Kiwifruit Allergy—Molecular Basis, Diagnostics and Treatment
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
3. Protein Families
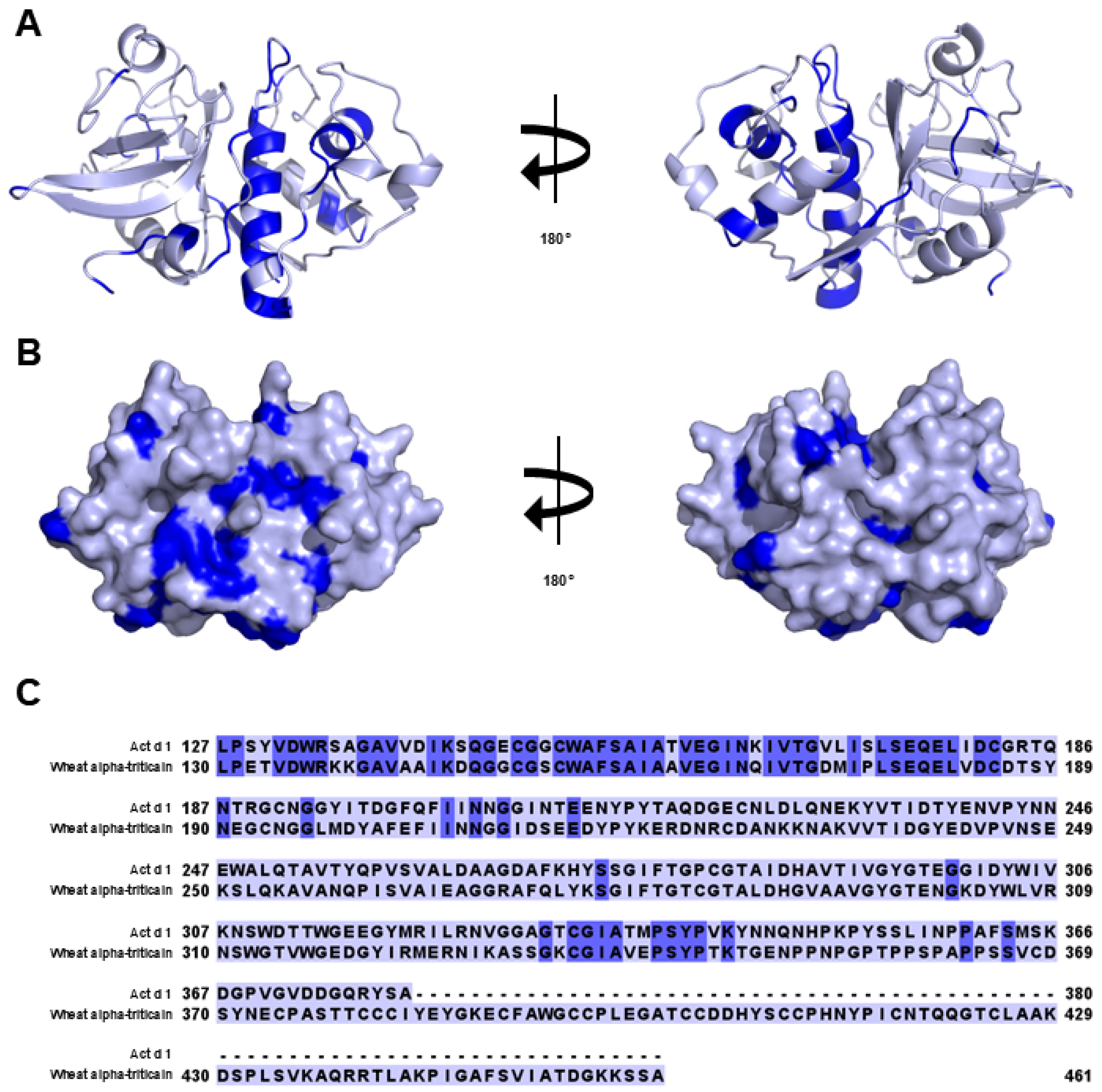
3.1. Cysteine Protease
3.2. Thaumatin-like Protein
3.3. Phytocystatin
3.4. Kiwellin
3.5. Pectin Methylesterase Inhibitor
3.6. Pectin Methylesterase
3.7. Pathogenesis-Related Class 10 Proteins
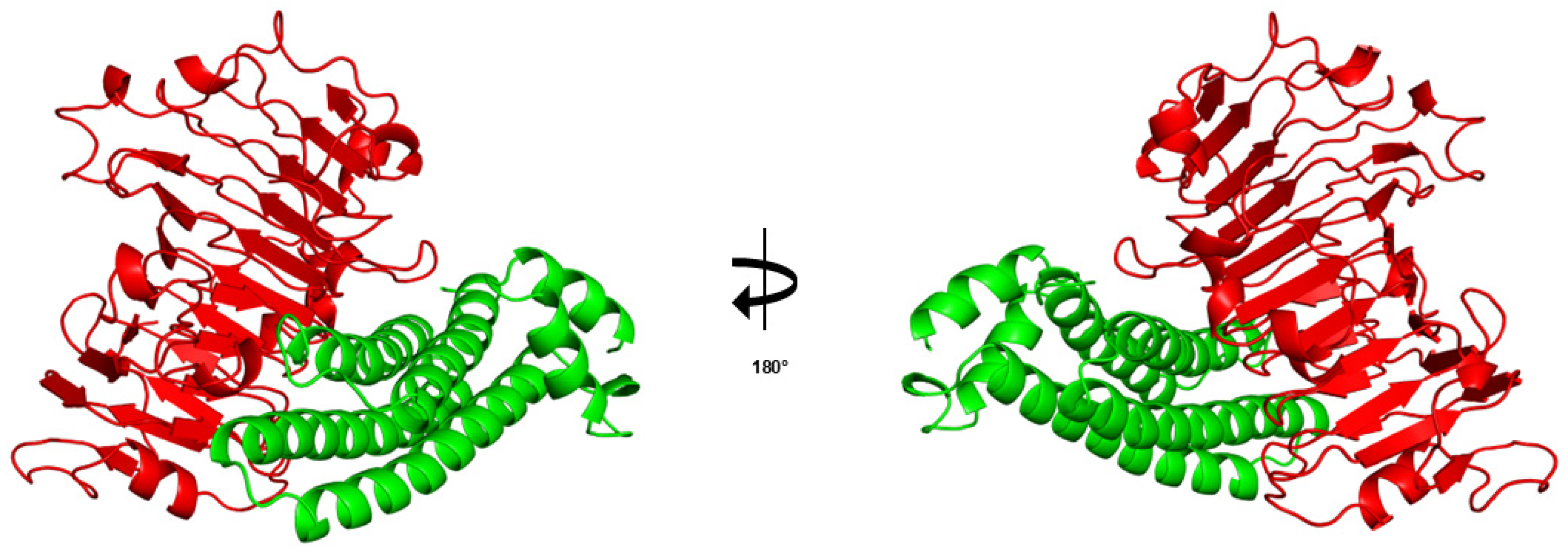
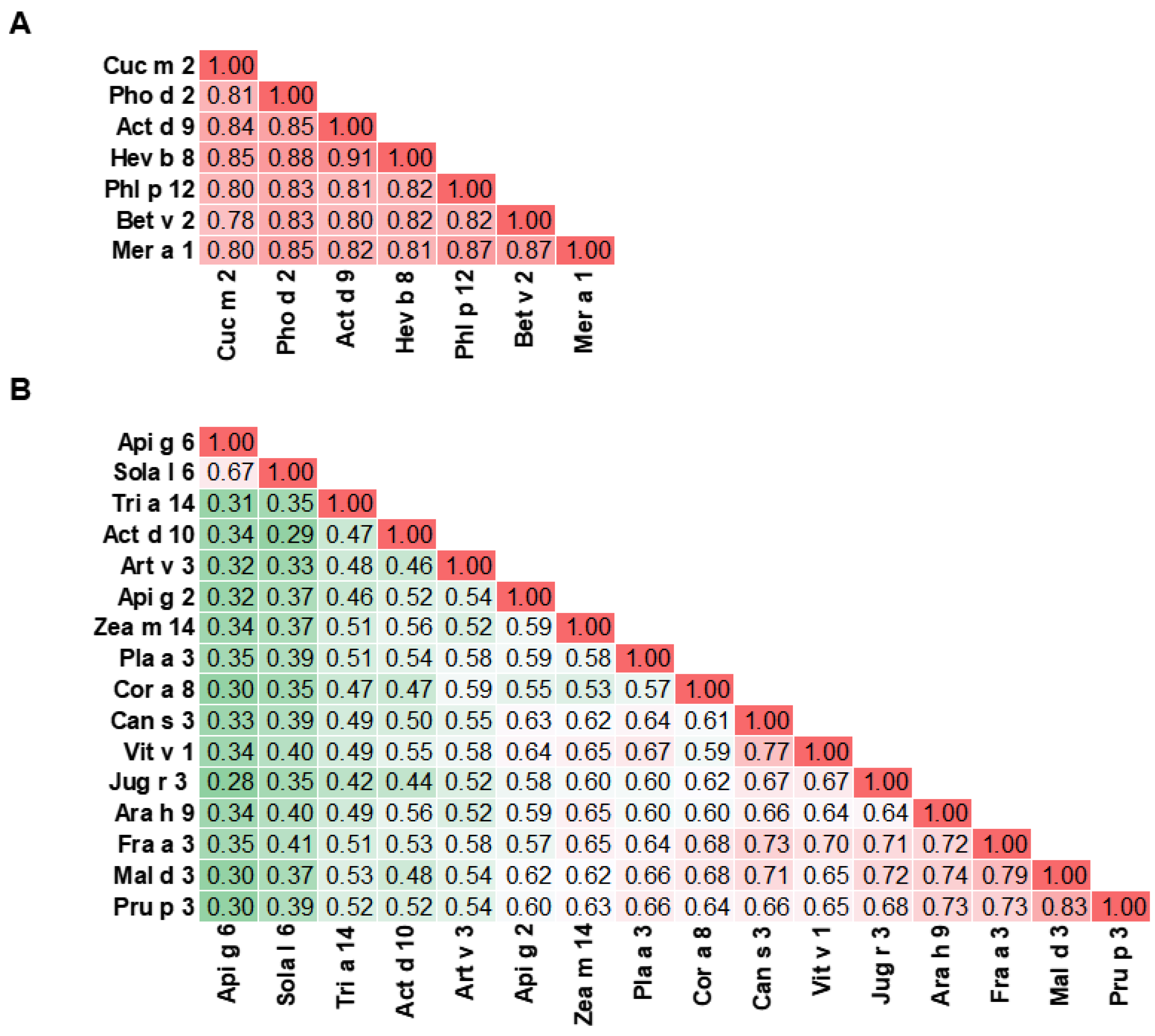
3.8. Profilin
3.9. Non-Specific Lipid Transfer Proteins
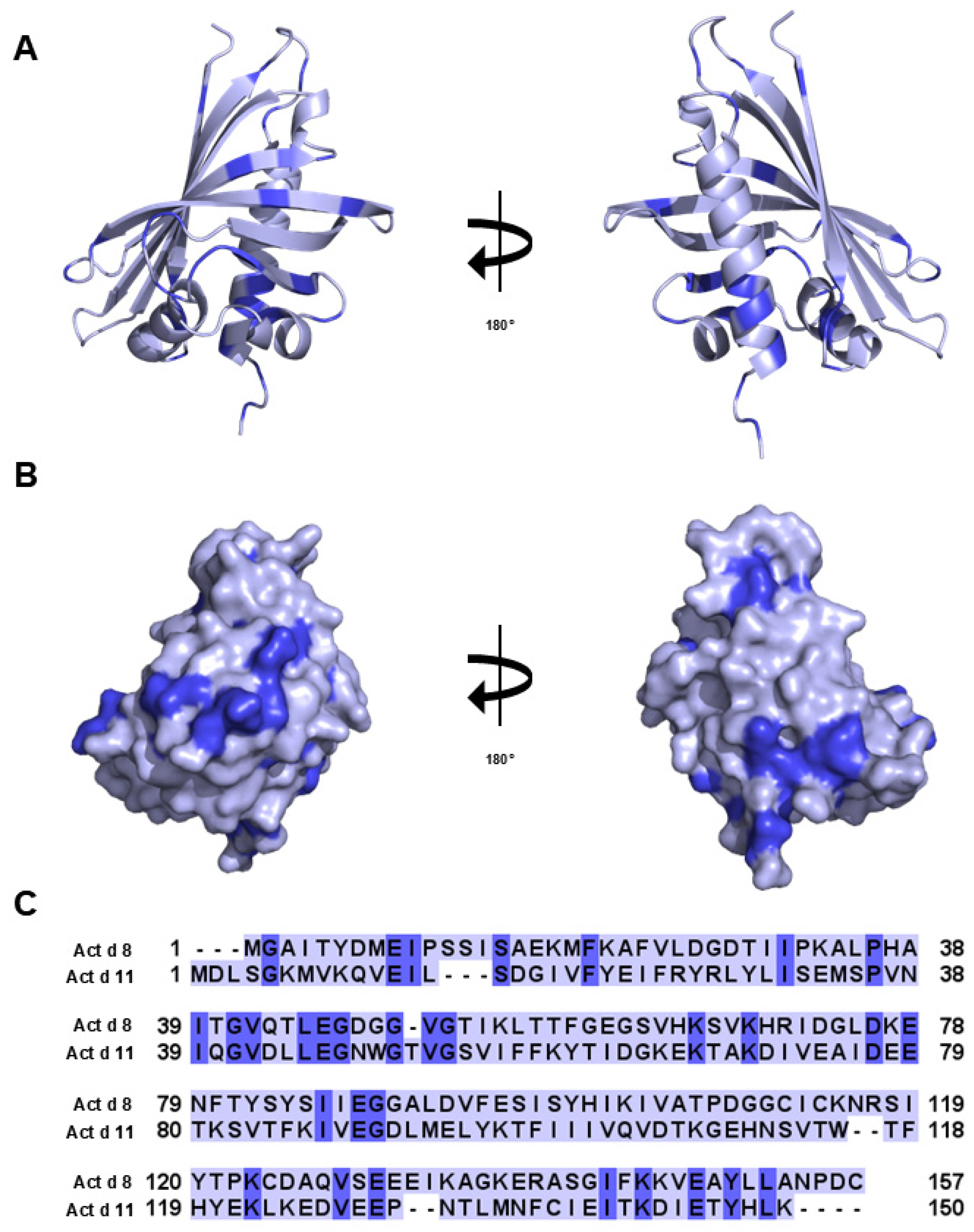
3.10. Major Latex Protein/Ripening-Related Protein
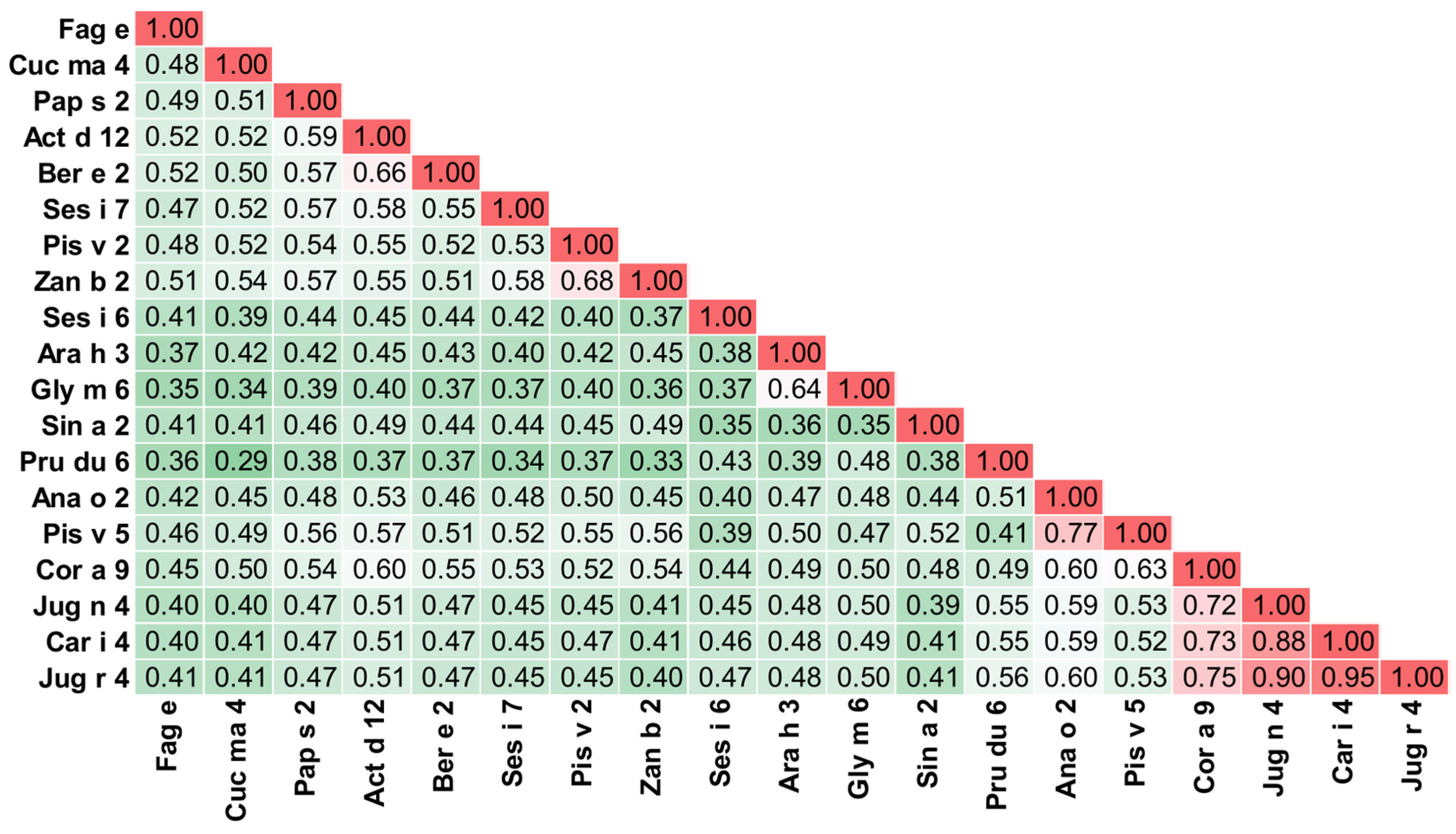
3.11. S Globulin and 2S Albumin
4. Diagnostics in Kiwifruit Allergy
4.1. Skin Prick Test
4.2. Prick-to-Prick Test
4.3. Oral Food Challenge Test
4.4. Allergen-Specific IgE in Serum
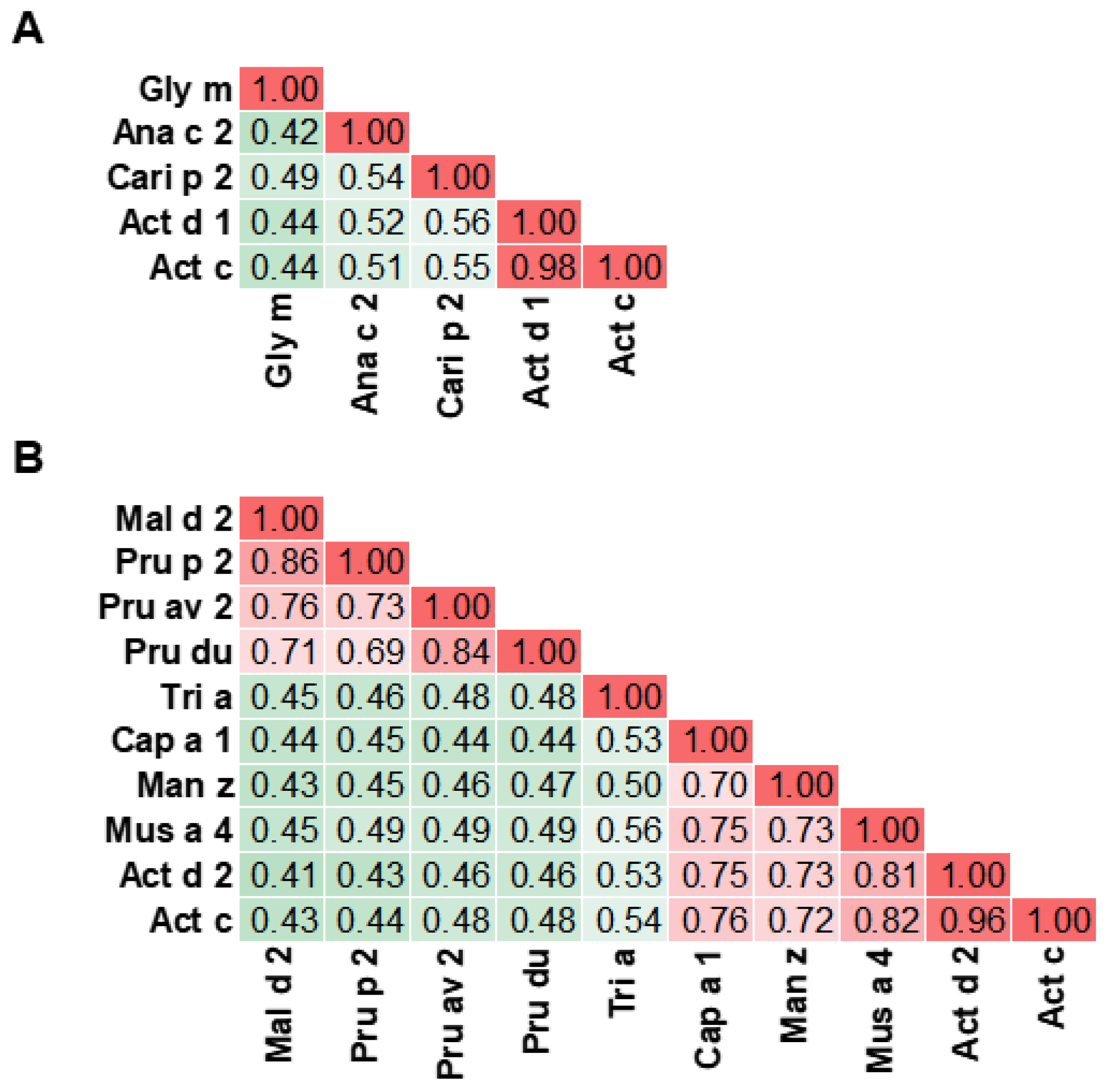
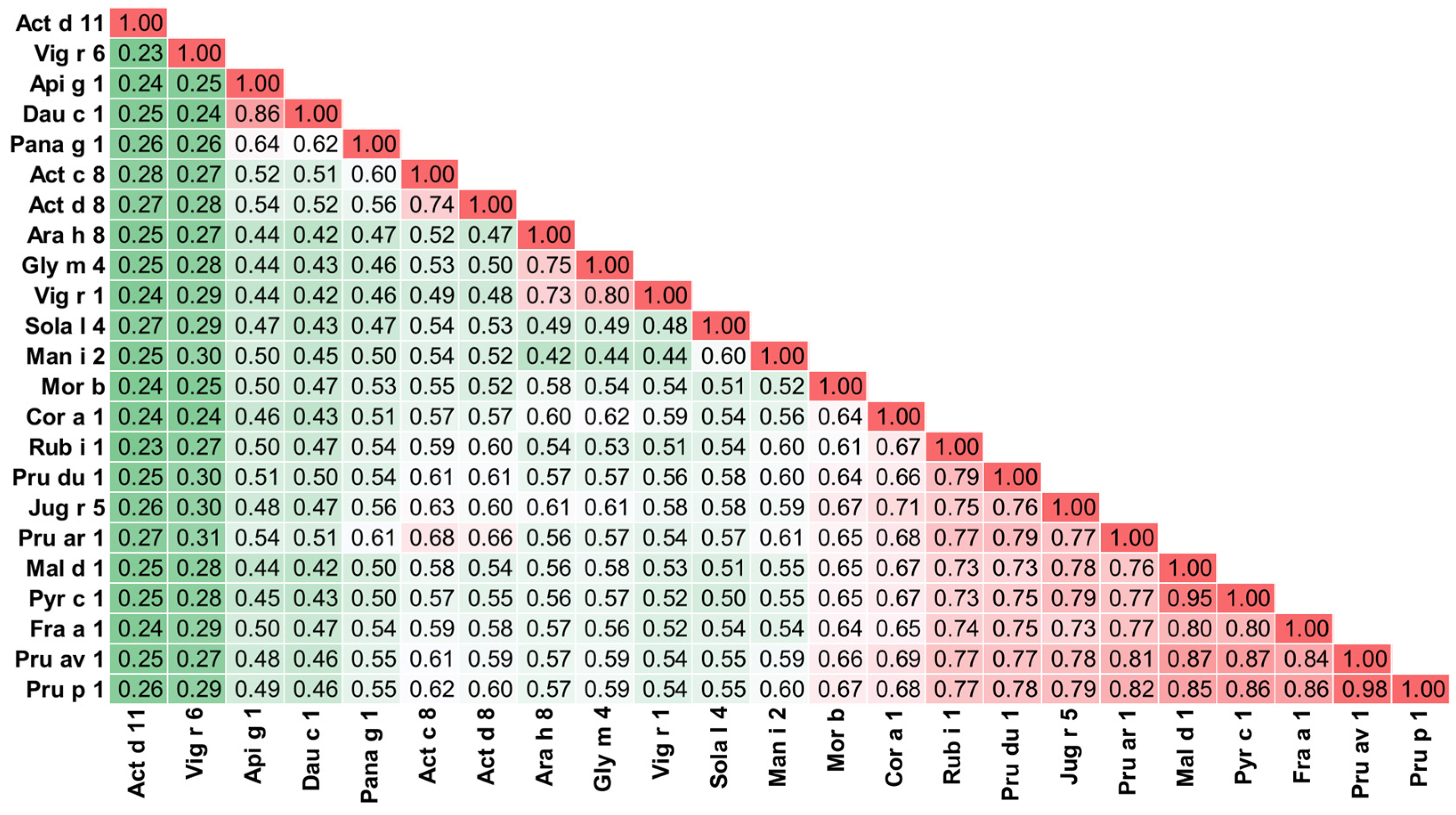
5. Cross-Reactivity of Kiwifruit Allergens
6. Patterns of Sensitization in Kiwifruit Allergy
7. Management of Kiwifruit Allergies
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| WHO | World Health Organization |
| IUIS | International Union of Immunological Societies |
| IgE | Immunoglobulin E |
| PFAS | Pollen food allergy syndrome |
| A-RISC | Allergens’-Relative Identity, Similarity and Cross-Reactivity |
| PDB | Protein Data Bank |
| ALEX2 | Allergy Explorer 2 |
| TLP | Thaumatin-like protein |
| PMEI | Pectin methylesterase inhibitor |
| PME | Pectin methylesterase |
| PR-10 | Pathogenesis-related proteins group 10 |
| nsLTP | Non-specific lipid transfer protein |
| MLP/RRP | Major latex protein/ripening-related protein |
| PR-5 | Pathogenesis-related proteins group 5 |
| SPT | Skin prick test |
| ISAC | Immune solid-phase allergen chip |
| ELISA | Enzyme-linked immunosorbent assay |
| NIAID | National Institute of Allergy and Infectious Diseases |
| PPT | Prick-to-prick testing |
| DBPCFC | Double-blind, placebo-controlled food challenge |
References
- Bringheli, I.; Brindisi, G.; Morelli, R.; Marchetti, L.; Cela, L.; Gravina, A.; Pastore, F.; Semeraro, A.; Cinicola, B.; Capponi, M.; et al. Kiwifruit’s Allergy in Children: What Do We Know? Nutrients 2023, 15, 3030. [Google Scholar] [CrossRef]
- Richardson, D.P.; Ansell, J.; Drummond, L.N. The nutritional and health attributes of kiwifruit: A review. Eur. J. Nutr. 2018, 57, 2659–2676. [Google Scholar] [CrossRef] [PubMed]
- James, C.A.; Welham, S.; Rose, P. Update on the global prevalence and severity of kiwifruit allergy: A scoping review. Int. J. Food Sci. Tech. 2023, 58, 6158–6181. [Google Scholar] [CrossRef]
- Pomes, A.; Davies, J.M.; Gadermaier, G.; Hilger, C.; Holzhauser, T.; Lidholm, J.; Lopata, A.L.; Mueller, G.A.; Nandy, A.; Radauer, C.; et al. WHO/IUIS Allergen Nomenclature: Providing a common language. Mol. Immunol. 2018, 100, 3–13. [Google Scholar] [CrossRef]
- Uberti, F.; Penas, E.; Manzoni, Y.; di Lorenzo, C.; Ballabio, C.; Fiocchi, A.; Terracciano, L.; Restani, P. Molecular characterization of allergens in raw and processed kiwifruit. Pediatr. Allergy Immunol. 2015, 26, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Lucas, J.S.; Lewis, S.A.; Trewin, J.B.; Grimshaw, K.E.; Warner, J.O.; Hourihane, J.O. Comparison of the allergenicity of Actinidia deliciosa (kiwi fruit) and Actinidia chinensis (gold kiwi). Pediatr. Allergy Immunol. 2005, 16, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Bublin, M.; Mari, A.; Ebner, C.; Knulst, A.; Scheiner, O.; Hoffmann-Sommergruber, K.; Breiteneder, H.; Radauer, C. IgE sensitization profiles toward green and gold kiwifruits differ among patients allergic to kiwifruit from 3 European countries. J. Allergy Clin. Immunol. 2004, 114, 1169–1175. [Google Scholar] [CrossRef]
- Mari, A.; Rasi, C.; Palazzo, P.; Scala, E. Allergen databases: Current status and perspectives. Curr. Allergy Asthma Rep. 2009, 9, 376–383. [Google Scholar] [CrossRef]
- Fine, A.J. Hypersensitivity reaction to kiwi fruit (Chinese gooseberry, Actinidia chinensis). J. Allergy Clin. Immunol. 1981, 68, 235–237. [Google Scholar] [CrossRef]
- Venkataraman, D.; Erlewyn-Lajeunesse, M.; Kurukulaaratchy, R.J.; Potter, S.; Roberts, G.; Matthews, S.; Arshad, S.H. Prevalence and longitudinal trends of food allergy during childhood and adolescence: Results of the Isle of Wight Birth Cohort study. Clin. Exp. Allergy 2018, 48, 394–402. [Google Scholar] [CrossRef]
- Vieira, T.; Cunha, L.; Neves, E.; Falcao, H. Diagnostic usefulness of component-resolved diagnosis by skin prick tests and specific IgE to single allergen components in children with allergy to fruits and vegetables. Allergol. Immunopathol. 2014, 42, 127–135. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, S.H.; Park, H.W.; Cho, S.H.; Chang, Y.S. Oral Allergy Syndrome in Birch Pollen-Sensitized Patients from a Korean University Hospital. J. Korean Med. Sci. 2018, 33, e218. [Google Scholar] [CrossRef] [PubMed]
- Mattila, L.; Kilpelainen, M.; Terho, E.O.; Koskenvuo, M.; Helenius, H.; Kalimo, K. Food hypersensitivity among Finnish university students: Association with atopic diseases. Clin. Exp. Allergy 2003, 33, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Karakaya, E.; Uzundumlu, A.S. Kiwi Production Forecasts for the Leading Countries in the Period 1983–2027. Appl. Fruit Sci. 2025, 67, 66. [Google Scholar] [CrossRef]
- Lucas, J.S.; Grimshaw, K.E.; Collins, K.; Warner, J.O.; Hourihane, J.O. Kiwi fruit is a significant allergen and is associated with differing patterns of reactivity in children and adults. Clin. Exp. Allergy 2004, 34, 1115–1121. [Google Scholar] [CrossRef]
- Ukleja-Sokolowska, N.; Zacniewski, R.; Lis, K.; Zbikowska-Gotz, M.; Kuzminski, A.; Bartuzi, Z. Exercise induced anaphylaxis in kiwi allergic patient: Case report. Allergy Asthma Clin. Immunol. 2021, 17, 91. [Google Scholar] [CrossRef]
- Aleman, A.; Sastre, J.; Quirce, S.; de las Heras, M.; Carnes, J.; Fernandez-Caldas, E.; Pastor, C.; Blazquez, A.B.; Vivanco, F.; Cuesta-Herranz, J. Allergy to kiwi: A double-blind, placebo-controlled food challenge study in patients from a birch-free area. J. Allergy Clin. Immunol. 2004, 113, 543–550. [Google Scholar] [CrossRef]
- D’Amelio, C.M.; Bernad, A.; Garcia-Figueroa, B.E.; Garrido-Fernandez, S.; Azofra, J.; Beristain, A.; Bueno-Diaz, C.; Garrido-Arandia, M.; Gastaminza, G.; Ferrer, M.; et al. Unraveling the Diagnosis of Kiwifruit Allergy: Usefulness of Current Diagnostic Tests. J. Investig. Allergol. Clin. Immunol. 2022, 32, 206–212. [Google Scholar] [CrossRef]
- Gall, H.; Kalveram, K.J.; Forck, G.; Sterry, W. Kiwi fruit allergy: A new birch pollen-associated food allergy. J. Allergy Clin. Immunol. 1994, 94, 70–76. [Google Scholar] [CrossRef]
- Lucas, J.S.; Lewis, S.A.; Hourihane, J.O. Kiwi fruit allergy: A review. Pediatr. Allergy Immunol. 2003, 14, 420–428. [Google Scholar] [CrossRef]
- Kashyap, R.R.; Kashyap, R.S. Oral Allergy Syndrome: An Update for Stomatologists. J. Allergy 2015, 2015, 543928. [Google Scholar] [CrossRef] [PubMed]
- Chruszcz, M.; Ciardiello, M.A.; Osinski, T.; Majorek, K.A.; Giangrieco, I.; Font, J.; Breiteneder, H.; Thalassinos, K.; Minor, W. Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins. Mol. Immunol. 2013, 56, 794–803. [Google Scholar] [CrossRef]
- Chruszcz, M.; Kapingidza, A.B.; Dolamore, C.; Kowal, K. A robust method for the estimation and visualization of IgE cross-reactivity likelihood between allergens belonging to the same protein family. PLoS ONE 2018, 13, e0208276. [Google Scholar] [CrossRef]
- Radauer, C.; Bublin, M.; Wagner, S.; Mari, A.; Breiteneder, H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J. Allergy Clin. Immunol. 2008, 121, 847–852.e847. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, 3.0; Schrödinger, LLC: New York, NY, USA, 2025; Available online: https://pymol.org/support.html (accessed on 23 June 2025).
- Bublin, M.; Pfister, M.; Radauer, C.; Oberhuber, C.; Bulley, S.; Dewitt, A.M.; Lidholm, J.; Reese, G.; Vieths, S.; Breiteneder, H.; et al. Component-resolved diagnosis of kiwifruit allergy with purified natural and recombinant kiwifruit allergens. J. Allergy Clin. Immunol. 2010, 125, 687–694.e1. [Google Scholar] [CrossRef]
- Pastorello, E.A.; Conti, A.; Pravettoni, V.; Farioli, L.; Rivolta, F.; Ansaloni, R.; Ispano, M.; Incorvaia, C.; Giuffrida, M.G.; Ortolani, C. Identification of actinidin as the major allergen of kiwi fruit. J. Allergy Clin. Immunol. 1998, 101, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Kaur, L.; Mao, B.; Bailly, J.; Oladeji, O.; Blatchford, P.; McNabb, W.C. Actinidin in Green and SunGold Kiwifruit Improves Digestion of Alternative Proteins-An In Vitro Investigation. Foods 2022, 11, 2739. [Google Scholar] [CrossRef]
- Nieuwenhuizen, N.J.; Beuning, L.L.; Sutherland, P.W.; Sharma, N.N.; Cooney, J.M.; Bieleski, L.R.F.; Schroder, R.; MacRae, E.A.; Atkinson, R.G. Identification and characterisation of acidic and novel basic forms of actinidin, the highly abundant cysteine protease from kiwifruit. Funct. Plant Biol. 2007, 34, 946–961. [Google Scholar] [CrossRef]
- Giangrieco, I.; Ciardiello, M.A.; Tamburrini, M.; Tuppo, L.; Mari, A.; Alessandri, C. Plant and Arthropod IgE-Binding Papain-like Cysteine Proteases: Multiple Contributions to Allergenicity. Foods 2024, 13, 790. [Google Scholar] [CrossRef] [PubMed]
- Grozdanovic, M.M.; Ostojic, S.; Aleksic, I.; Andjelkovic, U.; Petersen, A.; Gavrovic-Jankulovic, M. Active actinidin retains function upon gastro-intestinal digestion and is more thermostable than the E-64-inhibited counterpart. J. Sci. Food Agric. 2014, 94, 3046–3052. [Google Scholar] [CrossRef]
- Grozdanovic, M.M.; Cavic, M.; Nesic, A.; Andjelkovic, U.; Akbari, P.; Smit, J.J.; Gavrovic-Jankulovic, M. Kiwifruit cysteine protease actinidin compromises the intestinal barrier by disrupting tight junctions. Biochim. Biophys. Acta 2016, 1860, 516–526. [Google Scholar] [CrossRef]
- Fukuoka, A.; Yoshimoto, T. Barrier dysfunction in the nasal allergy. Allergol. Int. 2018, 67, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Soh, W.T.; Zhang, J.; Hollenberg, M.D.; Vliagoftis, H.; Rothenberg, M.E.; Sokol, C.L.; Robinson, C.; Jacquet, A. Protease allergens as initiators-regulators of allergic inflammation. Allergy 2023, 78, 1148–1168. [Google Scholar] [CrossRef] [PubMed]
- Yazici, D.; Ogulur, I.; Pat, Y.; Babayev, H.; Barletta, E.; Ardicli, S.; Bel Imam, M.; Huang, M.; Koch, J.; Li, M.; et al. The epithelial barrier: The gateway to allergic, autoimmune, and metabolic diseases and chronic neuropsychiatric conditions. Semin. Immunol. 2023, 70, 101846. [Google Scholar] [CrossRef]
- Cavic, M.; Grozdanovic, M.M.; Bajic, A.; Jankovic, R.; Andjus, P.R.; Gavrovic-Jankulovic, M. The effect of kiwifruit (Actinidia deliciosa) cysteine protease actinidin on the occludin tight junction network in T84 intestinal epithelial cells. Food Chem. Toxicol. 2014, 72, 61–68. [Google Scholar] [CrossRef]
- Nesic, A.; Cavic, M.; Popovic, M.; Zlatanova, M.; Pieters, R.; Smit, J.; Gavrovic-Jankulovic, M. The Kiwifruit Allergen Act d 1 Activates NF-kappaB Signaling and Affects mRNA Expression of TJ Proteins and Innate Pro-Allergenic Cytokines. Biomolecules 2019, 9, 816. [Google Scholar] [CrossRef]
- Gavrovic-Jankulovic, M.; cIrkovic, T.; Vuckovic, O.; Atanaskovic-Markovic, M.; Petersen, A.; Gojgic, G.; Burazer, L.; Jankov, R.M. Isolation and biochemical characterization of a thaumatin-like kiwi allergen. J. Allergy Clin. Immunol. 2002, 110, 805–810. [Google Scholar] [CrossRef]
- Gavrovic-Jankulovic, M.; Spasic, M.; Cirkovic Velickovic, T.; Stojanovic, M.; Inic-Kanada, A.; Dimitrijevic, L.; Lindner, B.; Petersen, A.; Becker, W.M.; Jankov, R.M. Quantification of the thaumatin-like kiwi allergen by a monoclonal antibody-based ELISA. Mol. Nutr. Food Res. 2008, 52, 701–707. [Google Scholar] [CrossRef]
- Palazzo, P.; Tuppo, L.; Giangrieco, I.; Bernardi, M.L.; Rafaiani, C.; Crescenzo, R.; Tamburrini, M.; Zuzzi, S.; Alessandri, C.; Mari, A.; et al. Prevalence and peculiarities of IgE reactivity to kiwifruit pectin methylesterase and its inhibitor, Act d 7 and Act d 6, in subjects allergic to kiwifruit. Food Res. Int. 2013, 53, 24–30. [Google Scholar] [CrossRef]
- Wang, H.; Ng, T.B. Isolation of an antifungal thaumatin-like protein from kiwi fruits. Phytochemistry 2002, 61, 1–6. [Google Scholar] [CrossRef]
- Bublin, M.; Radauer, C.; Knulst, A.; Wagner, S.; Scheiner, O.; Mackie, A.R.; Mills, E.N.C.; Breiteneder, H. Effects of gastrointestinal digestion and heating on the allergenicity of the kiwi allergens Act d 1, actinidin, and Act d 2, a thaumatin-like protein. Mol. Nutr. Food Res. 2008, 52, 1130–1139. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Vanga, S.K.; Raghavan, V. Structural responses of kiwifruit allergen Act d 2 to thermal and electric field stresses based on molecular dynamics simulations and experiments. Food Funct. 2020, 11, 1373–1384. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Kranthi Vanga, S.; Raghavan, V. Influence of high-intensity ultrasound on the IgE binding capacity of Act d 2 allergen, secondary structure, and In-vitro digestibility of kiwifruit proteins. Ultrason. Sonochem. 2021, 71, 105409. [Google Scholar] [CrossRef] [PubMed]
- Palacin, A.; Rodriguez, J.; Blanco, C.; Lopez-Torrejon, G.; Sanchez-Monge, R.; Varela, J.; Jimenez, M.A.; Cumplido, J.; Carrillo, T.; Crespo, J.F.; et al. Immunoglobulin E recognition patterns to purified Kiwifruit (Actinidinia deliciosa) allergens in patients sensitized to Kiwi with different clinical symptoms. Clin. Exp. Allergy 2008, 38, 1220–1228. [Google Scholar] [CrossRef]
- Palacin, A.; Rivas, L.A.; Gomez-Casado, C.; Aguirre, J.; Tordesillas, L.; Bartra, J.; Blanco, C.; Carrillo, T.; Cuesta-Herranz, J.; Bonny, J.A.; et al. The involvement of thaumatin-like proteins in plant food cross-reactivity: A multicenter study using a specific protein microarray. PLoS ONE 2012, 7, e44088. [Google Scholar] [CrossRef]
- Smole, U.; Bublin, M.; Radauer, C.; Ebner, C.; Scheiner, O.; Breiteneder, H. Allergenic fruit TLPs possess different degrees of IgE cross-reactivity. J. Allergy Clin. Immun. 2006, 117, S49. [Google Scholar] [CrossRef]
- Maddumage, R.; Nieuwenhuizen, N.J.; Bulley, S.M.; Cooney, J.M.; Green, S.A.; Atkinson, R.G. Diversity and relative levels of actinidin, kiwellin, and thaumatin-like allergens in 15 varieties of kiwifruit (Actinidia). J. Agric. Food Chem. 2013, 61, 728–739. [Google Scholar] [CrossRef]
- Martinez, M.; Santamaria, M.E.; Diaz-Mendoza, M.; Arnaiz, A.; Carrillo, L.; Ortego, F.; Diaz, I. Phytocystatins: Defense Proteins against Phytophagous Insects and Acari. Int. J. Mol. Sci. 2016, 17, 1747. [Google Scholar] [CrossRef]
- Popovic, M.M.; Milovanovic, M.; Burazer, L.; Vuckovic, O.; Hoffmann-Sommergruber, K.; Knulst, A.C.; Lindner, B.; Petersen, A.; Jankov, R.; Gavrovic-Jankulovic, M. Cysteine proteinase inhibitor Act d 4 is a functional allergen contributing to the clinical symptoms of kiwifruit allergy. Mol. Nutr. Food Res. 2010, 54, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Popovic, M.; Grozdanovic, M.; Gavrovic-Jankulovic, M. Kiwifruit as a food allergen source. J. Serb. Chem. Soc. 2013, 78, 333–352. [Google Scholar] [CrossRef]
- Tamburrini, M.; Cerasuolo, I.; Carratore, V.; Stanziola, A.A.; Zofra, S.; Romano, L.; Camardella, L.; Ciardiello, M.A. Kiwellin, a novel protein from kiwi fruit. Purification, biochemical characterization and identification as an allergen*. Protein J. 2005, 24, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Hamiaux, C.; Maddumage, R.; Middleditch, M.J.; Prakash, R.; Brummell, D.A.; Baker, E.N.; Atkinson, R.G. Crystal structure of kiwellin, a major cell-wall protein from kiwifruit. J. Struct. Biol. 2014, 187, 276–281. [Google Scholar] [CrossRef]
- Offermann, L.R.; Giangrieco, I.; Perdue, M.L.; Zuzzi, S.; Santoro, M.; Tamburrini, M.; Cosgrove, D.J.; Mari, A.; Ciardiello, M.A.; Chruszcz, M. Elusive Structural, Functional, and Immunological Features of Act d 5, the Green Kiwifruit Kiwellin. J. Agric. Food Chem. 2015, 63, 6567–6576. [Google Scholar] [CrossRef]
- Ciacci, C.; Russo, I.; Bucci, C.; Iovino, P.; Pellegrini, L.; Giangrieco, I.; Tamburrini, M.; Ciardiello, M.A. The kiwi fruit peptide kissper displays anti-inflammatory and anti-oxidant effects in in-vitro and ex-vivo human intestinal models. Clin. Exp. Immunol. 2014, 175, 476–484. [Google Scholar] [CrossRef]
- Ciardiello, M.A.; Giangrieco, I.; Tuppo, L.; Tamburrini, M.; Buccheri, M.; Palazzo, P.; Bernardi, M.L.; Ferrara, R.; Mari, A. Influence of the natural ripening stage, cold storage, and ethylene treatment on the protein and IgE-binding profiles of green and gold kiwi fruit extracts. J. Agric. Food Chem. 2009, 57, 1565–1571. [Google Scholar] [CrossRef]
- Tuppo, L.; Giangrieco, I.; Palazzo, P.; Bernardi, M.L.; Scala, E.; Carratore, V.; Tamburrini, M.; Mari, A.; Ciardiello, M.A. Kiwellin, a Modular Protein from Green and Gold Kiwi Fruits: Evidence of in Vivo and in Vitro Processing and IgE Binding. J. Agric. Food Chem. 2008, 56, 3812–3817. [Google Scholar] [CrossRef]
- Bonavita, A.; Carratore, V.; Ciardiello, M.A.; Giovane, A.; Servillo, L.; D’Avino, R. Influence of pH on the Structure and Function of Kiwi Pectin Methylesterase Inhibitor. J. Agric. Food Chem. 2016, 64, 5866–5876. [Google Scholar] [CrossRef]
- Asturias, J.A.; Ibarrola, I.; Eraso, E.; Arilla, M.C.; Martinez, A. The major Platanus acerifolia pollen allergen Pla a 1 has sequence homology to invertase inhibitors. Clin. Exp. Allergy 2003, 33, 978–985. [Google Scholar] [CrossRef]
- Ciardiello, M.A.; D’Avino, R.; Amoresano, A.; Tuppo, L.; Carpentieri, A.; Carratore, V.; Tamburrini, M.; Giovane, A.; Pucci, P.; Camardella, L. The peculiar structural features of kiwi fruit pectin methylesterase: Amino acid sequence, oligosaccharides structure, and modeling of the interaction with its natural proteinaceous inhibitor. Proteins 2008, 71, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Bublin, M.; Dennstedt, S.; Buchegger, M.; Antonietta Ciardiello, M.; Bernardi, M.L.; Tuppo, L.; Harwanegg, C.; Hafner, C.; Ebner, C.; Ballmer-Weber, B.K.; et al. The performance of a component-based allergen microarray for the diagnosis of kiwifruit allergy. Clin. Exp. Allergy 2011, 41, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Oberhuber, C.; Bulley, S.M.; Ballmer-Weber, B.K.; Bublin, M.; Gaier, S.; DeWitt, A.M.; Briza, P.; Hofstetter, G.; Lidholm, J.; Vieths, S.; et al. Characterization of Bet v 1-related allergens from kiwifruit relevant for patients with combined kiwifruit and birch pollen allergy. Mol. Nutr. Food Res. 2008, 52 (Suppl. S2), S230–S240. [Google Scholar] [CrossRef]
- Zeindl, R.; Franzmann, A.L.; Fernandez-Quintero, M.L.; Seidler, C.A.; Hoerschinger, V.J.; Liedl, K.R.; Tollinger, M. Structural Basis of the Immunological Cross-Reactivity between Kiwi and Birch Pollen. Foods 2023, 12, 3939. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, H.; Michalska, K.; Sikorski, M.; Jaskolski, M. Structural and functional aspects of PR-10 proteins. FEBS J. 2013, 280, 1169–1199. [Google Scholar] [CrossRef]
- Carlsson, L.; Nystrom, L.E.; Sundkvist, I.; Markey, F.; Lindberg, U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J. Mol. Biol. 1977, 115, 465–483. [Google Scholar] [CrossRef]
- Breiteneder, H.; Radauer, C. A classification of plant food allergens. J. Allergy Clin. Immunol. 2004, 113, 821–830, quiz 831. [Google Scholar] [CrossRef]
- van Ree, R.; Voitenko, V.; van Leeuwen, W.A.; Aalberse, R.C. Profilin is a cross-reactive allergen in pollen and vegetable foods. Int. Arch. Allergy Immunol. 1992, 98, 97–104. [Google Scholar] [CrossRef]
- Wang, J.; Vanga, S.K.; McCusker, C.; Raghavan, V. A Comprehensive Review on Kiwifruit Allergy: Pathogenesis, Diagnosis, Management, and Potential Modification of Allergens Through Processing. Compr. Rev. Food Sci. Food Saf. 2019, 18, 500–513. [Google Scholar] [CrossRef]
- Teran, M.G.; Garcia-Ramirez, B.; Mares-Mejia, I.; Ortega, E.; O’Malley, A.; Chruszcz, M.; Rodriguez-Romero, A. Molecular Basis of Plant Profilins’ Cross-Reactivity. Biomolecules 2023, 13, 608. [Google Scholar] [CrossRef]
- Valenta, R.; Duchene, M.; Ebner, C.; Valent, P.; Sillaber, C.; Deviller, P.; Ferreira, F.; Tejkl, M.; Edelmann, H.; Kraft, D.; et al. Profilins constitute a novel family of functional plant pan-allergens. J. Exp. Med. 1992, 175, 377–385. [Google Scholar] [CrossRef]
- Pastorello, E.A.; Pravettoni, V.; Ispano, M.; Farioli, L.; Ansaloni, R.; Rotondo, F.; Incorvaia, C.; Asman, I.; Bengtsson, A.; Ortolani, C. Identification of the allergenic components of kiwi fruit and evaluation of their cross-reactivity with timothy and birch pollens. J. Allergy Clin. Immunol. 1996, 98, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Schuler, S.; Ferrari, G.; Schmid-Grendelmeier, P.; Harr, T. Microarray-based component-resolved diagnosis of latex allergy: Isolated IgE-mediated sensitization to latexprofilin Hev b8 may act as confounder. Clin. Transl. Allergy 2013, 3, 11. [Google Scholar] [CrossRef]
- O’Malley, A.; Pote, S.; Giangrieco, I.; Tuppo, L.; Gawlicka-Chruszcz, A.; Kowal, K.; Ciardiello, M.A.; Chruszcz, M. Structural Characterization of Act c 10.0101 and Pun g 1.0101-Allergens from the Non-Specific Lipid Transfer Protein Family. Molecules 2021, 26, 256. [Google Scholar] [CrossRef] [PubMed]
- Pasquato, N.; Berni, R.; Folli, C.; Folloni, S.; Cianci, M.; Pantano, S.; Helliwell, J.R.; Zanotti, G. Crystal structure of peach Pru p 3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens. J. Mol. Biol. 2006, 356, 684–694. [Google Scholar] [CrossRef]
- Skypala, I.J.; Asero, R.; Barber, D.; Cecchi, L.; Diaz Perales, A.; Hoffmann-Sommergruber, K.; Pastorello, E.A.; Swoboda, I.; Bartra, J.; Ebo, D.G.; et al. Non-specific lipid-transfer proteins: Allergen structure and function, cross-reactivity, sensitization, and epidemiology. Clin. Transl. Allergy 2021, 11, e12010. [Google Scholar] [CrossRef] [PubMed]
- Salminen, T.A.; Blomqvist, K.; Edqvist, J. Lipid transfer proteins: Classification, nomenclature, structure, and function. Planta 2016, 244, 971–997. [Google Scholar] [CrossRef]
- Bernardi, M.L.; Giangrieco, I.; Camardella, L.; Ferrara, R.; Palazzo, P.; Panico, M.R.; Crescenzo, R.; Carratore, V.; Zennaro, D.; Liso, M.; et al. Allergenic lipid transfer proteins from plant-derived foods do not immunologically and clinically behave homogeneously: The kiwifruit LTP as a model. PLoS ONE 2011, 6, e27856. [Google Scholar] [CrossRef]
- Marion, D.; Bakan, B.; Elmorjani, K. Plant lipid binding proteins: Properties and applications. Biotechnol. Adv. 2007, 25, 195–197. [Google Scholar] [CrossRef]
- Giangrieco, I.; Alessandri, C.; Rafaiani, C.; Santoro, M.; Zuzzi, S.; Tuppo, L.; Tamburrini, M.; D’Avino, R.; Ciardiello, M.A.; Mari, A. Structural features, IgE binding and preliminary clinical findings of the 7kDa Lipid Transfer Protein from tomato seeds. Mol. Immunol. 2015, 66, 154–163. [Google Scholar] [CrossRef]
- Scheurer, S.; Schulke, S. Interaction of Non-Specific Lipid-Transfer Proteins with Plant-Derived Lipids and Its Impact on Allergic Sensitization. Front. Immunol. 2018, 9, 1389. [Google Scholar] [CrossRef] [PubMed]
- Yeats, T.H.; Rose, J.K. The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs). Protein Sci. 2008, 17, 191–198. [Google Scholar] [CrossRef]
- Asero, R.; Pravettoni, V.; Villalta, D.; Cecchi, L.; Scala, E. IgE-mediated reactivity to non-specific lipid transfer protein (nsLTP): Clinical implications and management—A consensus document of the Association of Italian Territorial and Hospital Allergists and Immunologists (AAIITO). Eur. Ann. Allergy Clin. Immunol. 2024, 56, 176–182. [Google Scholar] [CrossRef]
- Mothes-Luksch, N.; Raith, M.; Stingl, G.; Focke-Tejkl, M.; Razzazi-Fazeli, E.; Zieglmayer, R.; Wohrl, S.; Swoboda, I. Pru p 3, a marker allergen for lipid transfer protein sensitization also in Central Europe. Allergy 2017, 72, 1415–1418. [Google Scholar] [CrossRef] [PubMed]
- Palacin, A.; Quirce, S.; Sanchez-Monge, R.; Fernandez-Nieto, M.; Varela, J.; Sastre, J.; Salcedo, G. Allergy to kiwi in patients with baker’s asthma: Identification of potential cross-reactive allergens. Ann. Allergy Asthma Immunol. 2008, 101, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, M.L.; Giangrieco, I.; Camardella, L.; Panico, M.R.; Zennaro, D.; Ferrara, R.; Palazzo, P.; Tuppo, L.; Tamburrini, M.; Carratore, V.; et al. Biochemical and immunological characterisation of Act d 10 a lipid transfer proteins from green kiwifruit. Clin. Transl. Allergy 2011, 1, O23. [Google Scholar] [CrossRef]
- D’Avino, R.; Bernardi, M.L.; Wallner, M.; Palazzo, P.; Camardella, L.; Tuppo, L.; Alessandri, C.; Breiteneder, H.; Ferreira, F.; Ciardiello, M.A.; et al. Kiwifruit Act d 11 is the first member of the ripening-related protein family identified as an allergen. Allergy 2011, 66, 870–877. [Google Scholar] [CrossRef]
- Mills, E.N.; Jenkins, J.; Marigheto, N.; Belton, P.S.; Gunning, A.P.; Morris, V.J. Allergens of the cupin superfamily. Biochem. Soc. Trans. 2002, 30, 925–929. [Google Scholar] [CrossRef] [PubMed]
- Beyer, K.; Bardina, L.; Grishina, G.; Sampson, H.A. Identification of sesame seed allergens by 2-dimensional proteomics and Edman sequencing: Seed storage proteins as common food allergens. J. Allergy Clin. Immunol. 2002, 110, 154–159. [Google Scholar] [CrossRef]
- Menéndez-Arias, L.; Moneo, I.; Domínguez, J.; Rodríguez, R. Primary structure of the major allergen of yellow mustard (Sinapis alba L.) seed, Sin. a I. Eur. J. Biochem. 1988, 177, 159–166. [Google Scholar] [CrossRef]
- Palomares, O.; Vereda, A.; Cuesta-Herranz, J.; Villalba, M.; Rodriguez, R. Cloning, sequencing, and recombinant production of Sin a 2, an allergenic 11S globulin from yellow mustard seeds. J. Allergy Clin. Immunol. 2007, 119, 1189–1196. [Google Scholar] [CrossRef]
- Sirvent, S.; Canto, B.; Gomez, F.; Blanca, N.; Cuesta-Herranz, J.; Canto, G.; Blanca, M.; Rodriguez, R.; Villalba, M.; Palomares, O. Detailed characterization of Act d 12 and Act d 13 from kiwi seeds: Implication in IgE cross-reactivity with peanut and tree nuts. Allergy 2014, 69, 1481–1488. [Google Scholar] [CrossRef]
- Dramburg, S.; Hilger, C.; Santos, A.F.; de Las Vecillas, L.; Aalberse, R.C.; Acevedo, N.; Aglas, L.; Altmann, F.; Arruda, K.L.; Asero, R.; et al. EAACI Molecular Allergology User’s Guide 2.0. Pediatr. Allergy Immunol. 2023, 34 (Suppl. S28), e13854. [Google Scholar] [CrossRef]
- Panel, N.I.-S.E.; Boyce, J.A.; Assa’ad, A.; Burks, A.W.; Jones, S.M.; Sampson, H.A.; Wood, R.A.; Plaut, M.; Cooper, S.F.; Fenton, M.J.; et al. Guidelines for the diagnosis and management of food allergy in the United States: Report of the NIAID-sponsored expert panel. J. Allergy Clin. Immunol. 2010, 126, S1–S58. [Google Scholar] [CrossRef]
- Bernardi, M.L.; Picone, D.; Tuppo, L.; Giangrieco, I.; Petrella, G.; Palazzo, P.; Ferrara, R.; Tamburrini, M.; Mari, A.; Ciardiello, M.A. Physico-chemical features of the environment affect the protein conformation and the immunoglobulin E reactivity of kiwellin (Act d 5). Clin. Exp. Allergy 2010, 40, 1819–1826. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, M.A.; Meleleo, D.; Saviano, G.; Crescenzo, R.; Carratore, V.; Camardella, L.; Gallucci, E.; Micelli, S.; Tancredi, T.; Picone, D.; et al. Kissper, a kiwi fruit peptide with channel-like activity: Structural and functional features. J. Pept. Sci. 2008, 14, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Terlouw, S.; van Boven, F.E.; Borsboom-van Zonneveld, M.; de Graaf-In‘t Veld, T.; Gerth van Wijk, R.; van Daele, P.L.A.; van Maaren, M.S.; Kuijpers, J.; Veenbergen, S.; de Jong, N.W. Comparison of skin prick test and prick-to-prick test with fruits and vegetables in the diagnosis of food allergy. Clin. Transl. Allergy 2024, 14, e12375. [Google Scholar] [CrossRef]
- Alessandri, C.; Ferrara, R.; Bernardi, M.L.; Zennaro, D.; Tuppo, L.; Giangrieco, I.; Tamburrini, M.; Mari, A.; Ciardiello, M.A. Diagnosing allergic sensitizations in the third millennium: Why clinicians should know allergen molecule structures. Clin. Transl. Allergy 2017, 7, 21. [Google Scholar] [CrossRef]
- Nosslinger, H.; Mair, E.; Oostingh, G.J.; Ahlgrimm-Siess, V.; Ringauf, A.; Lang, R. Multiplex Assays in Allergy Diagnosis: Allergy Explorer 2 versus ImmunoCAP ISAC E112i. Diagnostics 2024, 14, 976. [Google Scholar] [CrossRef]
- Tuppo, L.; Giangrieco, I.; Tamburrini, M.; Alessandri, C.; Mari, A.; Ciardiello, M.A. Detection of Allergenic Proteins in Foodstuffs: Advantages of the Innovative Multiplex Allergen Microarray-Based Immunoassay Compared to Conventional Methods. Foods 2022, 11, 878. [Google Scholar] [CrossRef]
- Wagner, S.; Breiteneder, H. The latex-fruit syndrome. Biochem. Soc. Trans. 2002, 30, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Ebo, D.G.; Bridts, C.H.; Mertens, C.H.; Sabato, V. Principles, potential, and limitations of ex vivo basophil activation by flow cytometry in allergology: A narrative review. J. Allergy Clin. Immunol. 2021, 147, 1143–1153. [Google Scholar] [CrossRef]
- Carlson, G.; Coop, C. Pollen food allergy syndrome (PFAS): A review of current available literature. Ann. Allergy Asthma Immunol. 2019, 123, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Giangrieco, I.; Ciardiello, M.A.; Tamburrini, M.; Tuppo, L.; Rafaiani, C.; Mari, A.; Alessandri, C. Comparative Analysis of the Immune Response and the Clinical Allergic Reaction to Papain-like Cysteine Proteases from Fig, Kiwifruit, Papaya, Pineapple and Mites in an Italian Population. Foods 2023, 12, 2852. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Yin, J.; Wen, L. Papain Induced Occupational Asthma with Kiwi and Fig Allergy. Allergy Asthma Immunol. Res. 2016, 8, 170–173. [Google Scholar] [CrossRef]
- van Odijk, J.; Sjolander, S.; Brostedt, P.; Borres, M.P.; Englund, H. High frequency of IgE sensitization towards kiwi seed storage proteins among peanut allergic individuals also reporting allergy to kiwi. Clin. Mol. Allergy 2017, 15, 18. [Google Scholar] [CrossRef]
- Le, T.-M.; Bublin, M.; Breiteneder, H.; Fernández-Rivas, M.; Asero, R.; Ballmer-Weber, B.; Barreales, L.; Bures, P.; Belohlavkova, S.; De Blay, F.; et al. Kiwifruit allergy across Europe: Clinical manifestation and IgE recognition patterns to kiwifruit allergens. J. Allergy Clin. Immun. 2013, 131, 164–171. [Google Scholar] [CrossRef]
- Asaumi, T.; Yanagida, N.; Sato, S.; Takahashi, K.; Ebisawa, M. Negative Act d 8 indicates systemic kiwifruit allergy among kiwifruit-sensitized children. Pediatr. Allergy Immunol. 2017, 28, 291–294. [Google Scholar] [CrossRef]
- Al-Shaikhly, T.; Cox, A.; Nowak-Wegrzyn, A.; Cianferoni, A.; Katelaris, C.; Ebo, D.G.; Konstantinou, G.N.; Brucker, H.; Yang, H.J.; Protudjer, J.L.P.; et al. An International Delphi Consensus on the Management of Pollen-Food Allergy Syndrome: A Work Group Report of the AAAAI Adverse Reactions to Foods Committee. J. Allergy Clin. Immunol. Pract. 2024, 12, 3242–3249.e1. [Google Scholar] [CrossRef]
- Martelli, A.; Calvani, M.; Foiadelli, T.; Tosca, M.; Pingitore, G.; Licari, A.; Marseglia, A.; Ciprandi, G.; Caffarelli, C. Component resolved diagnosis and risk assessment in food allergy. Acta Biomed. 2021, 92, e2021528. [Google Scholar] [CrossRef]
- Anagnostou, A.; Abrams, E.M.; Anderson, W.C.; Carver, M.; Eftekhari, S.; Golden, D.B.K.; Jaffee, H.; Lieberman, J.A.; Mack, D.P.; Mustafa, S.S.; et al. Development of a validated, updated North American pediatric food allergy anaphylaxis management plan. Ann. Allergy Asthma Immunol. 2025; online ahead of print. [Google Scholar] [CrossRef]
- Shin, M. Food allergies and food-induced anaphylaxis: Role of cofactors. Clin. Exp. Pediatr. 2021, 64, 393–399. [Google Scholar] [CrossRef]
- Mempel, M.; Rakoski, J.; Ring, J.; Ollert, M. Severe anaphylaxis to kiwi fruit: Immunologic changes related to successful sublingual allergen immunotherapy. J. Allergy Clin. Immunol. 2003, 111, 1406–1409. [Google Scholar] [CrossRef]
- Fiocchi, A.; Restani, P.; Bernardo, L.; Martelli, A.; Ballabio, C.; D’Auria, E.; Riva, E. Tolerance of heat-treated kiwi by children with kiwifruit allergy. Pediatr. Allergy Immunol. 2004, 15, 454–458. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, L.; Shi, J.; Vanga, S.K.; Raghavan, V. Effect of microwave processing on the nutritional properties and allergenic potential of kiwifruit. Food Chem. 2023, 401, 134189. [Google Scholar] [CrossRef] [PubMed]
- Cubells-Baeza, N.; Gomez-Casado, C.; Tordesillas, L.; Ramirez-Castillejo, C.; Garrido-Arandia, M.; Gonzalez-Melendi, P.; Herrero, M.; Pacios, L.F.; Diaz-Perales, A. Identification of the ligand of Pru p 3, a peach LTP. Plant Mol. Biol. 2017, 94, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Khatri, K.; O’Malley, A.; Linn, C.; Kowal, K.; Chruszcz, M. Role of Small Molecule Ligands in IgE-Mediated Allergy. Curr. Allergy Asthma Rep. 2023, 23, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Pomes, A.; Smith, S.A.; Chruszcz, M.; Mueller, G.A.; Brackett, N.F.; Chapman, M.D. Precision engineering for localization, validation, and modification of allergenic epitopes. J. Allergy Clin. Immunol. 2024, 153, 560–571. [Google Scholar] [CrossRef]
- Smith, S.A.; Chruszcz, M.; Chapman, M.D.; Pomes, A. Human Monoclonal IgE Antibodies-a Major Milestone in Allergy. Curr. Allergy Asthma Rep. 2023, 23, 53–65. [Google Scholar] [CrossRef]
- Kerzl, R.; Simonowa, A.; Ring, J.; Ollert, M.; Mempel, M. Life-threatening anaphylaxis to kiwi fruit: Protective sublingual allergen immunotherapy effect persists even after discontinuation. J. Allergy Clin. Immunol. 2007, 119, 507–508. [Google Scholar] [CrossRef]
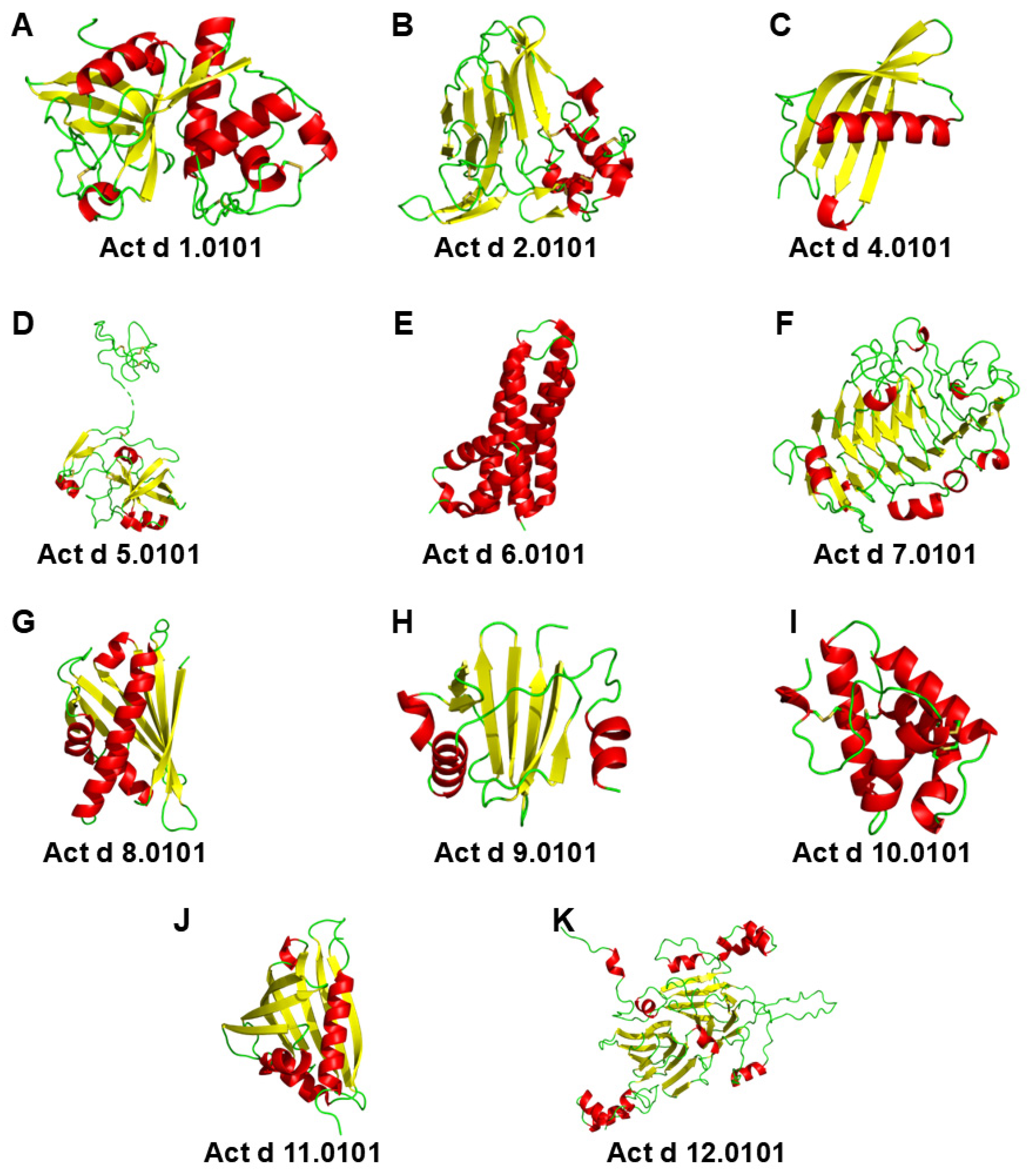
| Actinidia deliciosa (Green Kiwifruit) | Actinidia chinensis (Golden Kiwifruit) | Protein Family Biochemical Name |
|---|---|---|
| Act d 1 | Act c 1 * | Cysteine protease (actinidin) |
| Act d 2 | Act c 2 * | Thaumatin-like protein (TLP) |
| Act d 3 | -- | -- |
| Act d 4 | Act c 4 * | Phytocystatin |
| Act d 5 | Act c 5 | Kiwellin (kissper + KiTH) |
| Act d 6 | -- | Pectin methylesterase inhibitor (PMEI) |
| Act d 7 | -- | Pectin methylesterase (PME) |
| Act d 8 | Act c 8 | Pathogenesis-related protein (PR-10) |
| Act d 9 | -- | Profilin |
| Act d 10 | Act c 10 | Non-specific lipid transport protein (nsLTP1) |
| Act d 11 | -- | Major latex protein/ripening-related protein (MLP/RRP) |
| Act d 12 | -- | 11S globulin (cupin) |
| Act d 13 | -- | 2S albumin |
| IgE Titer (kUA/mL) | |||||
|---|---|---|---|---|---|
| Allergen | Protein | Patient 1 | Patient 2 | Patient 3 | Patient 4 |
| Act d 1 | Cysteine protease | 8.16 | |||
| Act d 2 | TLP | 10.32 | |||
| Act d 5 | Kiwellin | ||||
| Act d 10 | nsLTP | 3.47 | 16.88 | ||
| Aln g 1 | PR-10 | 1.54 | 35.63 | ||
| Alt a 1 | 5.45 | ||||
| Api g 1 | PR-10 | 0.77 | 6.91 | ||
| Api g 2 | nsLTP | 1.23 | |||
| Api g 6 | nsLTP | 1.57 | 0.72 | ||
| Ara h 1 | 7/8 globulin | 4.65 | |||
| Ara h 6 | 2S albumin | 0.97 | |||
| Ara h 8 | PR-10 | 1.59 | 8.26 | 26.13 | |
| Ara h 9 | nsLTP | 5.88 | 10.44 | ||
| Art v 3 | nsLTP | 10.45 | |||
| Bet v 1 | PR-10 | 23.79 | 39.79 | 38.32 | |
| Bet v 2 | Profilin | 7.99 | |||
| Can s 3 | nsLTP | 1.38 | |||
| Cor a 1.0103 | PR-10 | 4.64 | 17.33 | 38.08 | |
| Cor a 1.0401 | PR-10 | 2.99 | 5.51 | 27.13 | |
| Cor a 11 | 7/8 globulin | 3.29 | |||
| Cor a 8 | nsLTP | 4.08 | 1.31 | ||
| Cor a 9 | 11S globulin | 6.26 | |||
| Cuc m 2 | Profilin | 24.43 | |||
| Dau c 1 | PR-10 | 6.08 | |||
| Fag s 1 | PR-10 | 4.31 | 7.23 | 23.49 | |
| Fra a 1 + 3 | PR-10 + nsLTP | 1.04 | 0.89 | 9.12 | 10.48 |
| Gly m 4 | PR-10 | 3.18 | 9.83 | 26.68 | |
| Hev b 8 | Profilin | 6.26 | |||
| Jug r 3 | nsLTP | 0.57 | |||
| Jug r 4 | 11S globulin | 11.47 | |||
| Mal d 1 | PR-10 | 0.93 | 0.67 | 16.36 | |
| Mal d 3 | nsLTP | 4.03 | 2.24 | ||
| Mer a 1 | Profilin | 13.90 | |||
| Phl p 12 | Profilin | 8.32 | |||
| Pho d 2 | Profilin | 19.87 | |||
| Pis v 2 | 11S globulin | 1.84 | |||
| Pis v 3 | 7/8 globulin | 4.48 | |||
| Pla a 3 | nsLTP | 1.01 | |||
| Pru p 3 | nsLTP | 5.16 | 1.60 | ||
| Ses i 1 | 0.53 | ||||
| Sola l 6 | nsLTP | 2.16 | 2.01 | ||
| Tria a 14 | nsLTP | 1.17 | |||
| Vit v 1 | nsLTP | 4.06 | |||
| Zea m 14 | nsLTP | 10.58 | 15.68 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wright, E.M.; O’Malley, A.; Khatri, K.; Pittsley, R.; Offermann, L.R.; Covert, E.; Ruan, T.; Ciardiello, M.A.; Kowal, K.; Chruszcz, M. Kiwifruit Allergy—Molecular Basis, Diagnostics and Treatment. Appl. Sci. 2025, 15, 7182. https://doi.org/10.3390/app15137182
Wright EM, O’Malley A, Khatri K, Pittsley R, Offermann LR, Covert E, Ruan T, Ciardiello MA, Kowal K, Chruszcz M. Kiwifruit Allergy—Molecular Basis, Diagnostics and Treatment. Applied Sciences. 2025; 15(13):7182. https://doi.org/10.3390/app15137182
Chicago/Turabian StyleWright, Elaine M., Andrea O’Malley, Kriti Khatri, Rebekka Pittsley, Lesa R. Offermann, Emily Covert, Tiffany Ruan, Maria Antonietta Ciardiello, Krzysztof Kowal, and Maksymilian Chruszcz. 2025. "Kiwifruit Allergy—Molecular Basis, Diagnostics and Treatment" Applied Sciences 15, no. 13: 7182. https://doi.org/10.3390/app15137182
APA StyleWright, E. M., O’Malley, A., Khatri, K., Pittsley, R., Offermann, L. R., Covert, E., Ruan, T., Ciardiello, M. A., Kowal, K., & Chruszcz, M. (2025). Kiwifruit Allergy—Molecular Basis, Diagnostics and Treatment. Applied Sciences, 15(13), 7182. https://doi.org/10.3390/app15137182








