type of resolution, coordinates of the study area, temporal dates, and cloud service.
Module 4—Downloading the Data (download_modis_data): Once the query module identifies one of the granules in the specified ranges of date and location, the download function from “earthaccess” is used to fetch the data with the data_links generated for the specific granules.
Module 5—Opening the Downloaded Image Data (open_img_data): The inbuilt libraries rioxarray and xarray from Python are used to deal with the raster data. The inbuilt functions in the libraries are utilized to read and write the band images of the downloaded imagery.
Module 6—Clipping the Study Area (clipped_dataset): A single granule includes a larger area. The geometry library is used to crop the image to the desired study area. The LLC and URC are used as a bounding box to define the study area.
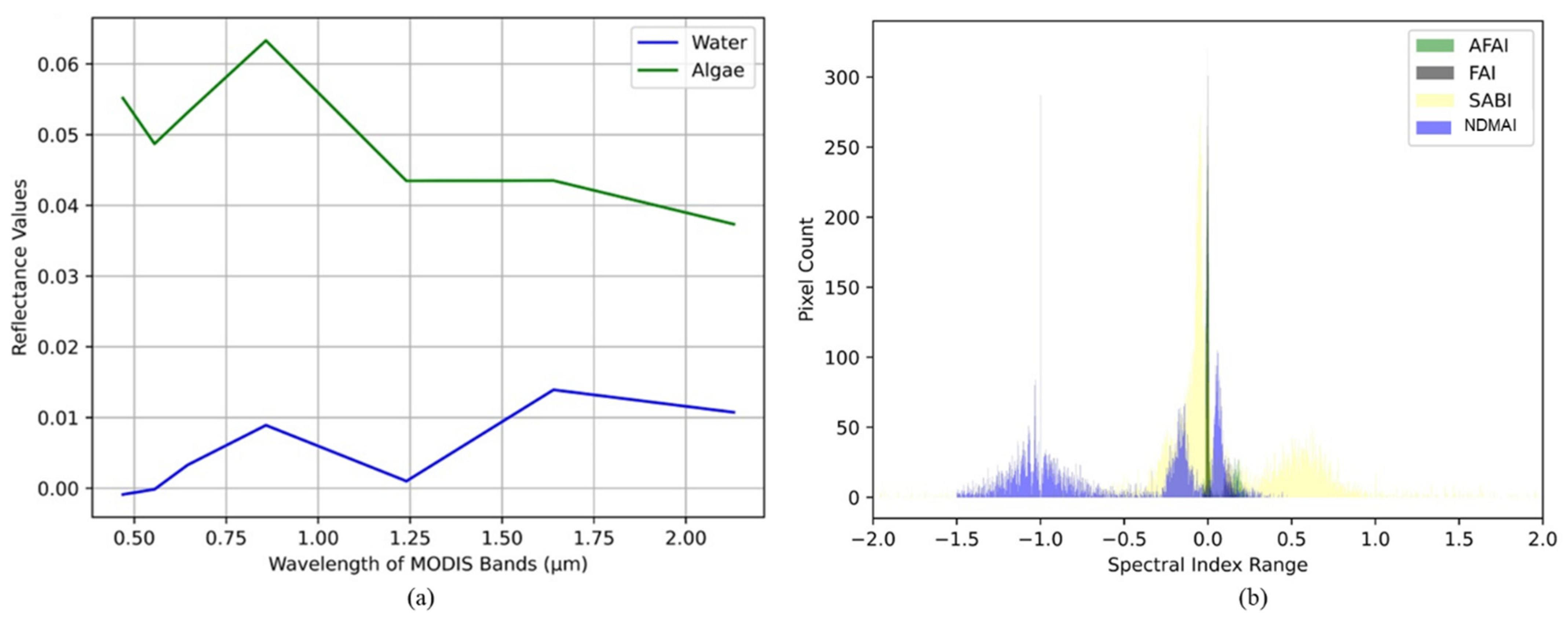
Module 7—Proposed Spectral Index for Algal Bloom: Spectral indices are applied to enhance the algal bloom in the water bodies. Both 500 m and 250 m spectral bands are analyzed and the spectral response curves are studied, as shown in
Figure 3.
Due to band limitations and coarser resolution, different spectral indices are proposed for both data sets, termed Normalized Difference MODIS Algae Indices (NDMAIs), for the data sets at both 500 m and 250 m spatial resolutions. The SRC is analyzed, and the following observations are made:
- i.
The spectral response is high in the VIS range and low in the SWIR range for algae, and vice versa for water.
- ii.
The reflectance in NIR is higher than in VIS for algae and water,
- iii.
The reflectance of the algae feature is lowest in SWIR2, whereas reflectance is comparatively high for the water feature.
The above facts compelled us to choose the NIR and SWIR2 bands; the difference between the two selected bands will give significantly positive values for algae features and negative values for water features. Consequently, this difference will enhance the feature of interest, i.e., algae, by suppressing the background, i.e., water. The NDMAI for 500 m resolution data is defined and implemented as mentioned in Equation (1):
The 250 m spatial resolution data give only two bands, red and NIR. The spectral index is defined and implemented through Equation (2).
Module 8—Threshold: The spectral index imagery is used to identify the threshold for the algal bloom features, and the enhanced blooming regions in the image are analyzed carefully to determine the range of the algal blooms. This range is then used to generate the threshold imagery.
The complete Python script for these modules has been uploaded to GitHub. The integration of the module provides a tool to download the MODIS data from Earthdata and process it for blooming region extraction and daily monitoring; the tool can be explored via the public access link
https://github.com/JamesFalconer/modis_data_download_and_process_tool (accessed on 14 May 2025).
4. Discussion
MODIS provides data at three spatial resolutions. This article uses data sets from 250 m and 500 m spatial resolutions to automate the process. MOD09GQ and MOD09GA files on the Earthdata website deal with MODIS Surface Reflectance Daily Level-2 Global data at 250 m and 500 m spatial resolutions, respectively. From the spectral response curve, it is observed that the spectral response is higher in the visible band than in the infrared bands for algal bloom features.
The 250 m data set has only two bands, red and NIR, which are formulated in the spectral index to extract algal features. In contrast, the 500 m data set has seven bands. An exhaustive study is performed using these bands; NIR and SWIR1 are identified as most suitable for the study of algal bloom features, as depicted in
Figure 10a. A novel index is proposed, the Normalized Difference MODIS Algae Index (NDMAI), and is also compared with the adjusted floating algae index (AFAI), floating algae index (FAI) and surface algal bloom index (SABI). The NDMAI outperforms these indices, as it discriminates water, land, and algal bloom features by creating a suitable threshold range for each without any feature mixing, which is depicted in
Figure 10b.
5. Performance Analysis and Validation of NDMAI
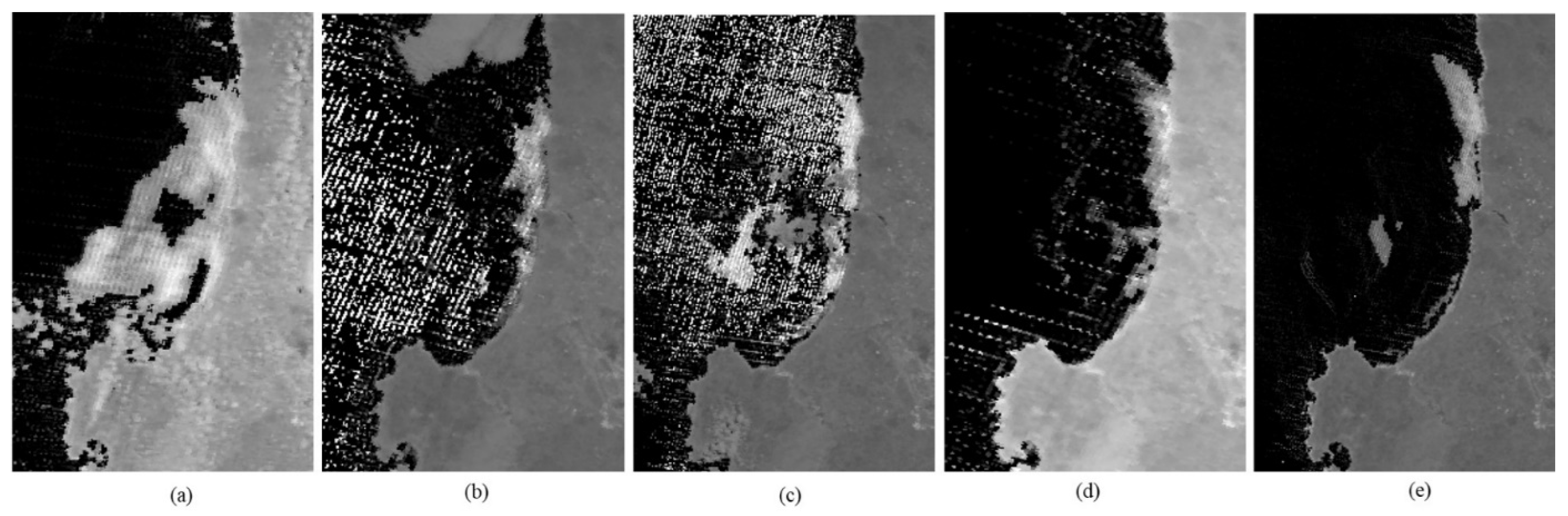
The proposed index, the NDMAI, is compared with existing indices like the FAI, AFAI, and SABI. The MODIS data from 500 m is utilized to apply the Band Math module of the ENvironment for Visualizing Images (ENVI) software (ENVI 5.1); the corresponding results are shown in
Figure 11.
It can be visualized from
Figure 11 that the proposed index is quite efficient in enhancing algal bloom features by suppressing the surrounding features of water and land.
Figure 11a shows a True Color Composite image of the MODIS 500 m resolution imagery from the data collected on 8 February 2023 in the region of the West Coast of South Africa. The bluish region shows the region covered by an algal bloom. A visualization of algal bloom extraction by the FAI and AFAI is shown in (b) and (c), whereas extraction by the SABI is shown in (c). The extraction by the proposed NDMAI is shown in (e); the visualization of (a) to (e) shows that the enhancement by the NDMAI perfectly matches with the RGB image.
Further, the spectral index imageries shown in
Figure 11b–e were classified using the Support Vector Machine (SVM) classifier from the Supervised Classification Module of ENVI. The chosen samples from the three regions, viz., algal bloom, water, and land, included 900 pixels from algal bloom, 1000 pixels from water, and 1700 pixels from land features. There were 142,186 pixels in the imagery, which were classified. The classified imagery is shown in
Figure 12, and the statistical results are shown in
Table 4.
It is observed from
Table 4 that the proposed index extracts algal blooms with 99.17% accuracy and outperforms the existing FAI, AFAI, and SABI. The stacked imagery gives the highest accuracy of 100%. However, it utilizes all seven bands from the MODIS 500 m data. In contrast, the proposed index only utilizes two bands, NIR and SWIR2, so it will have better computational efficiency due to the optimal use of bands compared to indices with a large number of bands.
Other parameters, like the confusion matrix, error of commission, and producer’s accuracy for the proposed NDMAI, are also analyzed and tabulated in
Table 5,
Table 6 and
Table 7.
6. Conclusions and Future Scope
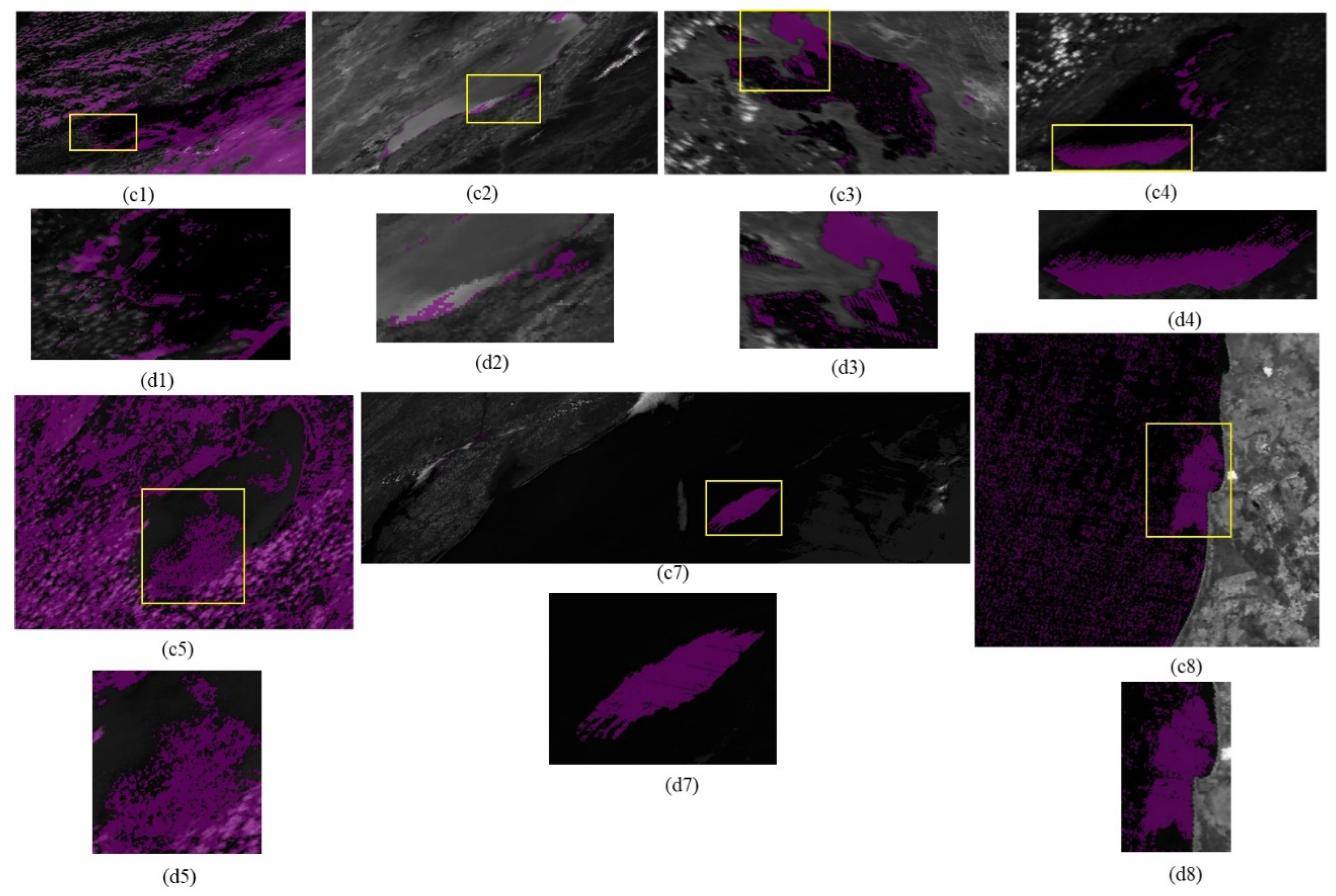
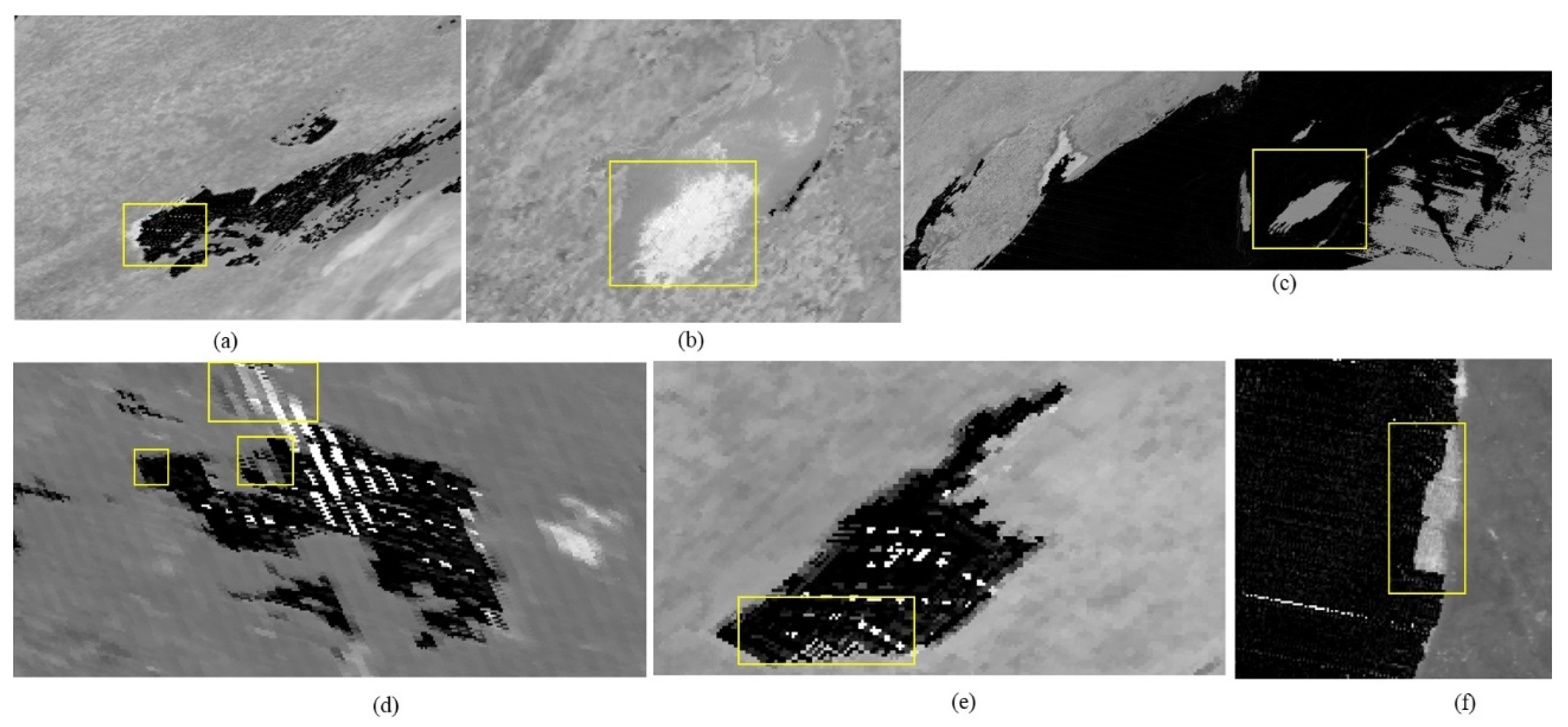
The proposed automation technique successfully extracts algal bloom features. Nine test sites were selected to extract algal bloom regions from NASA’s Earth Observatory reports. Out of eight test sites, the technique enhanced the blooming regions at seven sites. In the case of Lake Villarrica, the spectral index imagery had many dead pixels, which prohibited the identification of the threshold range. The blooming region was enhanced using both 250 m and 500 m spatial resolution data. The proposed technique requires two inputs for its execution: the latitude and longitude of the lower left coordinate and upper right coordinate of the test sites, and the date range (which could be greater than or equal to one day). After successfully enhancing the algal bloom features in the specific lakes on the date the NEO reported, further daily monitoring was also attempted on the West Coast of South Africa, which experienced a significant impact of red tide in April 2003, and was successfully performed for several consecutive days between 22 April and 26 April; the other days of the month were cloudy, and not successive, so they are not reported.
Since the spatial resolution of MODIS is coarser, dealing with small test sites and enhancing algal bloom features may not be possible. The NEO reported that most of the test sites used Landsat-8 and Landsat-9 sensors, which have a spatial resolution of 30 m. The other drawback of the proposed technique is that it deals with a single granule at a time. If the test site belongs to two granules, then the technique does not deal with the mosaicking of the granules. This mosaicking may be incorporated in the future. Furthermore, similarly to the Earthdata portal, there are portals like EarthExplorer and DataSpace Copernicus, which can be automated and integrated with the existing methods to establish a rich monitoring system and overcome the problems of dealing with small regions.
This automation has a vast potential for the future. Several other natural phenomena need daily monitoring, like floods. There are other spectral indices, such as vegetation indices for enhancing vegetation features, water indices for delineating water bodies, and urban indices for extracting urban features. MODIS provides 36 spectral bands; these multispectral bands can be exhaustively analyzed to delineate specific features by formulating the novel index. By changing the spectral index, several other features can be extracted, and daily monitoring is also possible with the proposed technique.