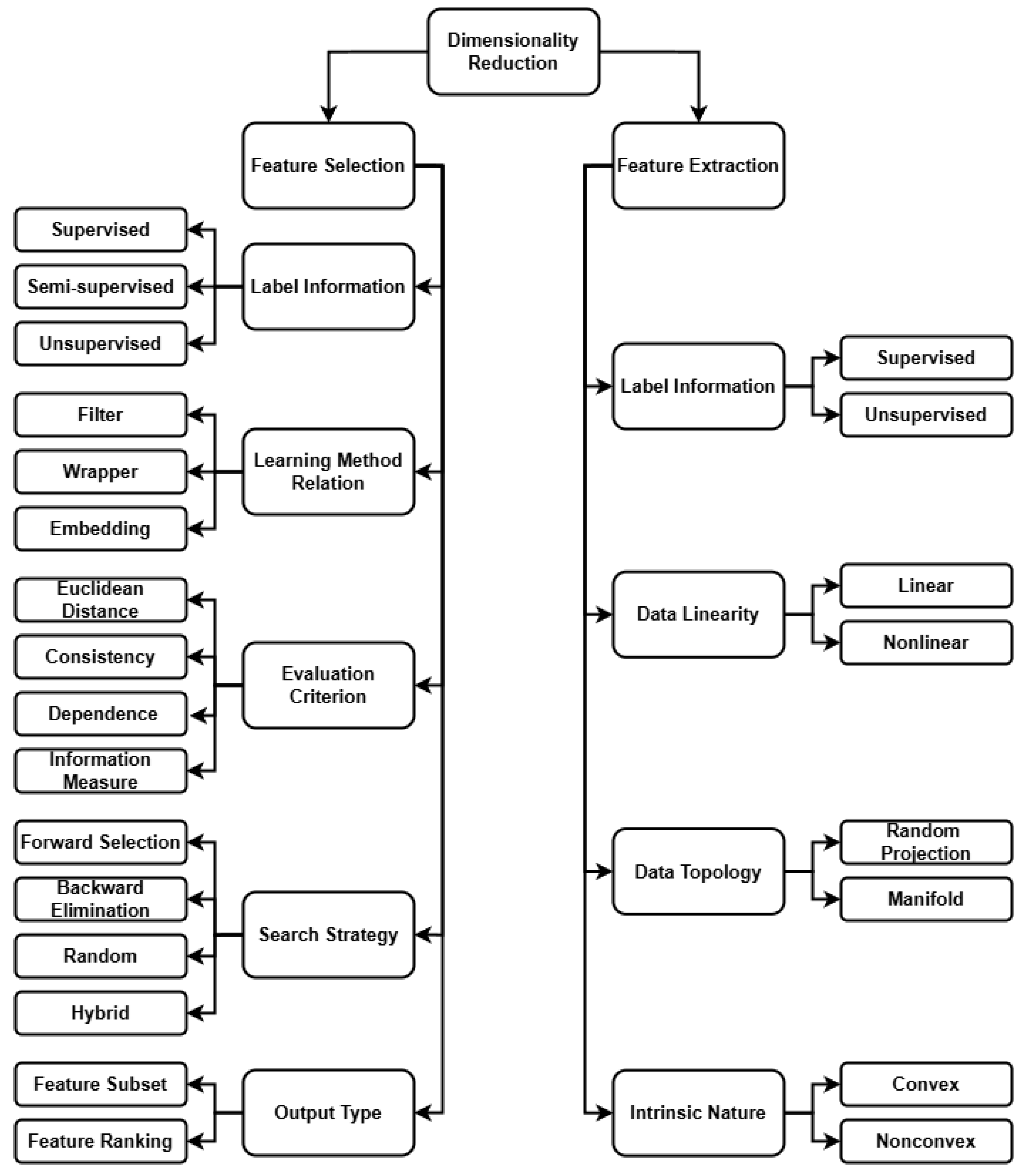
1. Introduction
Machine learning (ML) has achieved a prominent position in recent years. Much of its recent visibility may be attributed to large volumes of data (used for training purposes) as well as improved computing infrastructure [
1], giving rise to compelling applications like DALL-E
https://openai.com/index/dall-e/ (accessed on 13 January 2025) [
2] and ChatGPT
https://openai.com/index/chatgpt/ (accessed on 13 January 2025) [
3].
However, this also means greater importance is placed on this step of the ML pipeline than ever before. Data collecting is becoming one of the main bottlenecks in the field. The majority of the time for executing ML end-to-end is spent on preparing the data, which includes gathering, cleaning, analyzing, visualizing, and feature engineering [
1].
This only exacerbates already existing related issues, such as the cold-start problem that is observable in certain fields, such as the medical field [
4], e-commerce [
5], and online education [
6]. Manually labeling a dataset is very costly and complex. This is particularly true for research domains like healthcare that have limited access to labeled data [
4]. To abstract underlying patterns and create a proper model, approaches often require massive amounts of supervised training data per inherent concept [
7]. These learning processes can take days using specialized (and expensive) hardware [
7].
One should recognize that there are applications for which data are inherently expensive to acquire, whether due to financial constraints or time limitations, making the development of suitable models for these fields a significant challenge [
4].
These challenges mirror similar issues found in the field of digital marketing, where understanding human interaction across the physical and digital worlds requires selecting relevant tools and techniques that ensure flexibility and context-awareness [
8,
9]. Multi-modal artificial intelligence approaches have demonstrated the importance of integrating diverse data sources to analyze consumer sentiment and enhance engagement [
10,
11]. However, the complexity of integrating these modalities underscores the importance of efficient data pre-processing, including cost-effective feature selection, to reduce noise and improve the robustness of ML models [
12].
Furthermore, these are not the only problems present in this context. Even if valuable data are sourced to be used for one of these aforementioned fields, current features may be redundant or even irrelevant to the problem at hand, which means that they only add noise to the dataset [
12].
In order to deal with these problems, a data engineer may turn their attention to either the data or the algorithms that use them. That is, one may try to use approaches that require less data to obtain results, such as few or even zero-shot algorithms [
7]. Or one may dedicate more attention to the data and try to improve them by iteratively cleaning and analyzing them.
This work addresses the challenge of balancing effective feature exploration with computational efficiency through a novel data-centric approach. We propose a feature selection method that accelerates the traditionally time-intensive wrapper-based feature selection process by leveraging interim representations of feature combinations. These composite representations enable the method to approximate inter-feature dependencies while relying on filter-based evaluation, resulting in a substantial reduction in computational time without maintaining competitive performance. The core novelty of our approach lies in its ability to deliver wrapper-like effectiveness with filter-level efficiency, making it particularly suitable for resource-constrained machine learning scenarios. To support and contextualize our contribution, we also present a brief review of dimensionality reduction and its two key branches: feature selection and feature extraction.
The paper follows the following structure. Initially, a concise overview of fundamental principles related to dimensionality reduction is presented, as well as a short description of relevant feature selection approaches used in testing (
Section 2). Subsequently, the recommended solution is introduced and described (
Section 3). After that, the results generated by this solution are compared to other presented approaches in terms of both temporal cost and performance (
Section 4). Finally, conclusions are drawn, and future work is delineated (
Section 5).
3. Proposed Solution
The necessity of using effective methods of feature selection and dimensionality reduction across various fields is paramount. Applying feature selection appropriately makes further data-gathering processes cheaper while resulting in similar results. Furthermore, the use of dimensionality reduction greatly benefits high data volume throughput tasks since it counters the curse of dimensionality while maintaining as much relevant information as possible. In this section, an overview of the proposed solution will be put forward. This will be followed by a short discussion of its most salient problem of exponential combinatorial growth. Subsequently, the approach taken to implement it will be presented.
3.1. Proposal
In this particular case, there is an interest in countering the cold-start problem, that is, the fact that in certain domains, the amounts of data needed to complete a task is not always available, meaning there is a limited amount of data to work with [
65,
66]. One way to make this process cheaper/easier would be to properly select the optimal group of features needed to represent the original dataset, reaching a closer state to its intrinsic dimensionality.
Feature selection is a critical component in developing robust and interpretable machine learning models. Conventional evaluation standards typically classify feature selection algorithms into three main categories: filter, wrapper, and embedded approaches. Each of these paradigms presents its own set of advantages and trade-offs. In the context of the present work, our primary objective is twofold: to maintain the freedom of choice regarding the final learning algorithm and to ensure that the feature selection process is computationally efficient.
While wrapper methods are known for their ability to evaluate feature subsets in direct relation to the performance of a learning algorithm, they are often computationally intensive (as documented in [
17,
23,
24]). Given these constraints, our approach instead focuses on adapting a filter-based methodology. Traditional filter methods are prized for their speed and scalability; however, they typically overlook complex inter-dependencies between features. To address this limitation, an enhanced filter-based approach that partially accounts for inter-feature relationships is introduced. This modification enables the capture of interactions typically accessible only through the more computationally demanding wrapper methods.
Extending this line of reasoning, and in light of the capabilities presented by feature extraction approaches, it is theoretically viable to represent combinations of features through their intermediate representations. In doing so, one may apply feature selection not on the original variables, but instead on derived composite features that encapsulate higher-order interactions.
Consider, for instance, a dataset X of a given length. Our proposal envisions that this dataset can be approximately represented by a collection of n combinations, each comprising r features extracted from X. By employing such an interim representation, the modified filter-based selection method is afforded a broader and more nuanced feature space to explore—without incurring the high computational cost typically associated with wrapper techniques. This strategy is expected not only to significantly reduce the search time, but also to achieve performance metrics that are competitive with those of wrapper-based approaches.
In summation, the proposed framework offers a balanced solution by leveraging the computational efficiency of filter methods while simultaneously incorporating feature inter-dependencies to improve selection quality. This approach aims to allow for the rapid screening of a vast feature space, ensuring that the final selected set is both diverse and representative of the underlying data structure, thereby facilitating a more flexible and effective integration with subsequent learning algorithms.
The implementation of this approach relied on widely used Python libraries, including NumPy (1.23.5), Pandas (1.5.3), SciPy (1.11.3), Scikit-learn (1.2.1), and Py_FS (0.2.1). Among these, Scikit-learn and Py_FS played a particularly significant role in supporting the computational study discussed later in the article. Scikit-learn provided an extensive set of utility functions and facilitated the use of various filter-based feature selection and feature extraction techniques. Meanwhile, Py_FS was employed for its comprehensive suite of wrapper-based feature selection methods, which proved essential for the analysis.
3.2. Pitfalls
No truly foolproof approach exists; as such, it is to be expected that the proposed solution has its downsides, one of which may be self-evident even in a planning phase. This problem derives from its core concept, the use of interim features to represent combinations of the original ones, which is, in essence, a combinatorial problem.
As stated in [
67], a combination refers to an unordered arrangement of a set of objects. Repetitions may or not be allowed. The number of
r-combinations of a set with
n distinct elements may be expressed as seen in Equation (
1) [
68].
By fixing a value for
r (as 5 in this case) and then plotting the previous function,
Figure 2 may be drawn:
As depicted in the plot (
Figure 2), the number of combinations that can be created from a given group increases exponentially with the original sample size. For the proposed solution, it would become exponentially more expensive to use it for datasets with a large amount of features.
There are some ways to address this problem. One possibility would be to increase the number of features included in each combination group (r) in proportion to the increase in the total number of features (n). However, that would mean that for larger sets, each combination group would be equally broad, meaning that the effects of singular features and/or feature interactions would be greatly diluted.
A more permissible option would be to reduce the original feature set to a more manageable size. This can be accomplished by applying feature selection to the original set, and since the method should not be computationally expensive, a filter method is preferred.
3.3. Process
Given the proposed solution, a diagram of its intended progression is represented in
Figure 3.
Starting from a full set, the solution reduces the number of redundant features. It accomplishes this by performing dependency analysis over the provided features and disposing of highly dependent ones, which are reduced to a single representative. It then selects the top features via a filter feature-selection method. This outputs a reduced set for which all feature combinations of length r are computed. Since the number of features n has been reduced, this operation is not as computationally expensive. The resulting list of feature groups is then used to create an entirely new dataset composed of features extracted from each of them. The new interim features dataset is subsequently used with a filter method (which may or not be the same as the one previously used) to select the best of them. Any of the selected interim features directly correspond to a set of possible solutions for selection, all of which have a length of r.
It should be taken into account that this process is adaptable since there are steps in which the algorithm to be used may be chosen as needed, and, in some cases, steps may be skipped. During the entire process, up to two different filter-based algorithms for feature selection may be chosen, as well as one for feature extraction. Additionally, if the dataset is not too large, a reduced representation may not be needed. A representation of this solution’s algorithm may be seen in Algorithm 1.
| Algorithm 1: Proposed solution’s algorithm |
![Applsci 15 08196 i001]() |
The solution takes as input the dataset to be treated (D), its categorical features (if any) (), two filter-based feature selection methods (which may be the same) (), the chosen feature extraction method (), the max value allowed for feature redundancy/dependency (), the max feature set length to work with (), and the target feature set length (r).
Starting from the full feature set, our approach addresses feature redundancy through dependency analysis to identify groups of highly dependent features. Specifically, features with a dependence value exceeding the threshold are classified as highly dependent. Each resulting group is then represented by a single feature that preserves the core information shared among its members. Although redundant features may contain subtle complementary signals, this consolidation strategy prioritizes unique, non-redundant features to maximize distinct information and minimize overlap. This initial dimensionality reduction step alleviates the computational complexity of subsequent analyses and reduces the risk of overfitting, thereby promoting the development of more robust predictive models.
Following this reduction, if the dataset’s number of dimensions still surpasses , a filter-based feature selection method is employed to further identify and preserve the most promising features from the previously pruned set. This targeted selection ensures that only the most relevant features are carried forward, thereby streamlining the process.
Subsequently, the algorithm computes all possible feature combinations of a predetermined length, r, using the reduced feature set. These combinations are then used to generate a new dataset, wherein each derived feature corresponds to the extraction of each unique combination created using the chosen feature extraction method (). A second round of filter-based selection (potentially utilizing a different algorithm, ) is applied to this interim dataset to isolate its optimal features. The resultant features correspond directly to potential solutions, each comprising a feature set of length r. Going through its various steps returns the provided dataset reduced to its most relevant features (D).
4. Computational Study
In the following section, the results extracted from the proposed solution are presented and compared to those of other wrapper-based feature selection methods. These results and their meaning are then further discussed.
Taking into account the main problem referred to before, a test regarding the computation time necessary to create the interim features for all combinations of a dataset was carried out.
To illustratively validate the concern regarding the exponential growth in the number of feature combinations, an experiment was conducted. The length of combinations (
r) was fixed at five, a value chosen to strike a balance between computational feasibility and representativeness of the underlying trend. This test was carried out using the proposed method without applying any preliminary feature reduction steps, thereby isolating the impact of the combinatorial explosion. The results may be observed in the plot depicted in
Figure 4.
As expected, the exponential growth in potential combinations also significantly affects the temporal efficiency of the proposed algorithm, which only validates the importance of the first feature reduction steps proposed to counter it. Once that counter-measure was implemented, an experiment was designed to compare the performance of the proposed solution when compared to other feature selection methods.
To evaluate the effectiveness of our proposed solution, we compare its performance against nine established wrapper-based feature selection methods. This set of algorithms was chosen to provide a diverse and challenging benchmark, as wrapper methods are known to produce high-performing feature subsets. The selection represents a wide array of state-of-the-art metaheuristic strategies commonly used for optimization and feature selection. Furthermore, all comparative algorithms were implemented using the Py_FS package, which provides a standardized and fair framework for evaluating performance and computational cost. The chosen methods were:
- 1.
Binary Bat Algorithm (BBA)
- 2.
Cuckoo Search Algorithm (CSA)
- 3.
Equilibrium Optimizer (EO)
- 4.
Genetic Algorithm (GA)
- 5.
Gravitational Search Algorithm (GSA)
- 6.
Harmony Search (HS)
- 7.
Mayfly Algorithm (MA)
- 8.
Red Deer Algorithm (RDA)
- 9.
Whale Optimization Algorithm (WOA)
To conduct meaningful testing, a substantial number of data points are required. Consequently, thirty distinct datasets were utilized for both binary and multi-classification tasks, which may be reviewed in
Table 1.
The framework was designed as follows:
- 1.
All datasets are pre-processed in order to guarantee no missing rows and numerical features are standardized
- 2.
A cycle iterates over each combination of existing datasets and methods. In each step, the following occurs (seen in Algorithm 2):
- (a)
The dataset is read.
- (b)
For datasets exceeding 10,000 rows, a stratified sample of 10,000 instances was used to reduce the computational load while preserving the original class distribution. This approach minimizes the loss of informative patterns and ensures that the sampled subset remains representative of the full dataset.
- (c)
Its categorical features are encoded by using target encoding (these features are identified beforehand).
- (d)
This dataset is used with the current method, which iterates over the proposed solution and the remaining methods (these use 30 agents, and a max of 50 iterations, as these are the chosen defaults of Py_FS) to find a solution set.
- (e)
Given the solution set arrived at by the feature selection method, the best sets of features found for target lengths [2–5] are retrieved.
- (f)
For each of the target lengths, the best set found (if any) is used to train a default random forest classifier.
- (g)
Performance metrics are then retrieved from the trained classifier.
- 3.
These metrics are then saved while being associated with the feature selection method and dataset involved in the current step.
| Algorithm 2: Testing cycle’s algorithm |
![Applsci 15 08196 i002]() |
For this test, PCA was the chosen extraction method. This was the case since PCA has been extensively utilized in various experimental settings, particularly in the fields of biometrics, image processing, and fault detection, due to its effectiveness in feature extraction. Some notable examples are its usage in the realm of iris and face recognition [
129,
130,
131]. However, one limitation of PCA is that it may fail to capture complex non-linear structures that are often present in real-world datasets. To address this limitation, the approach chosen was to use Random Fourier Features (RFF). RFF approximates non-linear kernel methods (specifically those based on shift-invariant kernels like the Radial Basis Function) by mapping the input data into a randomized low-dimensional feature space where linear techniques can then be applied [
132].
Additionally, Mutual Information (MI) was employed as the filter-based feature selection algorithm for this experiment. This choice was motivated by several factors. First, MI has consistently demonstrated effectiveness across a wide range of feature selection scenarios, making it a robust and well-validated method [
133,
134]. Second, MI is computationally efficient, which aligns well with the broader objective of this study to maintain reasonable resource usage across large-scale evaluations. Finally, unlike correlation metrics, such as Pearson’s or Kendall’s coefficient, MI can capture both non-linear and non-monotonic dependencies between features and the target variable [
135]. This makes it particularly suitable for complex datasets where relationships among variables may not be simplistic.
Prior to applying any feature selection methods, all datasets used for testing underwent a standardized pre-processing pipeline. Rows with missing values were removed to ensure consistent input for downstream processing (although these were few in number due to this being a factor during dataset selection). Numerical features were standardized (using Z-score scaling) to ensure comparability across dimensions and to prevent features with larger magnitudes from dominating others.
Categorical features were encoded within the evaluation loop itself, using target encoding with smoothing to avoid introducing target leakage. This design was adopted in order to allow for easy substitution of encoding strategies if needed. Notably, one-hot encoding was avoided, as it would have prohibitively inflated the dimensionality of the feature space.
It is important to note that the pre-processing step was applied uniformly across all methods. Based on empirical observations during testing, the time spent on pre-processing was consistently negligible. While not formally benchmarked, this suggests that pre-processing does not materially affect the relative performance comparisons reported.
In this study, all feature selection methods and the downstream random forest classifier were evaluated using their default hyper-parameters. This decision was guided by considerations of fairness, computational feasibility, and reproducibility. Using default settings ensured a consistent baseline across all methods, allowing performance differences to reflect algorithmic behavior rather than hyperparameter tuning. Moreover, auto-tuning would have introduced a prohibitive computational overhead. Given the scale of the study—ten optimizers (the proposed approach plus nine wrapper approaches), sixty datasets, and multiple feature set sizes—exploring additional hyperparameter combinations would have multiplied the runtime significantly, rendering the analysis impractical.
Finally, default configurations reflect typical usage scenarios, making the results more accessible and easier to replicate. The chosen defaults are generally sensible values that offer a good balance between performance and abstraction of usage. While future work may explore optimized configurations, the use of defaults provided a balanced and efficient framework for large-scale comparison.
Due to the design of the wrapper-based feature selection methods (where the best feature subsets are selected independently of the target subset length), it is possible that fewer than thirty valid data points were obtained for certain target lengths
r. While this does not pose a methodological issue, it limits the extent to which these subsets can be assumed to follow a normal distribution under the central limit theorem [
136]. To address this, all wrapper-based methods were later aggregated into a single group during the statistical analysis, thereby ensuring a sufficient sample size and enabling a more reliable approximation to normality for comparative purposes.
The metrics used to measure the performance of the fitted classifiers were:
Weighted Precision—obtained by calculating the precision for each involved label, and finding their average weighted by the number of true instances for each label.
Weighted Recall—obtained by calculating the recall for each involved label, and finding their average weighted by the number of true instances for each label.
Weighted FBeta—obtained by calculating the fbeta score for each involved label, and finding their average weighted by the number of true instances for each label.
Time—obtained by measuring the elapsed time during the testing process
To further explore the collected results, and statistically test them, all data points were then divided into two groups “Proposed” and “Other”, which took into account the proposed method (PM) data points and all other data points, respectively. Thus, the central limit theorem was respected in both groups, meaning both should converge to a standard normal distribution. By plotting these new data groups according to the metrics mentioned earlier,
Figure 13,
Figure 14,
Figure 15,
Figure 16,
Figure 17,
Figure 18,
Figure 19 and
Figure 20 are produced. Specifically, in the case of the points relating to the elapsed time, the data regarding MA and RDA were excluded so as to not negatively impact the other approaches.
To statistically validate the observed performance differences between the proposed method and the baseline approaches, we applied a combination of Welch’s two-tailed t-tests and Generalized Least Squares (GLS) regression models. Welch’s t-test is appropriate for comparing two groups with potentially unequal variances and sample sizes, which suits our experimental setting where the number of valid data points varies across methods and feature set sizes. GLS modeling further allowed us to estimate the effect of the independent variables (specifically, the feature set length and method group) on each performance metric while controlling for task type (binary or multi-class classification). This dual approach ensures that the results are statistically sound and robust towards the structure and distribution of data.
The full summary statistics for the GLS models are presented in
Table 2,
Table 3,
Table 4,
Table 5,
Table 6,
Table 7,
Table 8 and
Table 9. The results from the Welch’s
t-tests (reported in
Table 10) confirm that the differences observed across several key metrics are statistically significant, particularly with respect to computational time and performance trends as the feature set length increases.
For reference, the hardware used to carry out these tests was a setup consisting of an MSI X370 mainboard, an AMD Ryzen 5 5600G CPU with a max clock speed of 4.4 GHz, a NVIDIA GeForce RTX 3070 GPU and 36 GBytes of DDR4 RAM running at 16 MHz.
Discussion
In
Figure 5,
Figure 6,
Figure 7,
Figure 8,
Figure 9,
Figure 10,
Figure 11,
Figure 12,
Figure 13,
Figure 14,
Figure 15,
Figure 16,
Figure 17,
Figure 18,
Figure 19 and
Figure 20, some outliers can be observed; as in any experiment, some amount of variability is expected, but some hypothesis should be put forward. Through observation of the presented boxplots, it is possible to see that most outliers are detected in the wrapper methods put forward to compare to the proposed solution. These outliers may result from various factors, either individually or in combination. Firstly, as was explained previously, these algorithms suffer a degree of randomness due to their design—it is possible that the initial solutions generated for these cases were simply extreme when compared to the norm. A second hypothesis could be that some of the datasets used in this experiment are not a good fit with these algorithms’ approach, meaning they inherently present a bad performance when compared to the average case. Observing the plots presented in
Figure 17 and
Figure 18, it can be discerned that the wrapper approaches seem to present some outliers referring to combination lengths of 2 under binary classification. This may be caused due to the datasets provided for testing in this context not being well represented by a small amount of features in most cases according to these methods’ approach.
In
Figure 5,
Figure 6,
Figure 7,
Figure 8,
Figure 9 and
Figure 10, the most clear trend present across the various plots is the improvement in performance of the PM as the length of the target feature set (combination_len) increases. The same trend seems to be visible in other methods such as EO and WOA, although it is less pronounced and more subtle. Through observation, it seems clear that PM lacks performance in smaller target set lengths; however, it rapidly catches the other methods with respect to performance when allowed to work with greater ones, even competing with most other feature selection methods. The loss in performance between binary and multi-classification tasks should also be pointed out. This decrease in performance is expected as multi-class classification is a distinctly more difficult task when compared to binary classification.
In
Figure 11 and
Figure 12, it is possible to observe the elapsed time distribution for each feature-selection method. It is important to note that due to how these other methods work (i.e., an agent-based architecture that has multiple agents search a solution space), a correction is taken into account regarding the max number of iterations and agents available for each one. This means that if, for example, the BBA method took 20,000 s in each of its tasks, the correct time to be compared to the PM would be as follows: 20,000 s/30 agents/50 iterations. This correction is applied to each data point collected from methods other than the PM. Despite that, as is observable, the PM still seemingly outperforms the other methods in all cases.
Turning to the results pertaining to the two grouped division depicted in
Figure 13,
Figure 14,
Figure 15,
Figure 16,
Figure 17,
Figure 18,
Figure 19 and
Figure 20. The overall conclusion from observing these plots matches those from the previous steps where other approaches were separate. The direct relation between the PM’s performance and the length of the target feature is still apparent. A clear difference is visible between the PM and other solutions; however, the more the target set grows the more both groups approach each other, while maintaining a significant disparity in computational time.
By analyzing the coefficients present in the GLS models represented in these tables, some conclusions can be drawn. Firstly, it is possible to reject the null hypothesis associated with the F-statistic for each GLS model—that is, the hypothesis that the independent variables and their interactions do not affect the dependent variable. This rejection is supported by the very low p-values (all below a significance threshold of , corresponding to a 95% confidence level). It is important to note, however, that the primary objective of these GLS models is not to achieve strong predictive power, but rather to explore and analyze the relationships between the independent variables and the dependent variable. Despite low R2 values, the statistical significance of the models ensures that the identified relationships are meaningful within the context of the analysis. Additionally, any conclusions derived from these models will later be compared to other analysis approaches, such as graphical analysis and two-tailed t-tests, to reinforce the robustness and consistency of the findings.
Focusing on the different dependent variables, some conclusions may be reached. Through comparison of coefficients relative to Group[T.Proposed] for precision, recall, and f-beta, it is clear that PM does not readily match the performance achieved by other approaches. This conclusion is very significant ( = 0.05 ≥p value) for all the cases presented. Counteracting the last point, the effect of the target feature set length (combination length) appears to impact the various dependent variables positively specifically for PM. This conclusion approaches significance for all cases except one, which means it is significant ( = 0.1 ≥ p value). The exception occurs for weighted precision in the case of multi-classification in which the relevant p-value is equal to 0.187, which means the calculated coefficient is only relevant for approximately 80% of cases.
The observations exposed above are largely reiterated when analyzing the results obtained for the
t-tests across the two groups (
Table 10). Taking into account weighted precision, weighted recall, and weighted F-beta, it is possible to conclude that the difference between the two groups, seems to become smaller and less relevant the more the combination length increases. This conclusion is reached since the test statistic (z) for these cases tends to decrease in absolute value, which denotes a smaller difference, and the
p-value on the other hand tends to increase, which allows for a stronger assurance relative to the methods null hypothesis; in this case
, meaning the difference between the two groups becomes less clear.
Concerning the elapsed time, in all cases, it is apparent that the PM is significantly faster than other solutions. This is observable through the coefficient values present for Group[T.Proposed]. This conclusion stands as strongly significant both for binary and multi-class classification, since
= 0.05 ≥
p value, as observed in
Table 8 and
Table 9. Additionally, the target feature set length appears to have little impact in regards to the PM, as may be observed through the confidence values for CombLength:Group[T.Proposed] on those same tables, as well as their
p-values, which do not allow for a reasonable significance level. Another conclusion one may make, although secondary, is that the group composed of other approaches is more affected by an increase in the elapsed time caused by an increase in the target feature set length; this may be interpreted through the difference between the value and the significance seen between the coefficients for CombLength and CombLength:Group[T.Proposed] in both cases.
The same conclusion may be reached through consultation of
Table 10, in which, no matter the combination length or task type involved, there appears to be a clear difference between the two groups. This is denoted by the very low
p-values and the negative values presented for the test statistic (z), which indicate an advantage for the proposed method regarding computational speed. This significant difference may be further observed by checking the confidence intervals involved in this dependent variable’s case.
These conclusions are, as above, consistent with the behavior demonstrated in
Figure 19 and
Figure 20.
Due to computational cost constraints, the method was not evaluated on datasets containing thousands of features. However, an analysis of descriptive statistics from datasets with 90 or more features indicates that the proposed approach remains competitive even at higher dimensionalities. Specifically, the method achieves a mean elapsed time of 10.6 s with a standard deviation of 12 s for this subset. This outperforms the average computational time observed across all datasets for other approaches, which exhibit a mean of 34.5 s and a standard deviation of 45.6 s. Although the average processing time for datasets with over 90 features is higher compared to those with fewer features (4.6 s mean, 6.3 s standard deviation), the method still demonstrates efficient performance, which is expected to stabilize further due to the previously discussed countermeasures.
5. Conclusions
Machine learning has been under a state of constant evolution in recent years—a good part of this may be attributed to great amounts of training data associated with better infrastructure [
1]. However, certain fields and applications do not have this same degree of data availability, be it due to monetary costs, temporal costs, or simple scarcity of data relating to the subject matter [
4]. This makes it especially important to know the most relevant features for a given task, as they have an associated cost. For this purpose, and to improve over use of wrapper or filter-based feature selection techniques, an approach using feature extraction to represent interim combinations of features from the original dataset was proposed.
This approach aims to counter downfalls present in regular feature selection alternatives [
22], namely, temporal cost in the case of wrapper methods, and disregard for feature interaction in the case of filter methods. The method operates by generating interim representations of feature combinations, which are then scored and selected using lightweight filter metrics. This structure is designed to mitigate the primary limitations of classical approaches while remaining adaptable across a variety of contexts.
The computational study demonstrated that the proposed method offers notable improvements in efficiency, particularly when selecting medium-length feature subsets. In these cases, its predictive performance was found to be comparable to that of several state-of-the-art wrapper-based methods, while requiring significantly less time. Statistical analyses using GLS models and Welch’s t-tests reinforced these observations, highlighting consistent trends across dataset types.
Designed with modularity in mind, the method allows for user-defined configurations of feature extractors and filter strategies, making it highly adaptable to different modeling scenarios. These characteristics, combined with its empirical performance, suggest that the method can serve as a practical tool for feature selection in constrained environments.
5.1. Limitations
Despite its strengths, the proposed method is not without limitations. Chief among them is the combinatorial growth in the number of feature groups as the dimensionality of the dataset increases. While mitigated by initial redundant feature filtering and limiting the feature pool (e.g., max_n = 20), this complexity remains a core constraint of the approach.
Another important limitation relates to dataset suitability. The method performs best when the target feature set size is of moderate size and when the dataset has sufficient structure for shallow transformations to capture informative patterns. It may be less effective in scenarios where:
Non-linear or hierarchical interactions dominate the data,
Very large numbers of features ( ) are present without the possibility of effective pre-filtering;
Real-time constraints require ultra-low latency beyond what even this method can accommodate;
The application demands deeply contextual or sequential understanding of features (e.g., natural language or temporal series);
The dataset is naturally very sparce, or there are a very low number of samples (in these cases, approaches such as meta-learning or few-shot learning would be better suited).
Furthermore, the use of default hyper-parameters in the experimental setup, while beneficial for reproducibility, may limit the method’s relative performance under different modeling configurations.
5.2. Future Work
Several promising research directions can be pursued to further refine and expand the proposed method.
First, to address the issue of combinatorial complexity, an iterative pruning strategy could be explored. A potential strategy would be to iteratively create interim feature sets with only one less element than the total available feature set, until reaching the desired length. As an example, the amount of combinations involved in a group of 100 elements for a sample size of 5 is 75,287,520; however, the amount of interim feature sets needed by iteratively eliminating the worse-performing ones is merely 5035. This happens since, in each step, the amount of interim feature sets created is equal to the length of the available feature set. The total amount of interim sets created is equal to the sum of all elements between the maximum feature set length and the target feature length, as may be seen in Equation (
2).
This process largely decreases the amount of interim sets needed, which may contribute to better performance [
69].
Second, the integration of additional feature extraction techniques could further increase the robustness of the proposed method. A systematic comparison of such extractors under different data contexts would help establish optimal configurations for diverse application areas.
Third, the method could include an AutoML pipeline, enabling automated exploration of filter criteria, transformation methods, as well as combination lengths. This would enhance usability for non-expert users and reduce the need for manual tuning.
Lastly, domain-specific applications represent a compelling area for deployment. The proposed algorithm encompasses several potential use cases for its application. One of these use cases is in bioinformatics, where datasets often contain a vast number of features, such as gene expression levels. Feature selection often leads to better results in such cases as is presented by Kasikci in [
137]. A different application of this method that may make use of its biggest advantage, its computational efficiency, is in settings where a faster (but not immediate) decision is needed, such as in medical diagnostics, and similarly to the previous case, medicine is no stranger to the use of feature selection be it due to excessive dimension or costs [
4].