Novel Probiotic Strain Lactiplantibacillus plantarum CNTA 628 Modulates Lipid Metabolism and Improves Healthspan in C. elegans
Abstract
1. Introduction
2. Materials and Methods
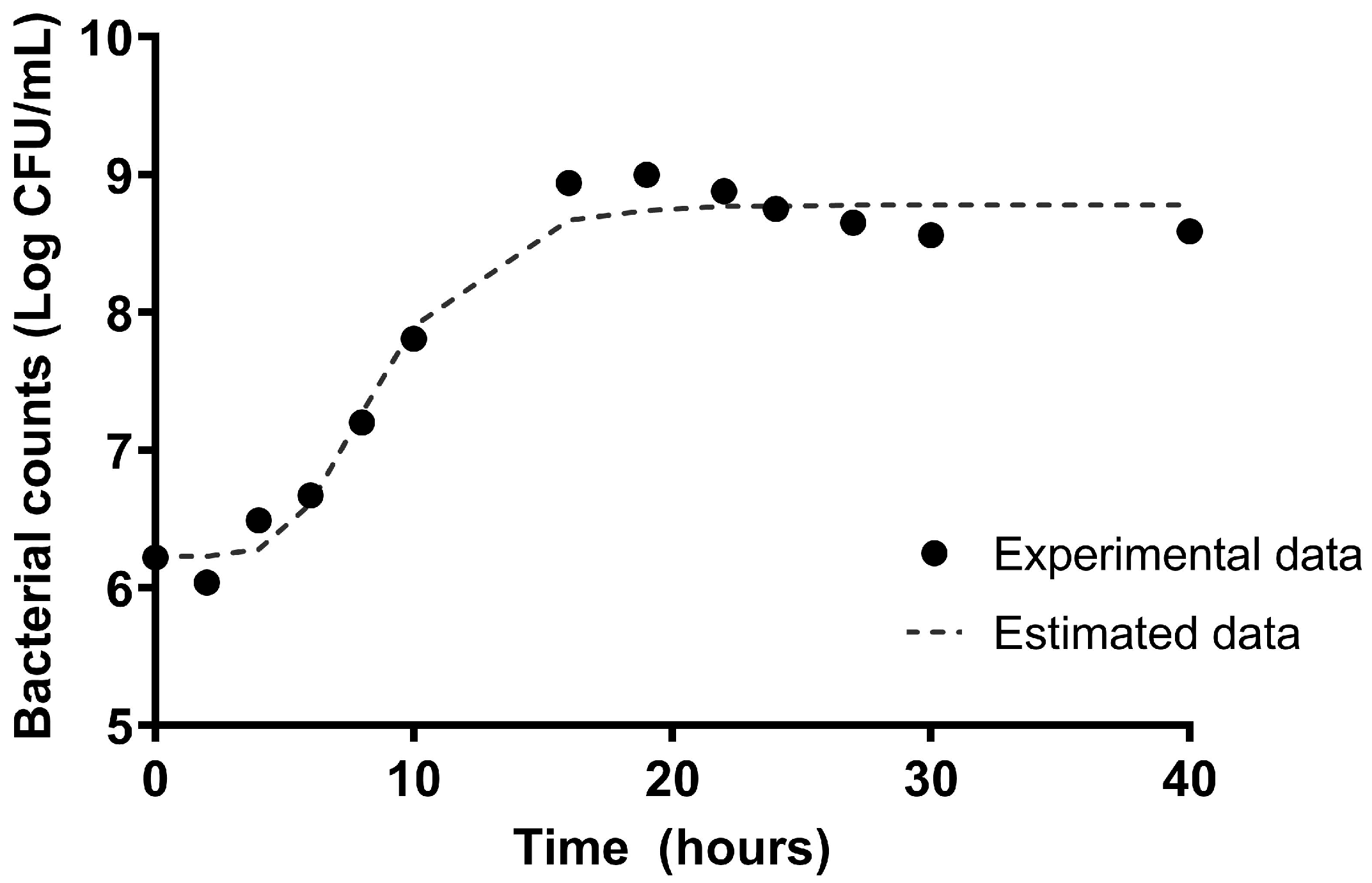
2.1. The Kinetic Characterization of the Microbial Growth of L. plantarum CNTA 628
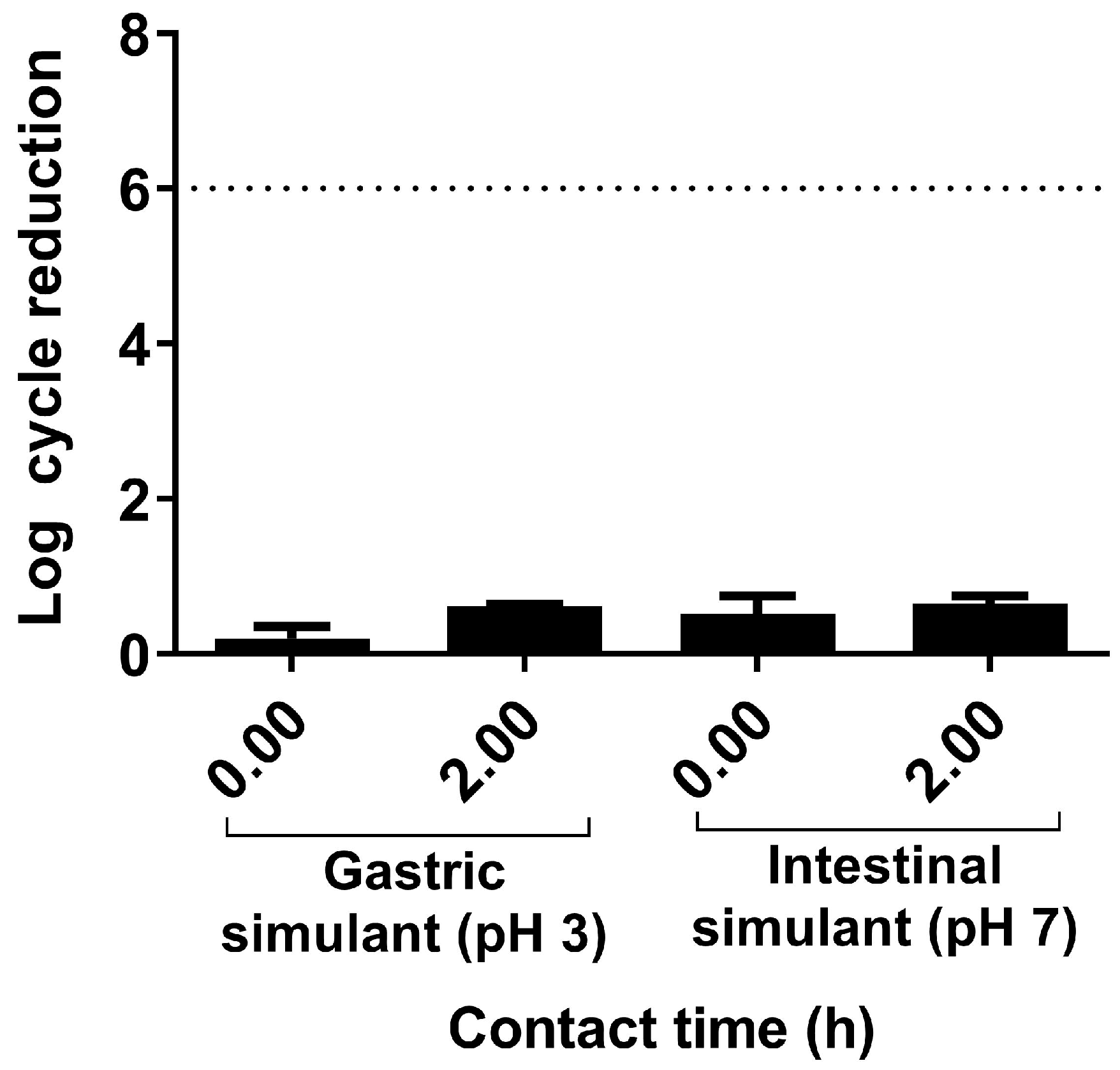
2.2. Resistance to Gastrointestinal Tract Conditions
2.3. Phenotypic Analysis of Antibiotic Resistance
2.4. Potencial Probiotics’capacity to Generate Short-Chain Fatty Acids (SCFAs)
2.5. Assessment of β-Galactosidase and Bile Salt Hydrolase (BSH) Activities
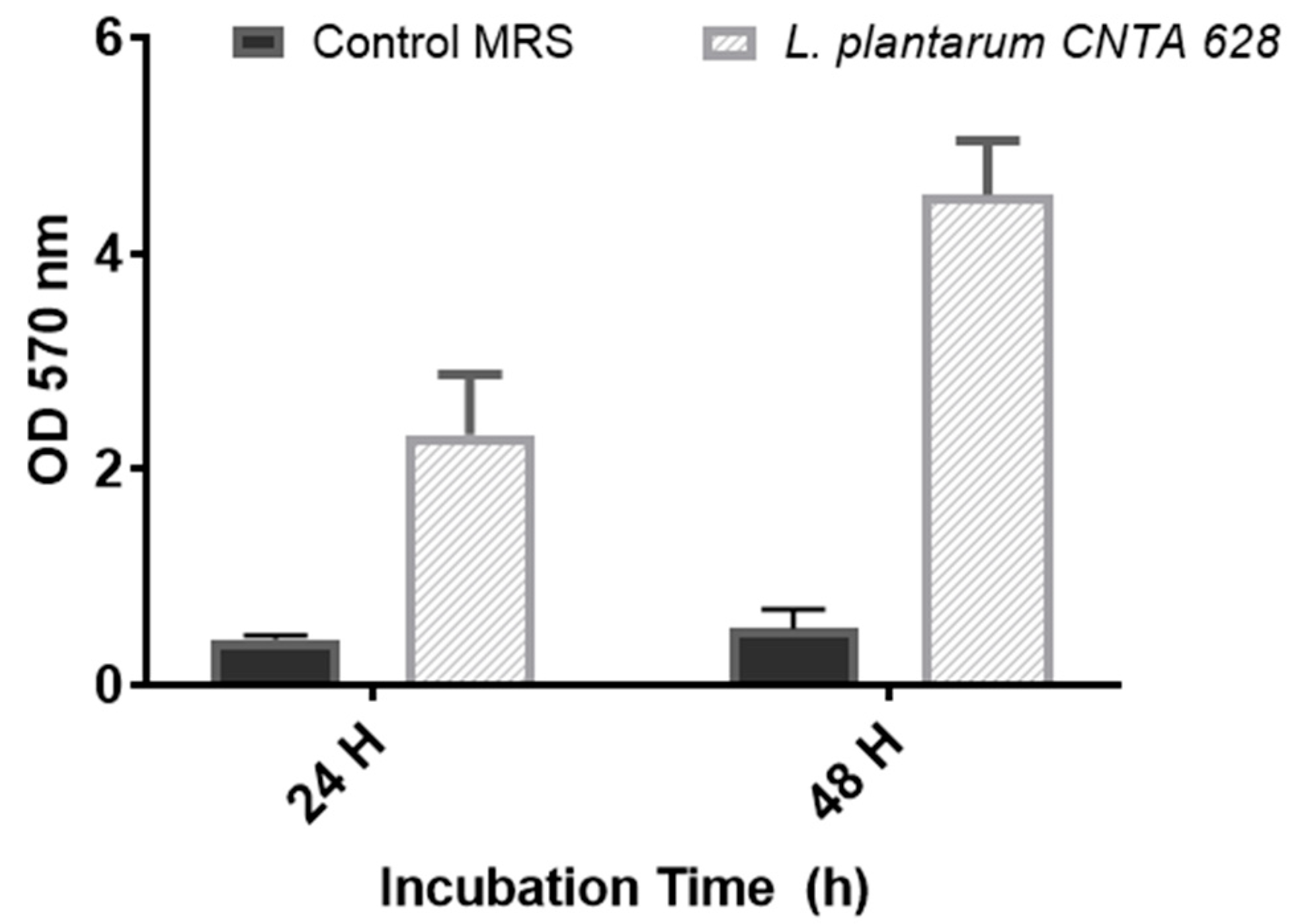
2.6. Capability to Generate a Biofilm
2.7. Adhesion, Exclusion and Competence Assays on Caco-2 Cells
2.8. Bacterial Culture, Cell Culture and Maintenance
2.9. Neutral Red Assays and Cytokine Quantification
2.10. Functional Evaluation of L. plantarum CNTA 628 in C. elegans
2.10.1. C. elegans Culture and Experimental Design
2.10.2. The Quantification of the Fat Accumulation, ROS Levels and Senescence in C. elegans
2.10.3. Lifespan and Egg-Laying Assays in C. elegans
2.10.4. RNA Extraction and qPCR Analyses
2.11. Statistical Analyses
3. Results
3.1. In Vitro Assessment of Probiotic Characteristics of L. plantarum CNTA 628
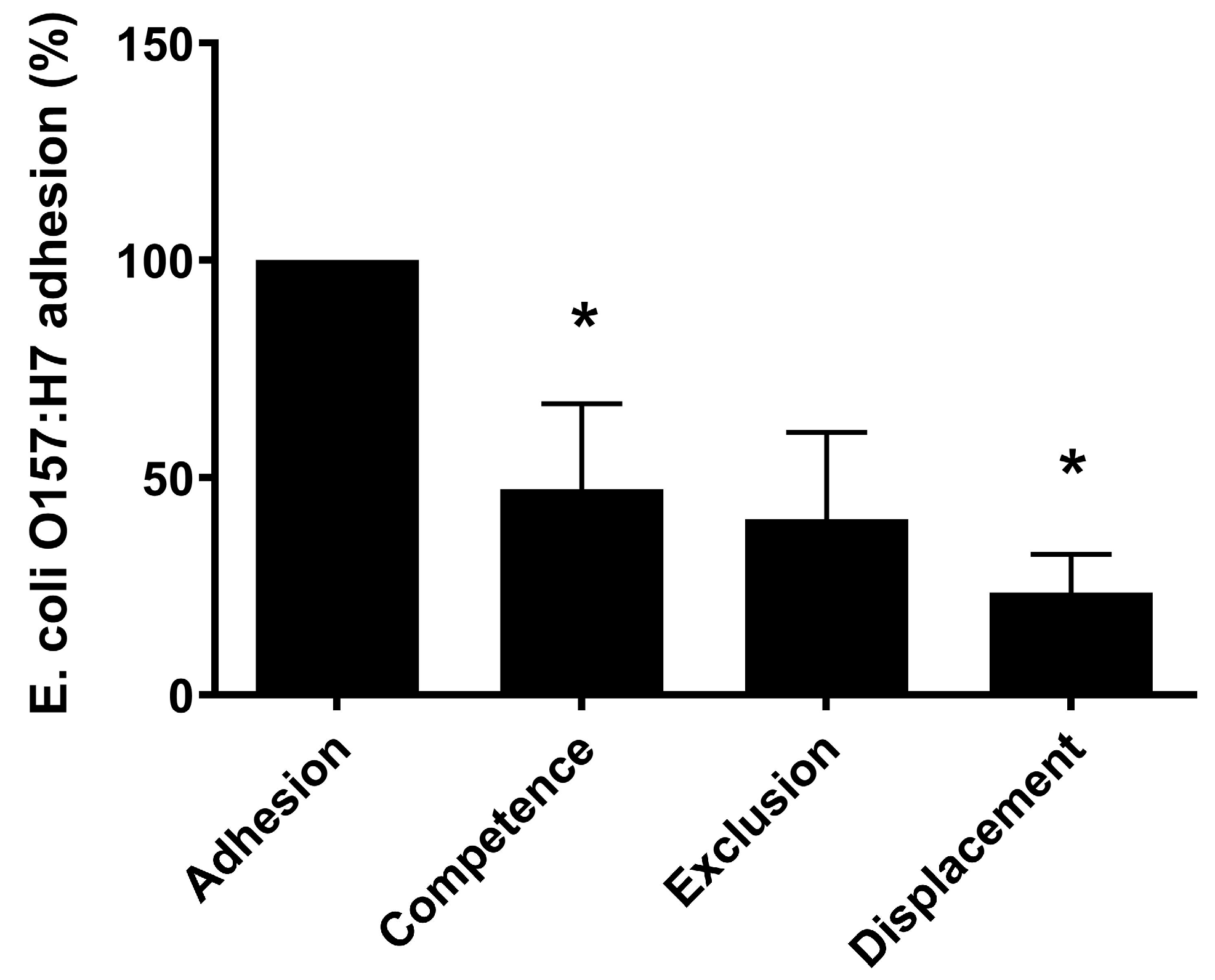
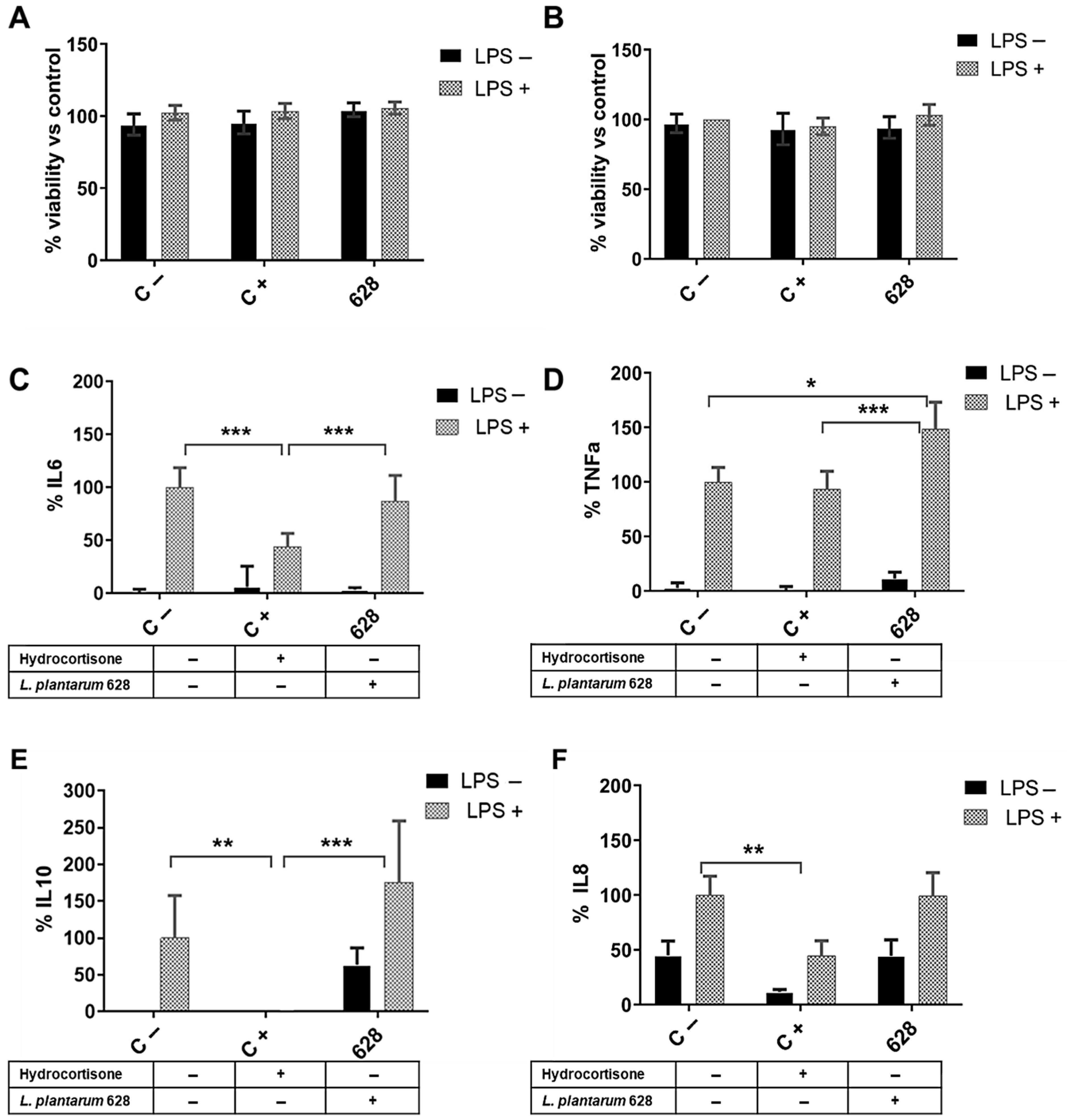
3.2. L. plantarum CNTA 628 Competes with Pathogenic Bacteria and Exhibits Immunomodulatory Capacity In Vitro
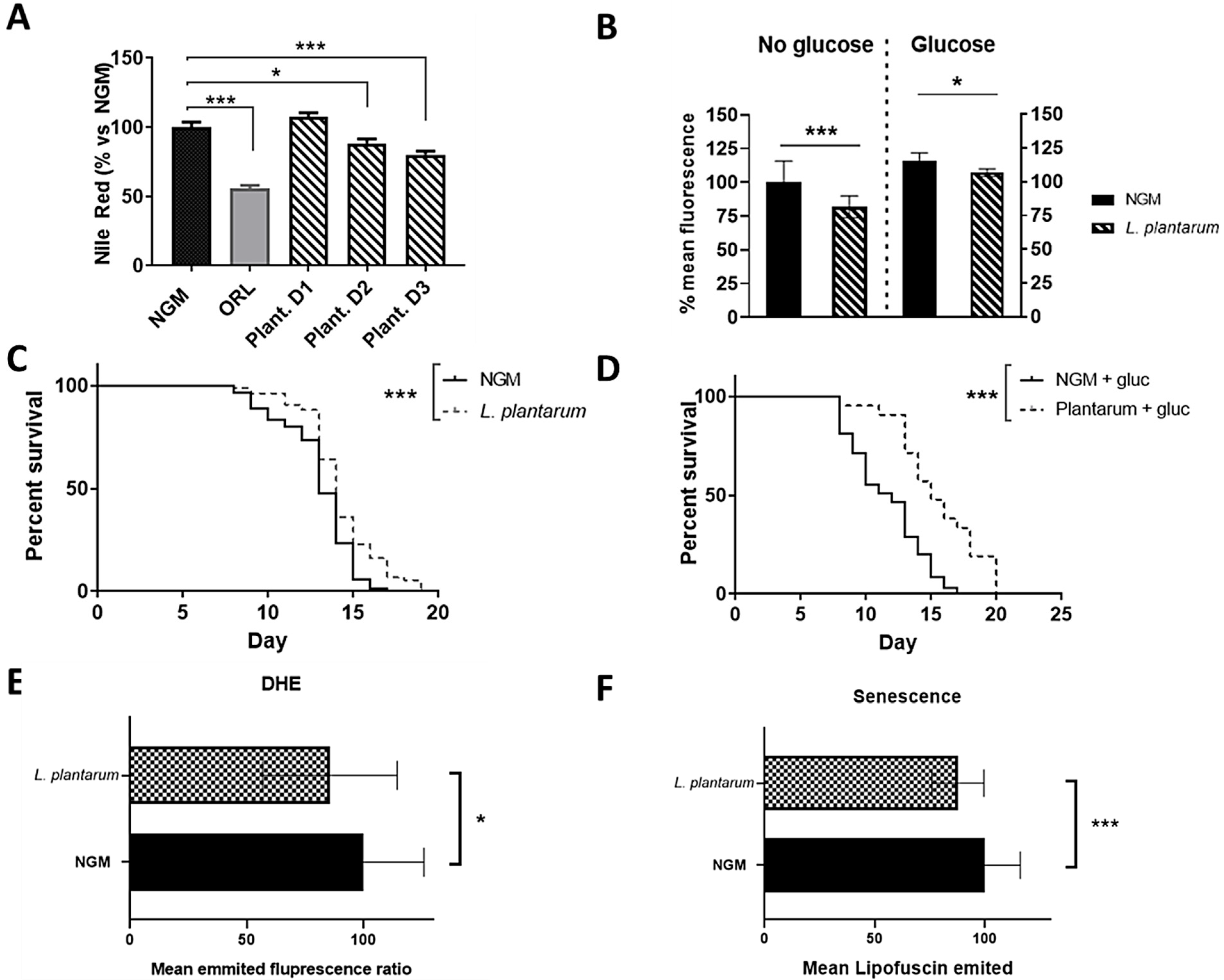
3.3. L. plantarum CNTA 628 Lowers Lipid Storage and Reduces Oxidative Stress and Senescence in C. elegans, Contributing to Lifespan Extension
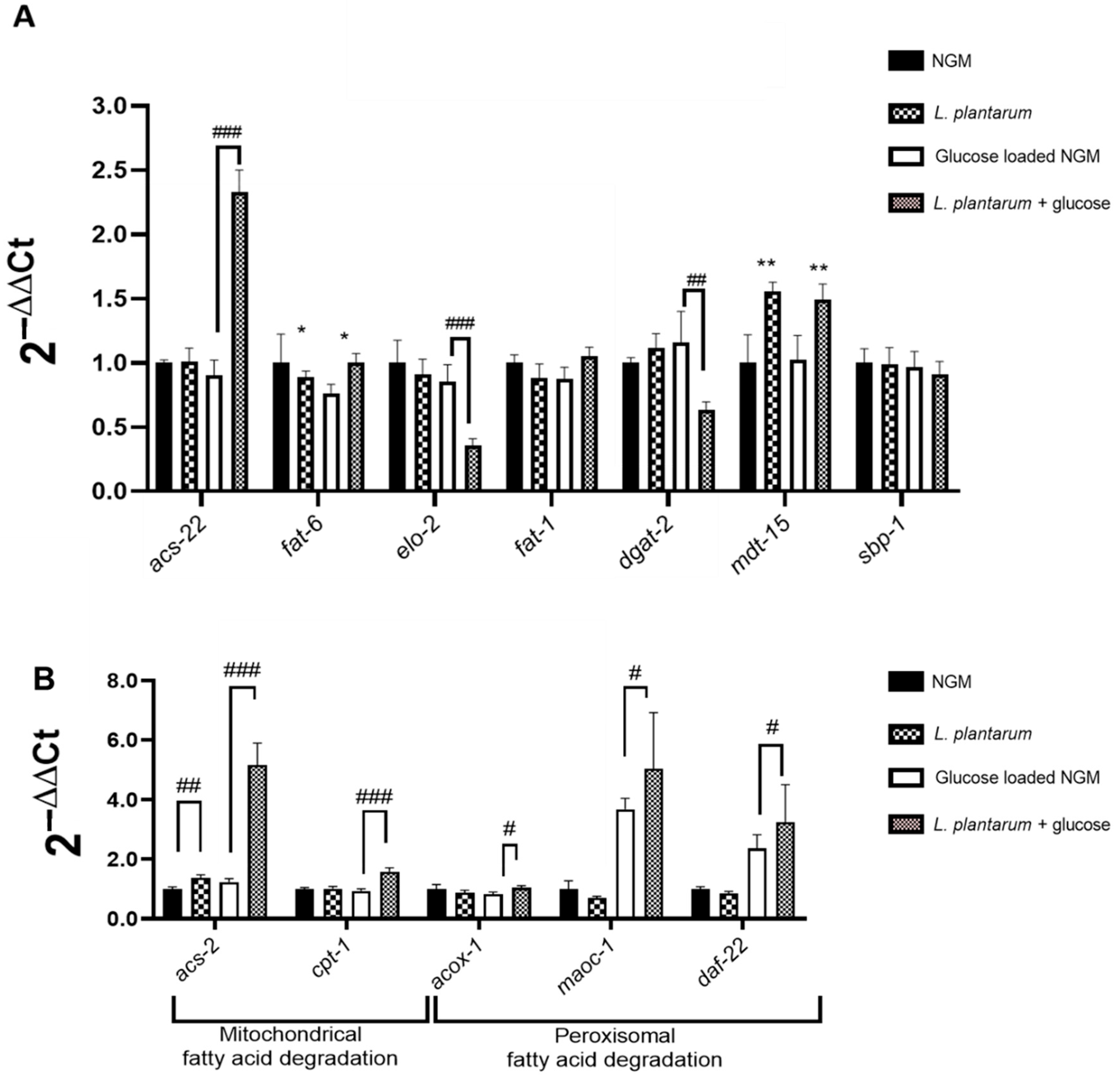
3.4. L. plantarum CNTA 628 Influences the Transcription of Essential Metabolic Genes
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NGM | Nematode Growth Medium |
| NGMg | Nematode Growth Medium with Glucose (10 mM) |
| CFU | Colony-Forming Unit |
| DHE | Dihydroethidium |
References
- Gomes, A.C.; Hoffmann, C.; Mota, J.F. The Human Gut Microbiota: Metabolism and Perspective in Obesity. Gut Microbes 2018, 9, 308. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Gou, Y.K.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The Gut Microbiota as an Environmental Factor That Regulates Fat Storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Castaner, O.; Goday, A.; Park, Y.M.; Lee, S.H.; Magkos, F.; Shiow, S.A.T.E.; Schröder, H. The Gut Microbiome Profile in Obesity: A Systematic Review. Int. J. Endocrinol. 2018, 2018, 4095789. [Google Scholar] [CrossRef] [PubMed]
- Gérard, P. Gut Microbiota and Obesity. Cell. Mol. Life Sci. 2015, 73, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Sun, B.; Yu, D.; Zhu, C. Gut Microbiota: An Important Player in Type 2 Diabetes Mellitus. Front. Cell. Infect. Microbiol. 2022, 12, 834485. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, B.; Delgado, S.; Blanco-Míguez, A.; Lourenço, A.; Gueimonde, M.; Margolles, A. Probiotics, Gut Microbiota, and Their Influence on Host Health and Disease. Mol. Nutr. Food Res. 2017, 61, 1600240. [Google Scholar] [CrossRef] [PubMed]
- Abenavoli, L.; Scarpellini, E.; Colica, C.; Boccuto, L.; Salehi, B.; Sharifi-Rad, J.; Aiello, V.; Romano, B.; De Lorenzo, A.; Izzo, A.A.; et al. Gut Microbiota and Obesity: A Role for Probiotics. Nutrients 2019, 11, 2690. [Google Scholar] [CrossRef] [PubMed]
- Million, M.; Angelakis, E.; Paul, M.; Armougom, F.; Leibovici, L.; Raoult, D. Comparative Meta-Analysis of the Effect of Lactobacillus Species on Weight Gain in Humans and Animals. Microb. Pathog. 2012, 53, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Zare, D.; Aryaee, H.; Mirdamadi, S.; Shirkhan, F. The Benefits and Applications of Lactobacillus plantarum in Food and Health: A Narrative Review. Iran. J. Public Health 2024, 53, 2201. [Google Scholar] [CrossRef] [PubMed]
- Behera, S.S.; Ray, R.C.; Zdolec, N. Lactobacillus plantarum with Functional Properties: An Approach to Increase Safety and Shelf-Life of Fermented Foods. BioMed Res. Int. 2018, 2018, 9361614. [Google Scholar] [CrossRef] [PubMed]
- Micioni Di Bonaventura, M.V.; Coman, M.M.; Tomassoni, D.; Micioni Di Bonaventura, E.; Botticelli, L.; Gabrielli, M.G.; Rossolini, G.M.; Di Pilato, V.; Cecchini, C.; Amedei, A.; et al. Supplementation with Lactiplantibacillus plantarum IMC 510 Modifies Microbiota Composition and Prevents Body Weight Gain Induced by Cafeteria Diet in Rats. Int. J. Mol. Sci. 2021, 22, 11171. [Google Scholar] [CrossRef] [PubMed]
- Jacouton, E.; Mondot, S.; Langella, P.; Bermúdez-Humarán, L.G. Impact of Oral Administration of Lactiplantibacillus plantarum Strain CNCM I−4459 on Obesity Induced by High-Fat Diet in Mice. Bioengineering 2023, 10, 1151. [Google Scholar] [CrossRef] [PubMed]
- Yoshitake, R.; Hirose, Y.; Murosaki, S.; Matsuzaki, G. Heat-Killed Lactobacillus plantarum L-137 Attenuates Obesity and Associated Metabolic Abnormalities in C57BL/6 J Mice on a High-Fat Diet. Biosci. Microbiota Food Health 2021, 40, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Fei, Y.; Han, X.; Liu, G.; Fang, J. Lactobacillus plantarum Alleviates Obesity by Altering the Composition of the Gut Microbiota in High-Fat Diet-Fed Mice. Front. Nutr. 2022, 9, 947367. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar, V.; Pramanik, J.; Rustagi, S.; Prajapati, B.; Jebreen, A.; Pande, R. Lactiplantibacillus plantarum as a Complementary Approach for Diabetes Treatment and Management. Curr. Nutr. Rep. 2025, 14, 72. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Joishy, T.; Das, S.; Kalita, M.C.; Mukherjee, A.K.; Khan, M.R. A Potential Probiotic Lactobacillus plantarum JBC5 Improves Longevity and Healthy Aging by Modulating Antioxidative, Innate Immunity and Serotonin-Signaling Pathways in Caenorhabditis elegans. Antioxidants 2022, 11, 268. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Liu, J.M.; Yang, F.E.; Ji, X.M.; Li, C.Y.; Lv, S.W.; Wang, S. A Novel Mediation Strategy of DSS-Induced Colitis in Mice Based on an Iron-Enriched Probiotic and in Vivo Bioluminescence Tracing. J. Agric. Food Chem. 2020, 68, 12028–12038. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhao, H.; Qi, X.; Sun, Y.; Ma, Y.; Li, Q. Lactobacillus plantarum HF02 Alleviates Lipid Accumulation and Intestinal Microbiota Dysbiosis in High-Fat Diet-Induced Obese Mice. J. Sci. Food Agric. 2023, 103, 4625–4637. [Google Scholar] [CrossRef] [PubMed]
- Li, C.P.; Chen, C.C.; Hsiao, Y.; Kao, C.H.; Chen, C.C.; Yang, H.J.; Tsai, R.Y. The Role of Lactobacillus plantarum in Reducing Obesity and Inflammation: A Meta-Analysis. Int. J. Mol. Sci. 2024, 25, 7608. [Google Scholar] [CrossRef] [PubMed]
- Palacios, T.; Vitetta, L.; Coulson, S.; Madigan, C.D.; Lam, Y.Y.; Manuel, R.; Briskey, D.; Hendy, C.; Kim, J.N.; Ishoey, T.; et al. Targeting the Intestinal Microbiota to Prevent Type 2 Diabetes and Enhance the Effect of Metformin on Glycaemia: A Randomised Controlled Pilot Study. Nutrients 2020, 12, 2041. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Gil-Campos, M.; Gil, A. Mechanisms of Action of Probiotics. Adv. Nutr. 2019, 10, S49–S66. [Google Scholar] [CrossRef] [PubMed]
- Goyache, I.; Yavorov-Dayliev, D.; Milagro, F.I.; Aranaz, P. Caenorhabditis elegans as a Screening Model for Probiotics with Properties against Metabolic Syndrome. Int. J. Mol. Sci. 2024, 25, 1321. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, Y.R.; Yang, H.; Park, C.H.; Yun, C.S.; Jang, B.C.; Hong, Y.; Park, D.S. Potential Probiotic Lactiplantibacillus plantarum DS1800 Extends Lifespan and Enhances Stress Resistance in Caenorhabditis elegans Model. Front. Physiol. 2024, 15, 1476096. [Google Scholar] [CrossRef] [PubMed]
- Marquez, A.; Andrada, E.; Russo, M.; Bolondi, M.L.; Fabersani, E.; Medina, R.; Gauffin-Cano, P. Characterization of Autochthonous Lactobacilli from Goat Dairy Products with Probiotic Potential for Metabolic Diseases. Heliyon 2022, 8, e10462. [Google Scholar] [CrossRef] [PubMed]
- Tjørve, K.M.C.; Tjørve, E. The Use of Gompertz Models in Growth Analyses, and New Gompertz-Model Approach: An Addition to the Unified-Richards Family. PLoS ONE 2017, 12, e0178691. [Google Scholar] [CrossRef] [PubMed]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST Static in Vitro Simulation of Gastrointestinal Food Digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef] [PubMed]
- Goyache, I.; Valdés-Varela, L.; Virto, R.; López-Yoldi, M.; López-Giral, N.; Sánchez-Vicente, A.; Milagro, F.I.; Aranaz, P. Anti-Obesity Properties of a Novel Probiotic Strain of Latilactobacillus sakei CNTA 173 in Caenorhabditis elegans. Int. J. Mol. Sci. 2025, 26, 3286. [Google Scholar] [CrossRef] [PubMed]
- ISO 10932:2010; Milk and Milk Products—Determination of the Minimal Inhibitory Concentration (MIC) of Antibiotics Applicable to Bifidobacteria and Non-Enterococcal Lactic Acid Bacteria (LAB). International Organization for Standardization: Geneva, Switzerland, 2010. Available online: https://www.iso.org/standard/46434.html (accessed on 21 January 2025).
- Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; de Lourdes Bastos, M.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; Gropp, J.; et al. Guidance on the Characterisation of Microorganisms Used as Feed Additives or as Production Organisms. EFSA J. 2018, 16, e05206. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Varela, L.; Hernández-Barranco, A.M.; Ruas-Madiedo, P.; Gueimonde, M. Effect of Bifidobacterium upon Clostridium difficile Growth and Toxicity When Co-Cultured in Different Prebiotic Substrates. Front. Microbiol. 2016, 7, 738. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, K.M.O.; Vieira, A.D.S.; Buriti, F.C.A.; do Nascimento, J.C.F.; de Melo, M.E.S.; Bruno, L.M.; de Fátima Borges, M.; Rocha, C.R.C.; de Souza Lopes, A.C.; de Melo Franco, B.D.G.; et al. Artisanal Coalho Cheeses as Source of Beneficial Lactobacillus plantarum and Lactobacillus rhamnosus Strains. Dairy Sci. Technol. 2015, 95, 209–230. [Google Scholar] [CrossRef]
- Dashkevicz, M.P.; Feighner, S.D. Development of a Differential Medium for Bile Salt Hydrolase-Active Lactobacillus spp. Appl. Environ. Microbiol. 1989, 55, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Kubota, H.; Senda, S.; Nomura, N.; Tokuda, H.; Uchiyama, H. Biofilm Formation by Lactic Acid Bacteria and Resistance to Environmental Stress. J. Biosci. Bioeng. 2008, 106, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, M.; Kheadr, E.E.; le Blay, G.; Fliss, I. In Vitro Inhibition of Escherichia coli O157:H7 by Bifidobacterial Strains of Human Origin. Int. J. Food Microbiol. 2004, 92, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Repetto, G.; del Peso, A.; Zurita, J.L. Neutral Red Uptake Assay for the Estimation of Cell Viability/Cytotoxicity. Nat. Protoc. 2008, 3, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Yavorov-Dayliev, D.; Milagro, F.I.; Ayo, J.; Oneca, M.; Aranaz, P. Pediococcus acidilactici CECT9879 (PA1c) Counteracts the Effect of a High-Glucose Exposure in C. elegans by Affecting the Insulin Signaling Pathway (IIS). Int. J. Mol. Sci. 2022, 23, 2689. [Google Scholar] [CrossRef] [PubMed]
- Escorcia, W.; Ruter, D.L.; Nhan, J.; Curran, S.P. Quantification of Lipid Abundance and Evaluation of Lipid Distribution in Caenorhabditis elegans by Nile Red and Oil Red O Staining. J. Vis. Exp. 2018, 2018, 57352. [Google Scholar] [CrossRef]
- Navarro-Herrera, D.; Aranaz, P.; Eder-Azanza, L.; Zabala, M.; Hurtado, C.; Romo-Hualde, A.; Martínez, J.A.; González-Navarro, C.J.; Vizmanos, J.L. Dihomo-Gamma-Linolenic Acid Induces Fat Loss in C. elegans in an Omega-3-Independent Manner by Promoting Peroxisomal Fatty Acid β-Oxidation. Food Funct. 2018, 9, 1621–1637. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Taki, F.A.; Zhang, Y.; Dobbins, D.L.; Pan, X. Evaluation and Identification of Reliable Reference Genes for Toxicological Study in Caenorhabditis elegans. Mol. Biol. Rep. 2014, 41, 3445–3455. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Oudat, Q.; Okour, A. The Role of Probiotics in Modulating Gut Microbiota and Metabolic Health for Weight Management: A Mini Review. Acta Microbiol. Hell. 2025, 70, 5. [Google Scholar] [CrossRef]
- da Silva, M.N.; Tagliapietra, B.L.; Flores, V.D.A.; Richards, N.S.P.d.S. In Vitro Test to Evaluate Survival in the Gastrointestinal Tract of Commercial Probiotics. Curr. Res. Food Sci. 2021, 4, 320. [Google Scholar] [CrossRef] [PubMed]
- Fara, A.; Sabater, C.; Palacios, J.; Requena, T.; Montilla, A.; Zárate, G. Prebiotic Galactooligosaccharides Production from Lactose and Lactulose by: Lactobacillus delbrueckii subsp. bulgaricus CRL450. Food Funct. 2020, 11, 5875–5886. [Google Scholar] [CrossRef] [PubMed]
- Caetano, M.A.F.; Castelucci, P. Role of Short Chain Fatty Acids in Gut Health and Possible Therapeutic Approaches in Inflammatory Bowel Diseases. World J. Clin. Cases 2022, 10, 9985. [Google Scholar] [CrossRef] [PubMed]
- Salas-Jara, M.J.; Ilabaca, A.; Vega, M.; García, A. Biofilm Forming Lactobacillus: New Challenges for the Development of Probiotics. Microorganisms 2016, 4, 35. [Google Scholar] [CrossRef] [PubMed]
- Vuotto, C.; Longo, F.; Donelli, G. Probiotics to Counteract Biofilm-Associated Infections: Promising and Conflicting Data. Int. J. Oral Sci. 2014, 6, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Geng, W.; Long, S.L.; Chang, Y.J.; Saxton, A.M.; Joyce, S.A.; Lin, J. Evaluation of Bile Salt Hydrolase Inhibitor Efficacy for Modulating Host Bile Profile and Physiology Using a Chicken Model System. Sci. Rep. 2020, 10, 4941. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Nakase, H. The Multifaceted Effects of Gut Microbiota on the Immune System of the Intestinal Mucosa. Immuno 2021, 1, 583–594. [Google Scholar] [CrossRef]
- Kostic, A.D.; Xavier, R.J.; Gevers, D. The Microbiome in Inflammatory Bowel Disease: Current Status and the Future Ahead. Gastroenterology 2014, 146, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Pinto, M.; Robine-Leon, S.; Appay, M.-D.; Kedinger, M.; Triadou, N.; Dussaulx, E.; Lacroix, B.; Simon-Assmann, P.; Haffen, K.; Fogh, J.; et al. Enterocyte-like Differentiation and Polarization of the Human Colon Carcinoma Cell Line Caco-2 in Culture. Biol. Cell 1983, 47, 323–330. [Google Scholar] [CrossRef]
- Patel, D.K.; Shah, K.R.; Pappachan, A.; Gupta, S.; Singh, D.D. Cloning, Expression and Characterization of a Mucin-Binding GAPDH from Lactobacillus acidophilus. Int. J. Biol. Macromol. 2016, 91, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Celebioglu, H.U.; Svensson, B. Dietary Nutrients, Proteomes, and Adhesion of Probiotic Lactobacilli to Mucin and Host Epithelial Cells. Microorganisms 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Chandhni, P.R.; Pradhan, D.; Sowmya, K.; Gupta, S.; Kadyan, S.; Choudhary, R.; Gupta, A.; Gulati, G.; Mallappa, R.H.; Kaushik, J.K.; et al. Ameliorative Effect of Surface Proteins of Probiotic Lactobacilli in Colitis Mouse Models. Front. Microbiol. 2021, 12, 679773. [Google Scholar] [CrossRef] [PubMed]
- Man, L.L.; Xiang, D.J. Characterization of a Broad Spectrum Bacteriocin Produced by Lactobacillus plantarum MXG-68 from Inner Mongolia Traditional Fermented Koumiss. Folia Microbiol. 2019, 64, 821–834. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, Y.; Nishizaki, Y.; Aida, K.; Yayama, K.; Ebisui, T.; Akiyama, T.; Nakamura, T. Lactobacillus plantarum Induces Innate Cytokine Responses That Potentially Provide a Protective Benefit against COVID-19: A Single-Arm, Double-Blind, Prospective Trial Combined with an in Vitro Cytokine Response Assay. Exp. Ther. Med. 2022, 23, 20. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Arce, L.; Tomotsune, K.; Albarracin, L.; Funabashi, R.; Vera, D.; Islam, M.A.; Vizoso-Pinto, M.G.; Takahashi, H.; Sasaki, Y.; et al. Lipoteichoic Acid Is Involved in the Ability of the Immunobiotic Strain Lactobacillus plantarum CRL1506 to Modulate the Intestinal Antiviral Innate Immunity Triggered by TLR3 Activation. Front. Immunol. 2020, 11, 571. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.J.; Paik, H.D.; Kim, J.Y. Immune-Enhancing Effects of Lactobacillus plantarum 200655 Isolated from Korean Kimchi in a Cyclophosphamide-Induced Immunocompromised Mouse Model. J. Microbiol. Biotechnol. 2021, 31, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Shen, Y.; Wang, Y.; Wang, L.; Zhang, L.; Zhao, Z.; Li, S. Lactobacillus plantarum S9 Alleviates Lipid Profile, Insulin Resistance, and Inflammation in High-Fat Diet-Induced Metabolic Syndrome Rats. Sci. Rep. 2022, 12, 15490. [Google Scholar] [CrossRef] [PubMed]
- Gallego, M.B.; Mazo, J.E.; Sánchez, M.M.; Carre, M.P.; Codina, A.F.; Brugué, S.A.; Sierra, M.A.B.; Castellana, J.C. Consumption of the Probiotic Lactobacillus planctarum CECT 7315/7316 Improves General Health in the Elderly Subjects. Nutr. Hosp. 2011, 26, 642–645. [Google Scholar]
- Lee, Y.D.; Hong, Y.F.; Jeon, B.; Jung, B.J.; Chung, D.K.; Kim, H. Differential Cytokine Regulatory Effect of Three Lactobacillus Strains Isolated from Fermented Foods. J. Microbiol. Biotechnol. 2016, 26, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Puertollano, E.; Puertollano, M.A.; Cruz-Chamorro, L.; de Cienfuegos, G.Á.; Ruiz-Bravo, A.; de Pablo, M.A. Orally Administered Lactobacillus plantarum Reduces Pro-Inflammatory Interleukin Secretion in Sera from Listeria Monocytogenes Infected Mice. Br. J. Nutr. 2008, 99, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.S.; Yang, H.R.; Chang, J.Y.; Seo, J.K. Lactobacillus plantarum Inhibits Epithelial Barrier Dysfunction and Interlukin-8 Secretion Induced by Tumor Necrosis Factor-α. World J. Gastroenterol. 2007, 13, 1962–1965. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Yue, Y.; Park, Y. A Living Model for Obesity and Aging Research: Caenorhabditis elegans. Crit. Rev. Food Sci. Nutr. 2018, 58, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.L. Fat Synthesis and Adiposity Regulation in Caenorhabditis elegans. Trends Endocrinol. Metab. 2009, 20, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.L.; Ristow, M. Lipid and Carbohydrate Metabolism in Caenorhabditis elegans. Genetics 2017, 207, 413–446. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.A.; Watts, J.L. Worms, Fat, and Death Caenorhabditis elegans Lipid Metabolites Regulate Cell Death. Metabolites 2021, 11, 125. [Google Scholar] [CrossRef] [PubMed]
- Franco-Juárez, B.; Gómez-Manzo, S.; Hernández-Ochoa, B.; Cárdenas-Rodríguez, N.; Arreguin-Espinosa, R.; de la Cruz, V.P.; Ortega-Cuellar, D. Effects of High Dietary Carbohydrate and Lipid Intake on the Lifespan of C. elegans. Cells 2021, 10, 2359. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Yue, Y.; Zheng, J.; Park, Y. Caenorhabditis elegans: A Convenient In Vivo Model for Assessing the Impact of Food Bioactive Compounds on Obesity, Aging, and Alzheimer’s Disease. Annu. Rev. Food Sci. Technol. 2018, 9, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Park, M.R.; Yun, H.S.; Son, S.J.; Oh, S.; Kim, Y. Short Communication: Development of a Direct in Vivo Screening Model to Identify Potential Probiotic Bacteria Using Caenorhabditis elegans. J. Dairy Sci. 2014, 97, 6828–6834. [Google Scholar] [CrossRef] [PubMed]
- Martorell, P.; Llopis, S.; González, N.; Chenoll, E.; López-Carreras, N.; Aleixandre, A.; Chen, Y.; Karoly, E.D.; Ramón, D.; Genovés, S. Probiotic Strain Bifidobacterium animalis subsp. lactis CECT 8145 Reduces Fat Content and Modulates Lipid Metabolism and Antioxidant Response in Caenorhabditis elegans. J. Agric. Food Chem. 2016, 64, 3462–3472. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Werlinger, P.; Cho, J.H.; Jang, N.; Choi, S.S.; Suh, J.W.; Cheng, J. Lactobacillus pentosus MJM60383 Inhibits Lipid Accumulation in Caenorhabditis elegans Induced by Enterobacter Cloacae and Glucose. Int. J. Mol. Sci. 2022, 24, 280. [Google Scholar] [CrossRef] [PubMed]
- Barathikannan, K.; Chelliah, R.; Elahi, F.; Tyagi, A.; Selvakumar, V.; Agastian, P.; Arasu, M.V.; Oh, D.H. Anti-Obesity Efficacy of Pediococcus acidilactici MNL5 in Caenorhabditis elegans Gut Model. Int. J. Mol. Sci. 2022, 23, 1276. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Murphy, C.T.; Kenyon, C. Glucose Shortens the Life Span of C. elegans by Downregulating DAF-16/FOXO Activity and Aquaporin Gene Expression. Cell Metab. 2009, 10, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Praneet, N.S.; Suchiang, K. Lactobacillus brevis MTCC 1750 Enhances Oxidative Stress Resistance and Lifespan Extension with Improved Physiological and Functional Capacity in Caenorhabditis elegans via the DAF-16 Pathway. Free Radic. Res. 2022, 56, 555–571. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Torres, I.; Castrejón-Téllez, V.; Soto, M.E.; Rubio-Ruiz, M.E.; Manzano-Pech, L.; Guarner-Lans, V. Oxidative Stress, Plant Natural Antioxidants, and Obesity. Int. J. Mol. Sci. 2021, 22, 1786. [Google Scholar] [CrossRef] [PubMed]
- Pompa, L.; Montanari, A.; Tomassini, A.; Bianchi, M.M.; Aureli, W.; Miccheli, A.; Uccelletti, D.; Schifano, E. In Vitro Probiotic Properties and In Vivo Anti-Ageing Effects of Lactoplantibacillus plantarum PFA2018AU Strain Isolated from Carrots on Caenorhabditis elegans. Microorganisms 2023, 11, 1087. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Fu, X.; Chen, J.; Ma, J. Application of Caenorhabditis elegans in Lipid Metabolism Research. Int. J. Mol. Sci. 2023, 24, 1173. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Zhang, S.O.; Cole, R.A.; McKinney, S.A.; Guo, F.; Haas, J.T.; Bobba, S.; Farese, R.V.; Mak, H.Y. The FATP1-DGAT2 Complex Facilitates Lipid Droplet Expansion at the ER-Lipid Droplet Interface. J. Cell Biol. 2012, 198, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.; Xu, K.; Li, Y.; Wong, G.; Wang, D. Using Acs-22 Mutant Caenorhabditis elegans to Detect the Toxicity of Nanopolystyrene Particles. Sci. Total Environ. 2018, 643, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Brandstädt, S.; Schmeisser, K.; Zarse, K.; Ristow, M. Lipid-Lowering Fibrates Extend C. elegans Lifespan in a NHR-49/PPARalpha-Dependent Manner. Aging 2013, 5, 270–275. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hu, Q.; D’Amora, D.R.; MacNeil, L.T.; Walhout, A.J.M.; Kubiseski, T.J. The Caenorhabditis elegans Oxidative Stress Response Requires the NHR-49 Transcription Factor. G3 Genes Genomes Genet. 2018, 8, 3857–3863. [Google Scholar] [CrossRef] [PubMed]
- Gissendanner, C.R.; Crossgrove, K.; Kraus, K.A.; Maina, C.V.; Sluder, A.E. Expression and Function of Conserved Nuclear Receptor Genes in Caenorhabditis elegans. Dev. Biol. 2004, 266, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Van Gilst, M.R.; Hadjivassiliou, H.; Yamamoto, K.R. A Caenorhabditis elegans Nutrient Response System Partially Dependent on Nuclear Receptor NHR-49. Proc. Natl. Acad. Sci. USA 2005, 102, 13496–13501. [Google Scholar] [CrossRef] [PubMed]
| Antibiotic MIC Values (mg/L) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gm | Km | Sm | Tc | Cl | Cm | Am | Em | |
| L. plantarum CNTA 628 | 0.5 | 2–16 | - | 32 | 0.063–1 | 2–4 | 0.5–1 | 0.032–0.5 |
| Medium | Acetic Acid |
|---|---|
| Defined medium without adding carbon source | 523.15 ± 54.57 |
| Defined medium with glucose (2%) | not detected |
| Defined medium with Synergy 1 (2%) | 500.50 ± 54.11 |
| Defined medium with P95 (2%) | 866.26 ± 62.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goyache, I.; Valdés-Varela, L.; Virto, R.; López-Yoldi, M.; López-Giral, N.; Sánchez-Vicente, A.; Milagro, F.I.; Aranaz, P. Novel Probiotic Strain Lactiplantibacillus plantarum CNTA 628 Modulates Lipid Metabolism and Improves Healthspan in C. elegans. Appl. Sci. 2025, 15, 8007. https://doi.org/10.3390/app15148007
Goyache I, Valdés-Varela L, Virto R, López-Yoldi M, López-Giral N, Sánchez-Vicente A, Milagro FI, Aranaz P. Novel Probiotic Strain Lactiplantibacillus plantarum CNTA 628 Modulates Lipid Metabolism and Improves Healthspan in C. elegans. Applied Sciences. 2025; 15(14):8007. https://doi.org/10.3390/app15148007
Chicago/Turabian StyleGoyache, Ignacio, Lorena Valdés-Varela, Raquel Virto, Miguel López-Yoldi, Noelia López-Giral, Ana Sánchez-Vicente, Fermín I. Milagro, and Paula Aranaz. 2025. "Novel Probiotic Strain Lactiplantibacillus plantarum CNTA 628 Modulates Lipid Metabolism and Improves Healthspan in C. elegans" Applied Sciences 15, no. 14: 8007. https://doi.org/10.3390/app15148007
APA StyleGoyache, I., Valdés-Varela, L., Virto, R., López-Yoldi, M., López-Giral, N., Sánchez-Vicente, A., Milagro, F. I., & Aranaz, P. (2025). Novel Probiotic Strain Lactiplantibacillus plantarum CNTA 628 Modulates Lipid Metabolism and Improves Healthspan in C. elegans. Applied Sciences, 15(14), 8007. https://doi.org/10.3390/app15148007







