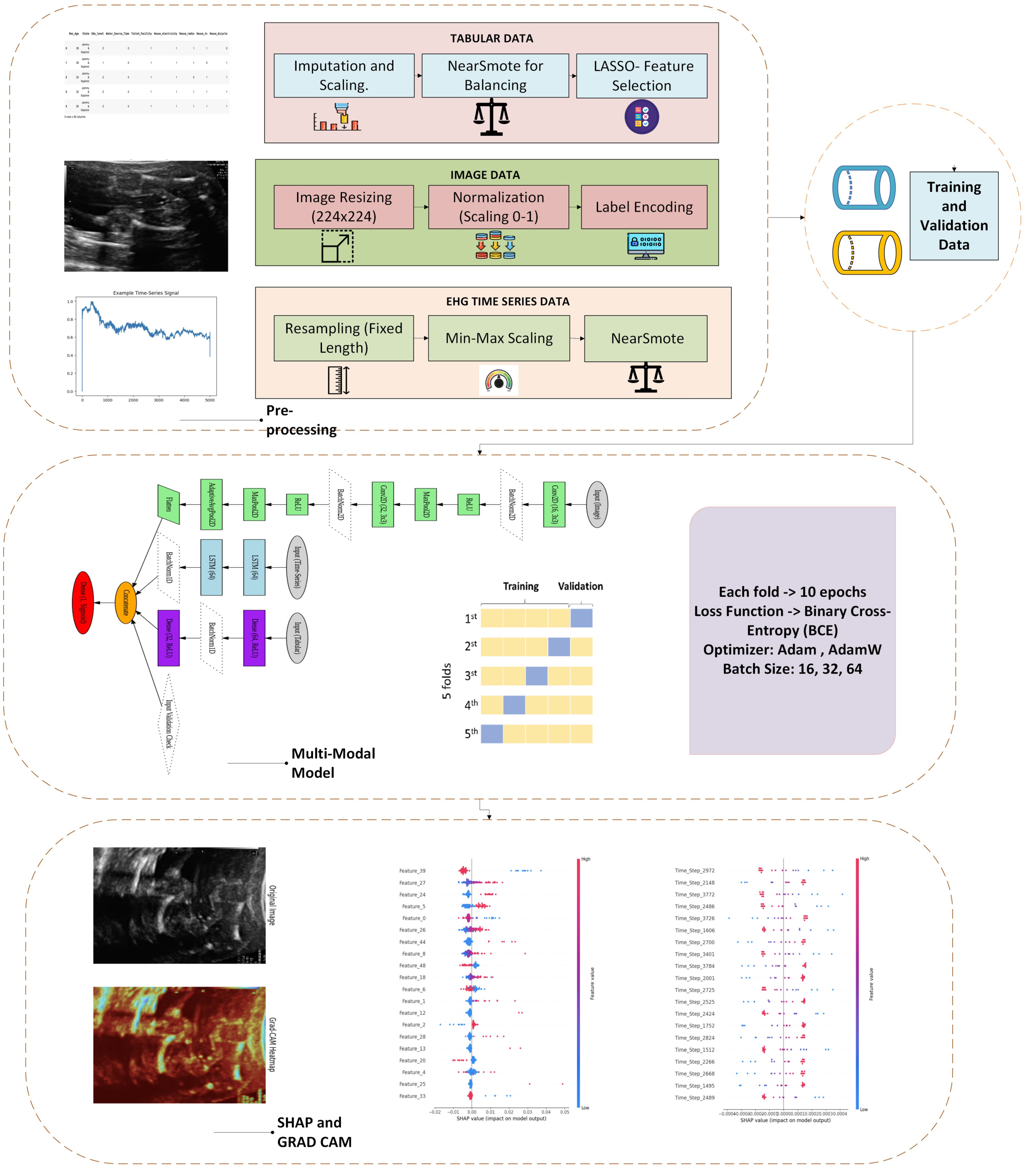
1. Introduction
Pregnancy alters the physiology and structure of a woman’s body in many ways, and it therefore requires close, thorough, and consistent supervision. During this period, mother and fetal health begins to be influenced by numerous clinical, demographic, and lifestyle factors, as depicted in
Figure 1.
However, pregnancy monitoring in today’s world uses rather rough and unsophisticated methods that do not seem to take into consideration the differences among patients. The absence of customized monitoring processes increases the chance of early indicators of conditions that could adversely affect both the mother and newborn. Thus, it becomes critical for modern forms of medical systems to require precise tracking technologies with individuality and prevention for maternal and fetal safety.
AI and particularly ML generate new possibilities to improve pregnancy surveillance systems. In the field of healthcare, the study of machine-learning-applicable systems towards analyzing complex patterns in extensive datasets has proved to be effective. In pregnancy monitoring, the data streams that can be analyzed using ML technologies include clinical scan data, devices with wearables, and demographic information. One of the major datasets available in this domain is the National Family and Health Survey-5 (NFHS-5), containing complete data of 136,136 instances and 95 features. The models that can be built with such an extensive dataset are predictive in the broadest sense of the term. Still, the dataset has its issues, such as class imbalance and feature redundancy, which greatly affect the model’s performance. To meet these challenges, data balancing has been conducted using NearSMOTE, and feature selection has been conducted using a method called the least absolute shrinkage and selection operator (LASSO) to obtain better and reliable prediction models [
1].
Although various features in any large dataset can improve the predictive power of a model, they can also increase complexity, making it more prone to overfitting. These challenges are avoided when we apply feature selection techniques such as the LASSO because it reduces the model complexity and thus its proficiency. This particular optimization is very important for constructing a surveillance system for maternal health. The least absolute shrinkage and selection operator model provides valid predictive modeling capabilities because its optimal selection patterns simplify the model [
1].
Currently, routine pregnancy monitoring remains limited to a great extent by traditional check-up appointments, the use of pencil and paper for data registration, and conventional instruments. These methods do not have real-time assessment of maternal and fetal condition and are worsened by limited resources and unequal access to healthcare. Therefore, the activation of artificial intelligence utilization in the field of maternal health causes positive outcomes regarding these challenges. This approach involves the use of advanced computation mechanisms that analyze various pregnancy-related factors to alert concerned medical practitioners on time [
2].
Deep learning, as a branch of AI, has extended these capabilities to a great extent. Neural networks can handle a huge amount of data and analyze complex data sources that include clinicians’ data of mother and fetus and all kinds of imaging data to provide a data-driven view of a mother’s health condition. The use of explainable AI (XAI) builds on these ideas and improves the utility of these systems by interpreting the results of model interpretation for healthcare providers [
3].
Modern methods of monitoring involve scanning electronic health record (EHR) data for such complications as pre-eclampsia by predicting the readings of blood pressure, proteinuria, and specific patients’ histories to recommend interventions appropriately [
4,
5].
All over the world, and despite the modern development and availability of medical resources, it is worth noting that maternal mortality is still rife and was estimated to be at 287,000 cases in 2020. Maternal health outcomes are influenced not only through biomedical factors but also social, environmental, and systemic determinants; therefore, holistic, multi-faceted, and advanced approaches are required for managing the condition beyond clinical medicine [
6]. Globally, the community healthcare system considers maternal health as one of the critical success factors for social and economic growth. However, complications during pregnancy remain the major cause of hospitalization and mortality among pregnant women, particularly in countries with low to middle incomes (LMICs). In 2017, approximately 295,000 women died following childbirth according to a WHO report. Statistics indicate that 94% of deaths belong to LMIC countries [
7]. The adoption of machine learning and deep learning in the management of maternal healthcare leads to the early prediction of potential high-risk complications, such as preterm labor. In [
8], the ability of ML algorithms to analyze the patient records and data from wearable sensors to predict early labor signs accurately has been demonstrated. However, there are currently systems used to monitor pregnancy that need several devices to be connected to a base unit to monitor the number of objectives, including blood oxygenation, temperature, blood pressure, heart rate, fetal heart rate, and uterine contractions [
9].
Another of the main application areas of maternal care includes the use of methods and tools to detect fetal and maternal diseases. Due to the innovative utility of deep learning, especially convolutional neural networks (CNNs), ultrasonic visualization and examination have improved to make precise diagnoses of fetal structural abnormalities and placental pathologies. Recall that CNNs have obtained accuracy rates of 92% in the task of defect detection in ultrasound images [
10].
A precise image database has been used to a great extent by conducting rigorous research to create procedures that can identify disorders such as spina bifida and cardiac defects. In the same manner, wearable sensor technologies that integrate deep learning extract the vital signs of the mother, such as blood pressure and heart rate, to monitor the clinical well-being of the mother [
11].
Owing to technological improvements in fetal MRI in recent years, the capabilities of describing the microstructures and patterns of fetal brain development in utero with higher spatial resolution have become more advanced. However, the manual segmentation process of fetal brain structure configuration is time-consuming and prone to variability. This has made it possible to segment fetal brain images through deep learning techniques such as CNNs and the U-Net family in dealing with aspects such as motion artifacts, low tissue contrast, and limited annotated samples [
12].
A supplementary aspect of maternal care is the mental health of the woman during her pregnancy. Naturally, recent works have been devoted to the integration of machine learning approaches with methods of prenatal mental health screening that can contribute to the early stages of clinical practice. For instance, Krishnamurti et al. investigated an interpretable model of using first-trimester structured patient-reported data for early-onset high-risk moderate to severe depression during pregnancy. By incorporating such psychosocial factors as food insecurity, the model performed well in terms of predictive accuracy and remained easy to implement, thus underlining the role of mobile-app-based tools in early mental health screening [
13].
The study conducted by Yang et al. [
14] is one of the few that applied the approach of machine learning for early pregnancy loss (EPL) prediction across women who had recurrent pregnancy loss (RPL) based on preconception data only. Hou et al. implemented a Gradient Boosting Machine (GBM) on their study’s dataset to develop a strong and efficient predictive model with an AUC of 0.805 depending on nine features, which include age, BMI, induced abortion, previous pregnancy losses, and homocysteine concentration. This approach ensures that working with AI-based models supports early risk assessment for high-risk pregnancies and clinical decision-making.
The emerging trends in deep learning indicate that multimodal approaches aid in clinical prediction. In [
15], Wang et al. presented MSTP-Net, which is a three-path framework that enhances medical image segmentation through uniting local and global parameters. For instance, the authors in [
16] pointed out that multimodal learning and the attention mechanism in the deep learning framework enable handling of limitations like scarce annotation and lesion variability in medical imaging. These advances also strengthen the importance of AI in improving maternal health monitoring methods and enriching recognition accuracy.
Some of the recent developments show that AI has a significant role in obstetric imaging. Bonnard et al. [
17] presented a Prior-Guided Attribution framework aimed at enhancing ultrasound diagnosis by using spatial priors learned during the dataset training phase. Csillag et al. [
18] designed AmnioML, a deep learning architecture for segmenting amniotic fluid and estimating volume from fetal MRIs, where the proposed network integrated uncertainty quantification. Wang et al. [
19] presented MiTU-Net as a transformer-based U-Net model for the segmentation of fetal head and pubic symphysis, which helped in accurate angle measurement of progression. Together, these milestones indicate surging reliance on AI in enhancing maternal care and reducing the time to diagnose complications.
However, contemporary research further discusses the issues of predictive approaches regarding the processes of maternal care. Bai et al. [
20] discussed sonographic thresholds to guide timely treatment for miscarriage. According to Lee et al. [
21], sperm DNA damage, characterized by double-strand breaks, is said to predict miscarriage in poor-prognosis couples. Qi et al. [
22] developed ML models of postpartum depression based on biopsychosocial information. Hunt et al. [
23] described emergency cesarean models as needing clinically useful tools for delivery planning. This work provides support for the previous research findings on the importance of improving the use of data-driven models to improve maternal health.
1.1. Contributions
The major contributions of this study are as follows:
1.1.1. Model Development
We propose and develop two deep learning architectures—ResRNN-Net (combining Bi-LSTM, CNN, and attention mechanisms) and MultiScaleFusion-Net (utilizing GRU and multiscale convolution)—designed initially for tabular clinical data.
Based on the knowledge from single-modality experiments, we introduce a multimodal deep learning model that integrates tabular features, fetal ultrasound images, and EHG time-series [
24] signals into a unified prediction system.
1.1.2. Methodological Innovations
A robust data preprocessing pipeline is implemented, addressing class imbalance through a hybrid NearMiss-SMOTE strategy and reducing feature redundancy using LASSO feature selection.
Explainability is prioritized through the application of SHAP values (for tabular and time-series data) and Grad-CAM (for imaging data), offering model interpretability that is essential for clinical adoption.
1.1.3. Experimental Rigor
Comprehensive 5-fold stratified cross-validation is used for robust evaluation.
Performance is compared against strong baseline models such as XGBoost, voting ensemble, TabNet, and MLPs.
An in-depth analysis of the trade-offs between precision, recall, computational efficiency, and interpretability is provided.
1.1.4. Real-World Applicability
The proposed models demonstrate strong generalizability, scalability, and transparency, making them viable candidates for integration into clinical maternal health monitoring systems.
2. Related Work
The research by Javed et al. presents two novel ensemble models, Echo Dense Inception Blending (EDI-Blend) and Dense Reservoir Inception Modular Network (DRIM-Net), aimed at predicting miscarriage risks by applying advanced data analytics techniques, particularly deep learning (DL). The study utilizes a balanced dataset processed through the hybrid balancing technique NearSMOTE and features selected using the least absolute shrinkage and selection operator (LASSO). The models are validated using 10-fold cross-validation, with EDI-Blend achieving notable performance metrics such as 0.732 accuracy and 0.721 ROC–AUC, while DRIM-Net outperformed with 0.769 accuracy and 0.837 ROC–AUC. The results highlight the potential for these models to aid healthcare professionals in early detection and intervention, ultimately improving patient outcomes [
1].
Through real-time data collection, healthcare providers obtain pregnancy-related health information about their patients that helps them to spot potential medical complications early. Wearable sensors possess the capability to send notifications about vital sign changes that indicate pre-eclampsia or gestational diabetes to both patients and healthcare staff [
11].
Bada et al. discuss a novel hypertension prediction model by merging deep learning (DL) with transfer learning (TL) techniques to escalate early detection and lessen the risk of cardiovascular diseases. Their perspective entails fine-tuning a pre-trained Feed-Forward Deep Neural Network (FFDNN), initially prospered for diabetes prediction using the PIMA dataset, and modifying it for hypertension prediction using the PPG-BP dataset. They link a hybrid feature selection strategy integrating statistical methods and domain expertise, choosing four major features—systolic and diastolic blood pressure, heart rate, and age—to upgrade the model performance. The TL strategy incorporates freezing pre-trained layers and re-training recently added layers to confirm efficient knowledge transfer while decreasing training time. The final model arrived at an accuracy of 81.34%, with 88% precision and 80% recall, evidencing a strong balance between identifying hypertensive patients and reducing false positives. This effort focuses on the potential of transfer learning to reshape the existing healthcare models for associated diagnostic tasks, especially in data-scarce environments [
25].
Barredo Arrieta et al. address the requirement to provide understandable explanations, which faces additional challenges because various stakeholders, including patients, clinicians, and developers, possess different needs. Implementing XAI into clinical operational systems becomes difficult due to the requirement of avoiding workflow disruptions [
26].
Gao, Osmundson et al. discuss various deep learning approaches that have multiple applications in pregnancy monitoring to solve crucial maternal and fetal health problems. The prediction of preterm birth stands as the most essential deep learning application because it prevents neonatal mortality and morbidity. Studies conducted by [
27] confirm DL models’ achievement to accurately predict risk of extreme preterm birth.
The research by Fiorentino, Villani et al. extensively assesses deep learning applications for fetal anomaly detection, where CNNs demonstrate high precision for anomaly recognition. The detection models achieve outstanding results in identifying minor structural anomalies, which leads to early identification of diseases and quick medical help [
28].
The research by Islam, Mustafina et al. presents an extensive review about machine learning systems for pregnancy outcome prediction while demonstrating their ability to improve maternal healthcare with data-based knowledge. The work integrates previous models into hierarchies that group together models based on clinical parameters and demographic factors, fetal health indicators, and more features. Additionally, the authors state that machine learning has improved early risk assessment for gestational diabetes and preterm birth, yet obstacles with balancing data and interpretation difficulties alongside generalization issues block its extensive adoption [
29].
Machine learning explainability techniques deliver information about the prediction processes used by models. In their research, Tempel, Groos et al. compare SHAP and Grad-CAM for human activity recognition through a comparative analysis. These researchers demonstrate that SHAP provides optimal feature-level explanations through its quantitative analysis, which lets users track individual input feature contributions; thus, it excels at structured and tabulated datasets. On the other hand, the visual nature of Grad-CAM produces clear heatmaps showing which image sections drive the model toward its decision outcome [
30].
Margret et al. introduce an extensive survey on the application of machine learning (ML) and deep learning (DL) box models in maternal healthcare, with an emphasis on reducing maternal mortality. The survey classifies models into white, gray, and black boxes based on interpretability and estimates their efficacy across numerous predictive tasks, such as fetal monitoring, pre-eclampsia detection, and childbirth outcome prediction. Composed of 71 studies, the review illuminates the growing role of AI-driven clinical decision support systems (CDSSs) while also observing their underutilization in the existing research. The researchers underscore the importance of manipulating diverse data sources, including electronic health records, imaging, and behavioral data, and advise to improve model transparency and ethical deployment, explicitly in resource-constrained settings. Medical imaging stands better due to the system that identifies the main differences in images above what traditional assessment tools can detect [
31,
32].
Sun, Zou et al. present a deep learning model that uses EHR data to forecast preterm birth delivery with excellent accuracy levels while creating time for necessary preventive healthcare actions for at-risk pregnancies [
33]. The research by Du, McNestry et al. discusses that the implementation of CDSSs adds multiple capabilities at different levels of health delivery, which minimizes errors in medication prescription and adverse incidents while enhancing clinical oversight, lowering costs, delivering administrative tasks, supporting diagnosis functions and decision systems, and simplifying medical operations [
34].
The research by Shaheen, Javed et al. examines a DL-based CDSS for gestational diabetes management that resulted in better patient results and decreased healthcare expenses. The research demonstrates that DL has the potential to advance pregnancy medical care through its ability to deliver prompt data-based insights. The studies showcase obstacles as well as benefits since healthcare systems require better interface design and adaptation of DL models into the current healthcare infrastructure [
35]. These comparative approaches are summarized in
Table 1, highlighting their respective outcomes, benefits, and limitations in prenatal care.
Ukrit et al. form a web-based maternal health risk forecasting model using machine learning classifiers and IoT-collected vital signs. Within the spectrum of models tested, an ensemble of random forest, AdaBoost, and Gradient Boosting with soft voting secures the highest accuracy of 81.5%. The research highlights the usefulness of ensemble learning in improving pregnancy risk classification for clinical decision support [
36].
Li et al. propose PregnancyLine, a visual analysis system that improves pregnancy care and risk communication by revealing temporal medical data through intuitive visual metaphors. The system assists doctor–patient interaction, promotes early detection of abnormalities, and aids pregnant women in understanding their examination results and associated risks. The evaluation results display improved comprehension and reduced anxiety among users [
37].
Continuous accumulation of data provides a broad array of information on maternal health and makes it easier for healthcare examiners to examine the changes in pregnancy health over a given period. The integration of DL and IoT systems improves the predictive analysis of societies while availing the early prenatal facilities that help organizations to achieve better health of new mothers and their babies [
38]. XAI develops methods to make artificial intelligence frameworks explain their choices in essential environments, including healthcare. However, various limitations block the widespread acceptance of XAI systems. The primary difficulty faced by interpretable artificial intelligence involves choosing between understandable prediction systems that provide limited precision and advanced prediction methods that offer enhanced precision [
39].
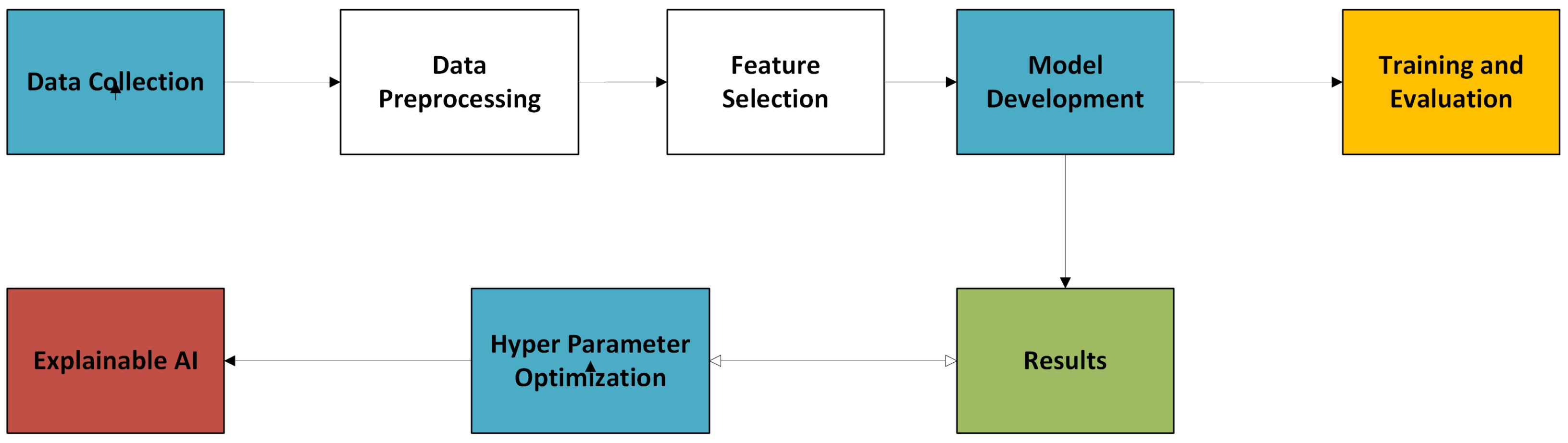
3. Architectural System Design
This section describes the overall approach used to create the deep learning framework for personalized pregnancy tracking. This includes dataset acquisition, preprocessing, feature selection, and model building.
3.1. Dataset Acquisition
The research utilizes the National Family Health Survey-5 (NFHS-5) dataset, which is publicly available and can be accessed via platforms such as Kaggle (
https://www.kaggle.com/datasets/ravisinghiitbhu/nfhs5, accessed on 1 February 2025). The dataset contains 136,136 samples and 95 variables. The variables include essential clinical, demographic, and lifestyle factors related to maternal health, such as BMI, hemoglobin concentration, blood pressure resolution, age, education, previous pregnancy, or if they have breastfed, etc. The target variable shows whether there is the presence or absence of pregnancy complications, modeled as a binary classification problem [
40].
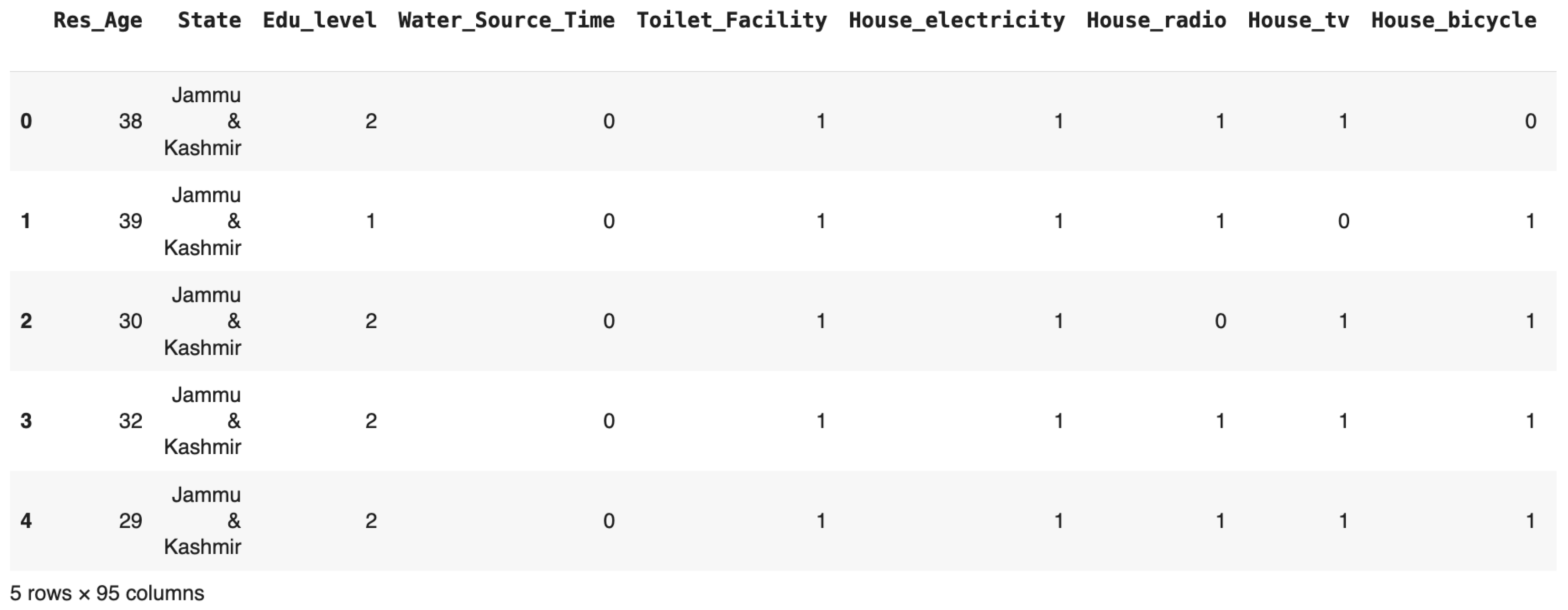
Figure 2 presents a snapshot of the NFHS-5 dataset, illustrating features such as respondent age, education level, household assets, and healthcare access. This tabular view highlights the diversity and richness of the collected data, forming the foundation for pregnancy complication prediction.
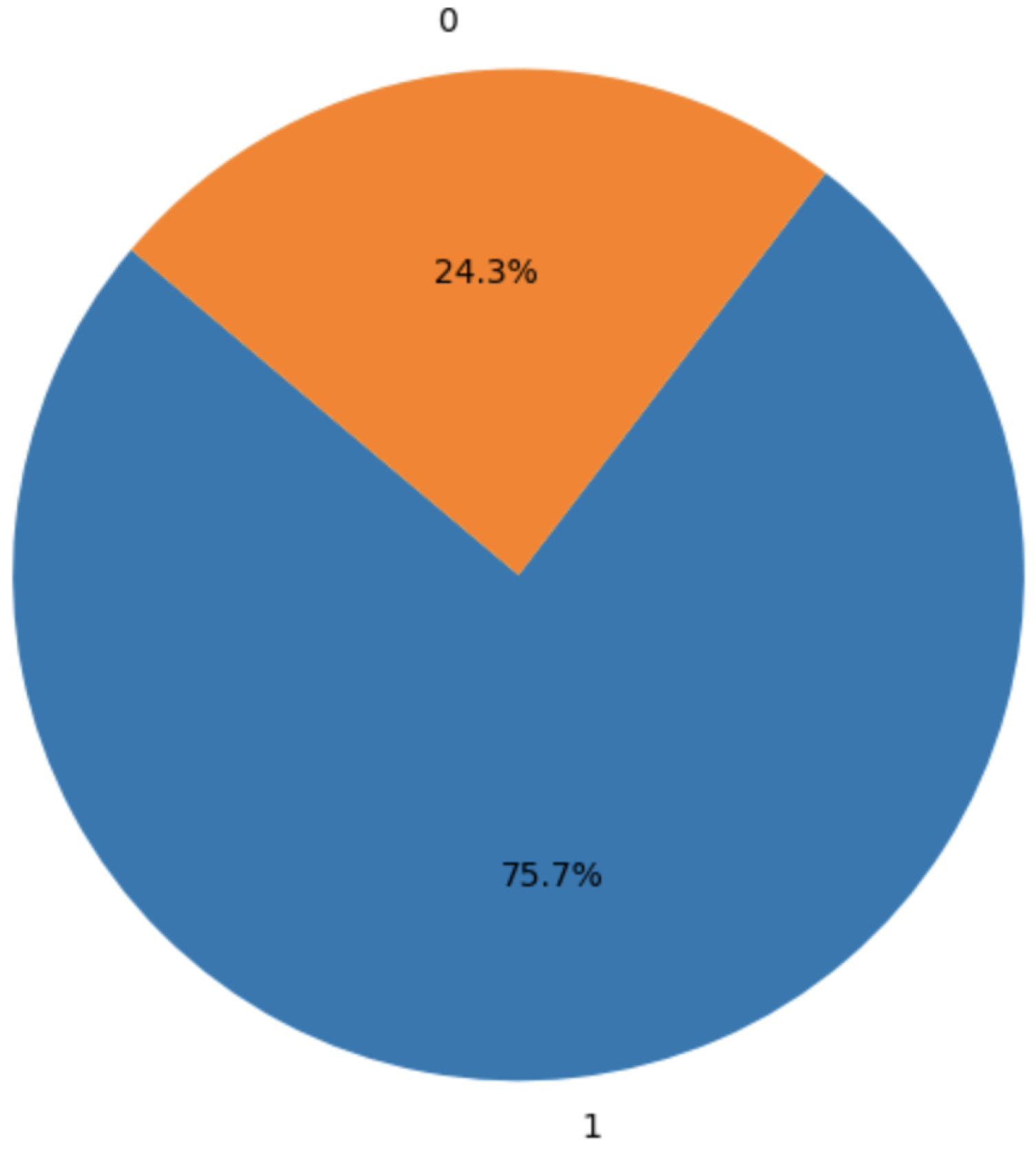
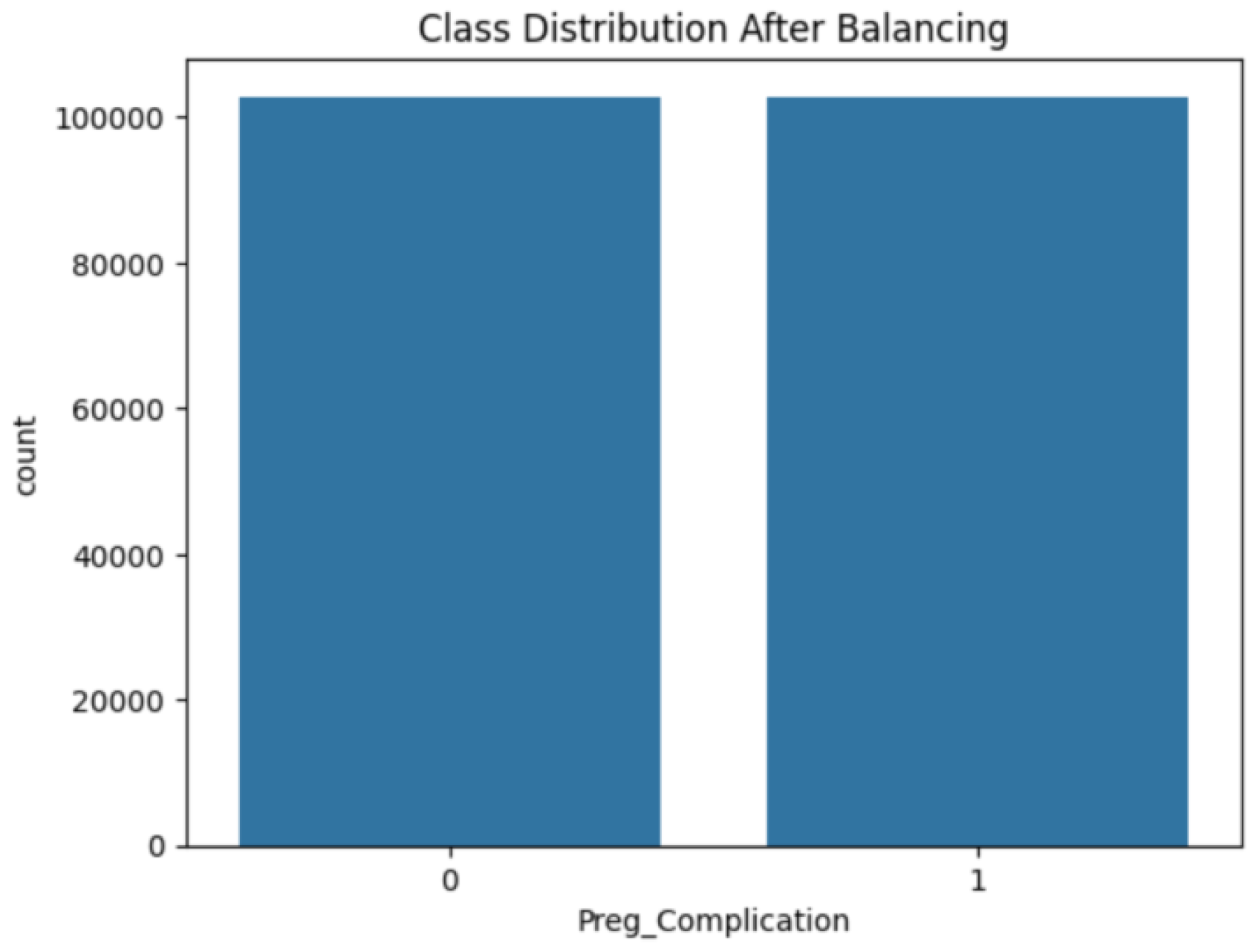
Figure 3 depicts the class imbalance in the target variable, with approximately 75.7% of instances indicating pregnancy complications and 24.3% representing non-complications. This imbalance was addressed using the NearMiss-SMOTE technique during preprocessing.
3.2. Data Preprocessing
To prepare the dataset, preprocessing steps were applied:
Missing values were imputed using mean imputation for numerical features and mode imputation for categorical variables.
Categorical features were one-hot-encoded for machine learning compatibility.
Numerical features were normalized using StandardScaler to ensure zero mean and unit variance.
A correlation heatmap (
Figure 4) was generated to identify redundant features.
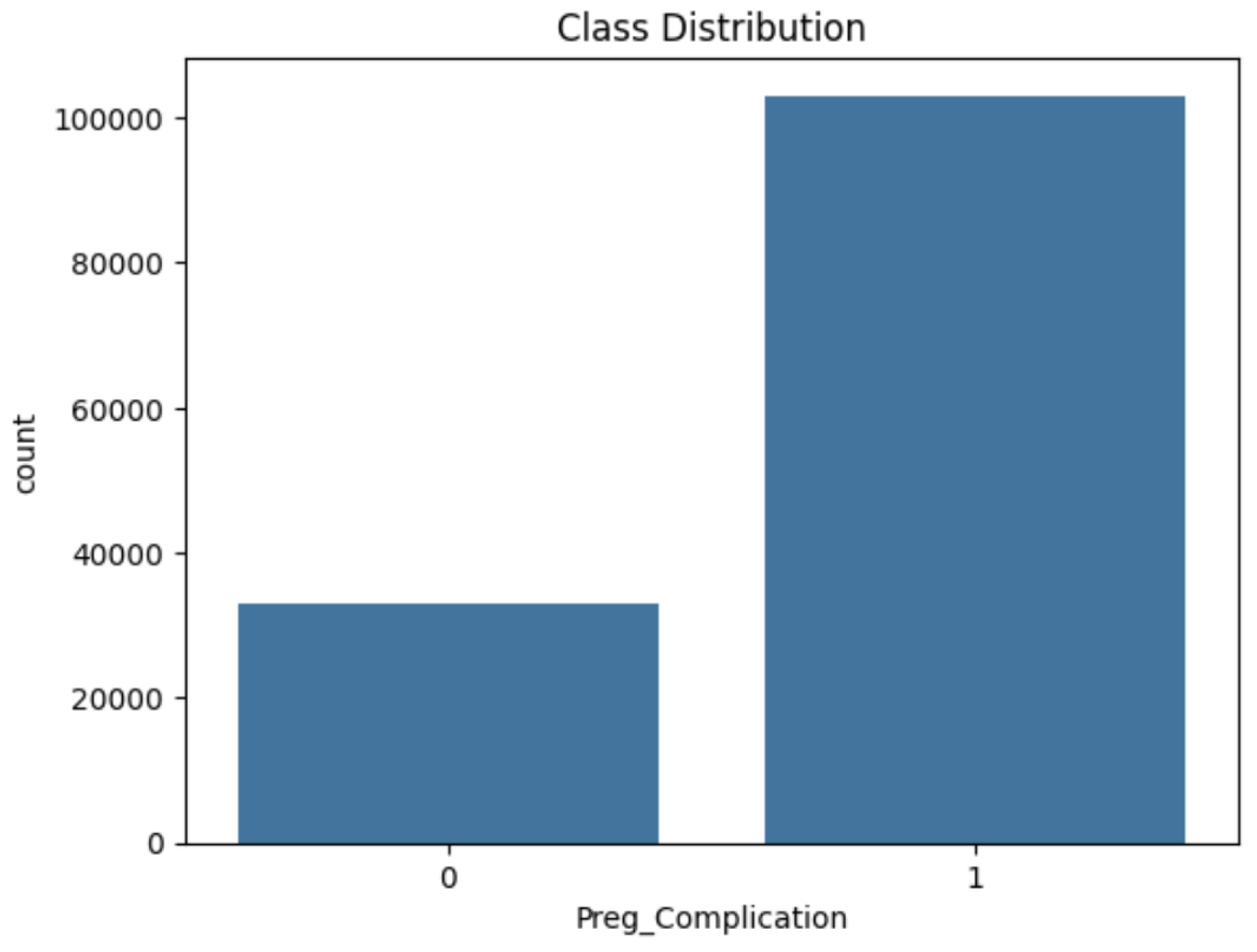
3.3. Addressing Class Imbalance
A hybrid approach combining NearMiss under-sampling and SMOTE over-sampling was applied to address class imbalance. The final class distribution is illustrated in
Figure 5 and
Figure 6.
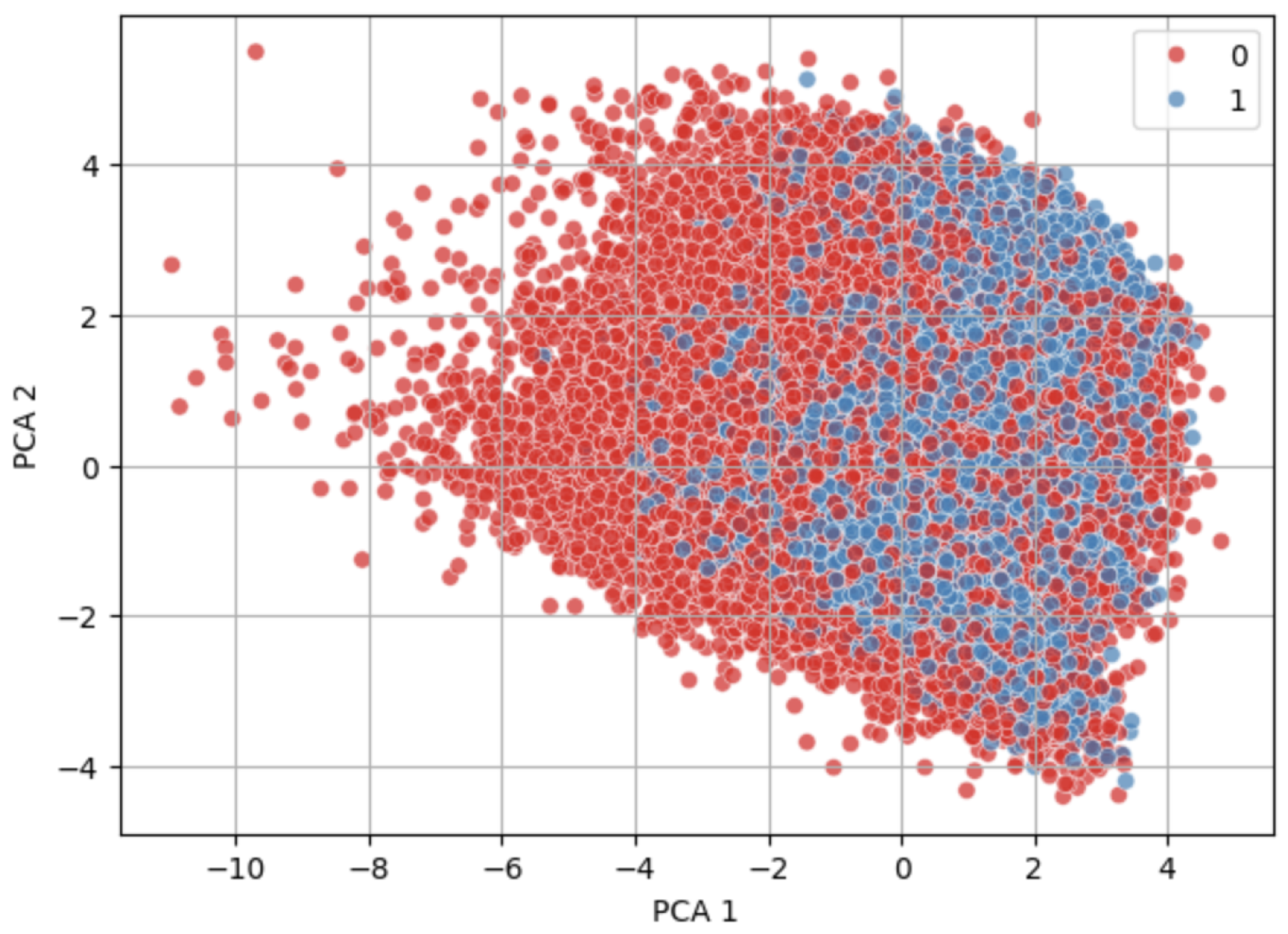
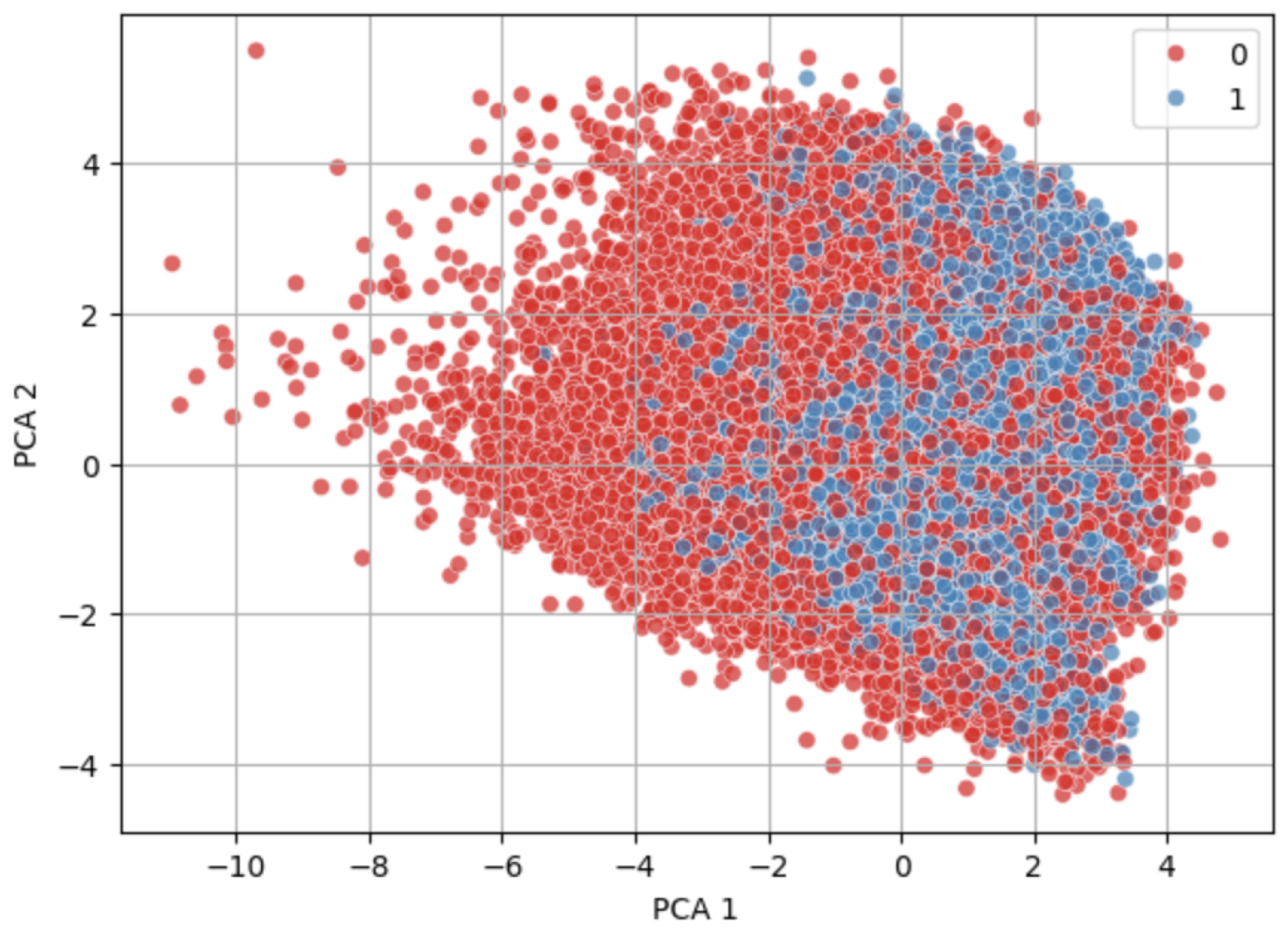
Figure 7 and
Figure 8 show visually similar distributions before and after NearMiss-SMOTE, indicating that the synthetic samples generated by NearMiss-SMOTE follow the original data’s structure without introducing significant outliers.
Table 2 presents a comparative evaluation of the model’s performance with and without the NearMiss-SMOTE technique, illustrating its impact on recall and F1-score.
3.4. Feature Selection Using LASSO
The LASSO (least absolute shrinkage and selection operator) was applied to reduce high-dimensional features to the most relevant predictors. The regularization parameter () was selected using 5-fold cross-validation, automatically optimized via LassoCV from scikit-learn. The cross-validation procedure ensures a balance between underfitting and overfitting, enabling selection of the most predictive yet sparse feature subset.
After extensive tuning, a value of was found to produce the best trade-off between model sparsity and performance. This value effectively shrank irrelevant coefficients to zero while preserving the most informative features for predicting pregnancy complications.
The objective function minimized by the LASSO is
Here, controls the regularization strength, and denotes the coefficients. Features with zero coefficients are excluded.
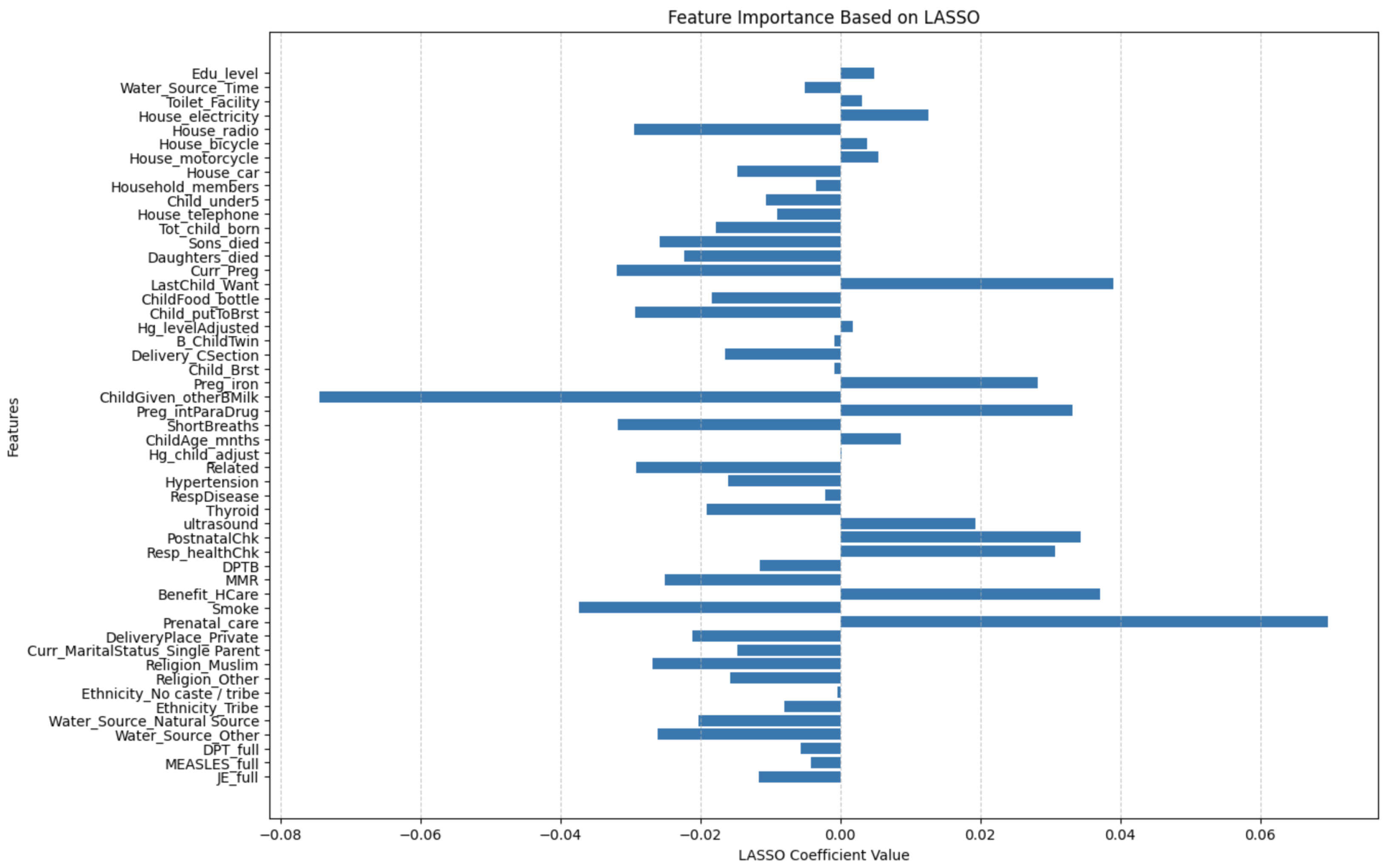
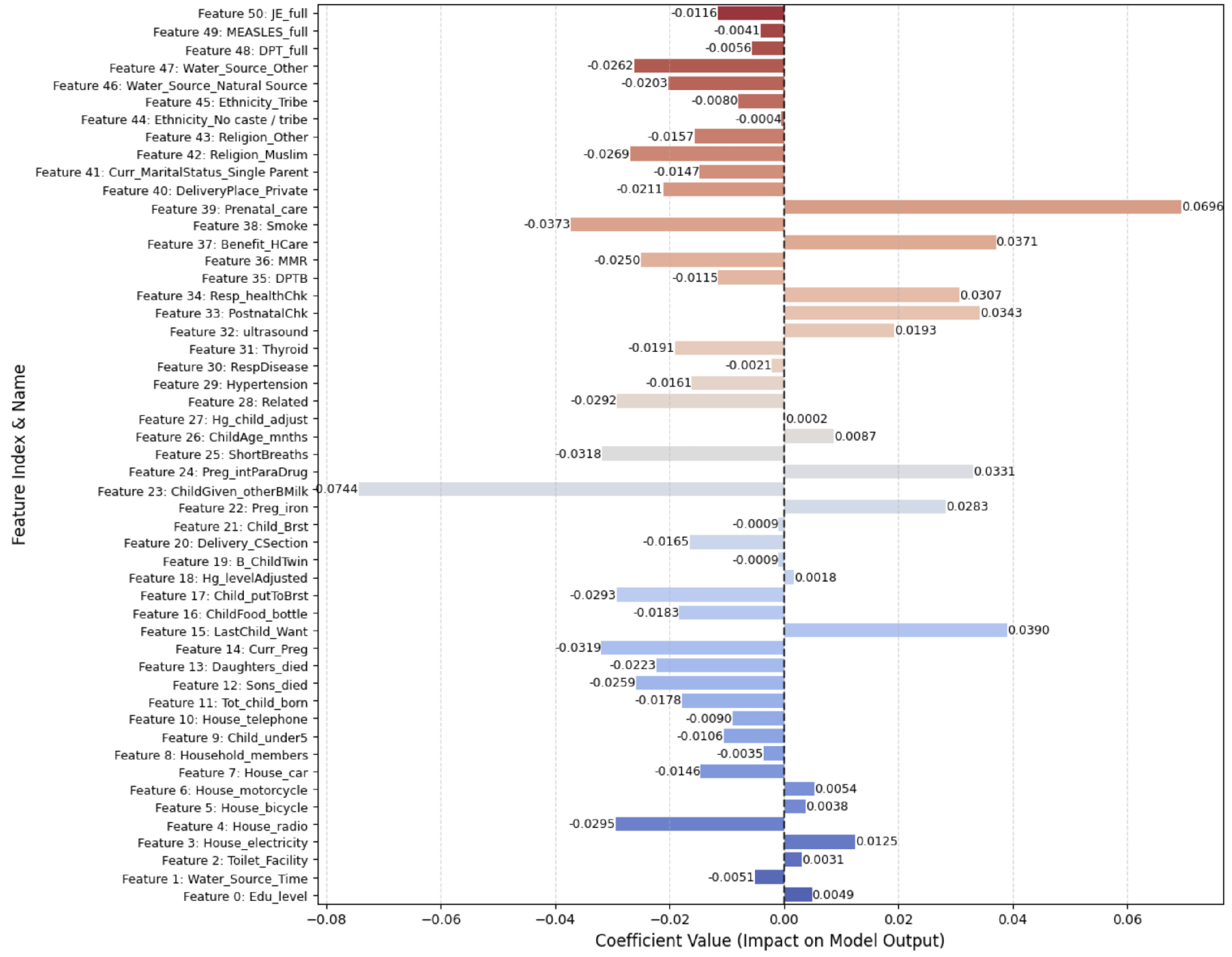
Figure 9 visualizes the non-zero (most impactful) features identified after regularization, reducing the dataset to 51 key features, some of which include
Sons_died, Daughters_died, Curr_Preg, LastChild_Want, ChildFood_bottle, Child_ putToBrst, Hg_levelAdjusted, B_ChildTwin, Delivery_CSection, Child_Brst, Preg_iron, ChildGiven_otherBMilk, Preg_intParaDrug, ShortBreaths, ChildAge_mnths, Hg_child_adjust, Related, Hypertension, RespDisease, Thyroid, ultrasound, PostnatalChk, Resp_healthChk, DPTB, MMR, Benefit_HCare, Smoke, Prenatal_care, DeliveryPlace_Private, Water_Source_ Natural Source, Water_Source_Other, DPT_full, MEASLES_full, JE_full.
3.5. Model Development
Three deep learning architectures were implemented:
ResRNN-Net: Combines Bi-LSTM, CNN, and an attention mechanism to capture sequential dependencies.
MultiScaleFusion-Net: Utilizes GRU and multiscale convolutions for robust feature extraction.
Multimodal Model: Integrates tabular, imaging, and time-series data for comprehensive pregnancy monitoring.
3.6. Baseline Machine Learning Models
To set a performance baseline for deep learning models, some of the conventional machine learning models were deployed and tested. These models provided an idea of how classical methods could forecast pregnancy complications based on the NFHS-5 dataset.
XGBoost (Extreme Gradient Boosting): XGBoost is a powerful ensemble-based boosting algorithm known for its efficiency and accuracy. It builds decision trees sequentially, with each tree correcting errors from the previous one. The model was optimized using hyperparameter tuning to balance bias and variance effectively.
Random Forest: This ensemble method builds several decision trees at training time and returns the mode of the classes (classification) or the average prediction (regression). Its power is in preventing overfitting while having high predictive accuracy by randomized feature selection and bootstrapped sets.
Logistic Regression: Logistic regression, being a baseline model, offered a simple method of binary classification. It predicts the probability of a class given a linear combination of input features and played a critical role in establishing the minimum attainable performance. Each model was trained on the preprocessed dataset, with the hyperparameters tuned via randomized search.
3.7. System Overview: High-Level Architecture of the Pregnancy Monitoring Framework
The pregnancy monitoring framework was designed as an end-to-end system integrating state-of-the-art deep learning techniques with advanced data processing and explainability tools. The system follows a sequential modular pipeline, as illustrated in
Figure 10.
The system starts with the collection of the National Family and Health Survey-5 (NFHS-5) dataset. This dataset contains 136,136 records and 95 features spanning clinical, demographic, and lifestyle aspects of maternal health. In stage two, missing values are addressed through mean imputation for numerical features and mode imputation for categorical features. Numerical features are standardized to provide even scaling, whereas the sole categorical feature, ‘State’, receives one-hot encoding. To improve model efficacy and readability, the LASSO (least absolute shrinkage and selection operator) was used to scale down the dataset into 51 vital features responsible for making accurate predictions of pregnancy complications. Next, three deep neural network models were constructed: ResRNN-Net based on Bi-LSTM, CNN, and attention techniques, MultiScaleFusion-Net using GRU and multiscale convolution blocks for stronger feature fusion, and the multimodal model, which integrates images through CNN, time series by LSTM, and tabular by MLP.
Models were trained with 5-fold stratified cross-validation to have data splits balanced for all folds. Accuracy, precision, recall, F1-score, and ROC–AUC were used as the evaluation metrics. Then, search was used to optimize the learning rate, batch size, and dropout rate to achieve optimal model performance. For model explanation, SHAP (SHapley Additive exPlanation) and Grad-CAM (gradient-weighted class activation mapping) were used, providing global and local understanding of feature importance. The final outcomes were assessed according to model performance criteria, providing robust predictions and explainable insights to inform healthcare decisions. This modular structure guarantees the system’s scalability, stability, and clinical validity, making it flexible for operational deployment in real-world healthcare environments.
3.8. System Architecture
The system architecture comprises five main elements: data preprocessing, feature selection, model development, training and evaluation, and performance metrics with explainable AI.
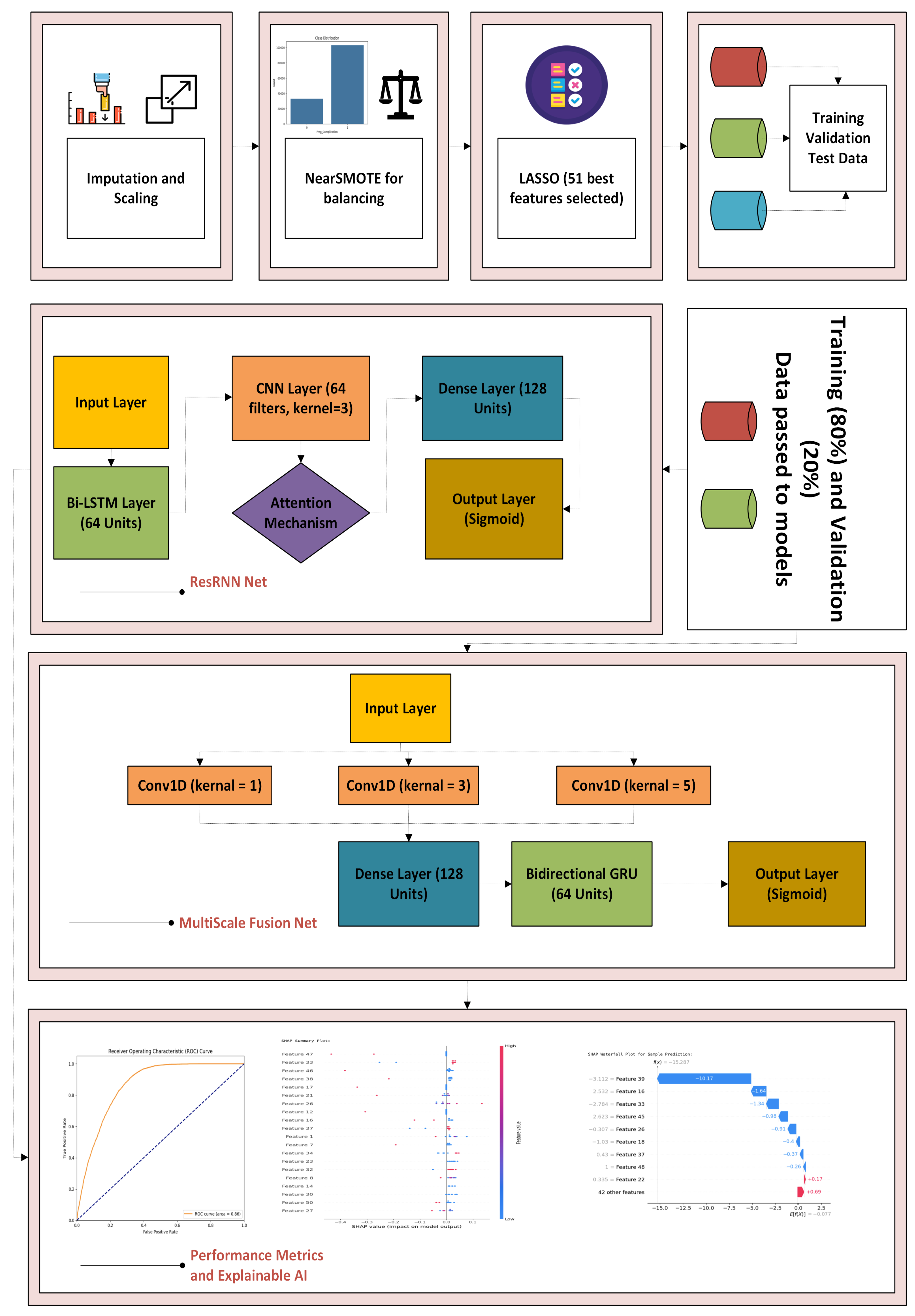
3.8.1. ResRNN-Net and MultiScaleFusion-Net
Data Preprocessing and Balancing: The NFHS-5 dataset was processed to handle inconsistencies. Missing values were imputed using mean (numerical) and mode (categorical). StandardScaler was applied for feature normalization. Class imbalance was addressed using a hybrid NearMiss under-sampling and SMOTE over-sampling technique.
Feature Selection: The LASSO was applied to select 51 highly predictive features, reducing dimensionality and improving model efficiency.
Model Development: Two deep learning architectures were constructed:
ResRNN-Net: A hybrid architecture combining Bi-LSTM (64 units) for sequential learning, CNN (64 filters, kernel size = 3) for spatial feature extraction, an attention mechanism for feature prioritization, and a dense layer (128 units) for final predictions.
MultiScaleFusion-Net: This model has a Bidirectional GRU (64 units), and it captures multiscale dependencies using Conv1D layers with kernel sizes of 1, 3, and 5. Outputs are concatenated and processed through a dense layer (128 units) followed by a sigmoid classifier.
The architectural choices of ResRNN-Net and MultiScaleFusion-Net are based on the nature and complexity of the NFHS-5 dataset. While the data are tabular, several variables exhibit logical progressions or temporal dependencies—such as antenatal visit sequences, immunization coverage, and time-related lifestyle changes. To exploit these implicit patterns, ResRNN-Net integrates Bi-LSTM for capturing sequential patterns, CNN for extracting local feature interactions, and attention mechanisms for prioritizing influential signals. On the other hand, MultiScaleFusion-Net employs GRU units for lightweight sequence modeling and leverages multiscale Conv1D filters to capture patterns at varying granularities across feature subsets. These architectures were selected to balance performance, interpretability, and computational efficiency for personalized maternal health prediction tasks.
Training and Evaluation: The data were divided into training (80%) and validation subsets (20%). Deep learning models were trained utilizing the Adam optimizer with a learning rate of 0.0005 and a batch size of 64. Overfitting was avoided with early stopping (patience = 3 epochs) and also learning rate reduced dynamically on plateau in performance by using the ReduceLROnPlateau callback. For rigorous evaluation, 5-fold stratified cross-validation was utilized. Metrics used to evaluate models included accuracy, precision, recall, F1-score, and ROC–AUC, thereby ensuring balanced analysis across all aspects of performance.
Algorithm 1 illustrates the steps of ResRNN-Net and MultiScaleFusion-Net framework.
| Algorithm 1: ResRNN-Net and MultiScaleFusion-Net with Ensemble Learning |
Input: Scaled input features , binary labels y, input shape s, number of dense blocks L, growth rate k, selected block indices Output: Binary prediction
1: Perform stratified train/validation split 2: Apply SMOTE on training data: 3: Compute class weights: 4: Define ResRNN-Net: Reshape: Apply Bi-LSTM: CNN path: Attention vector: Final representation: Output: 5: Define MultiScaleFusion-Net: Reshape: for each to L do if then Add Dense Block with growth rate k Add Transition Layer end if end for Apply multiscale conv: Apply Dense Block: GRU block: Final output: 6: Train XGBoost: Use early stopping with validation set to obtain 7: Train Voting Classifier: Combine random forest and logistic regression (soft voting) Output: , 8: Return: Predictions from all models: |
3.8.2. Multimodal Model
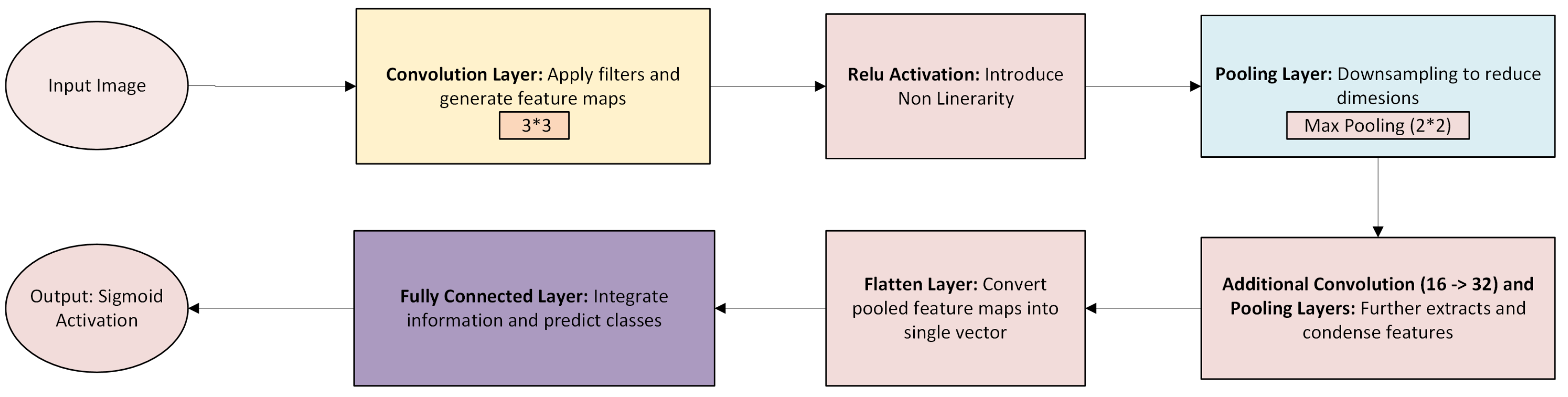
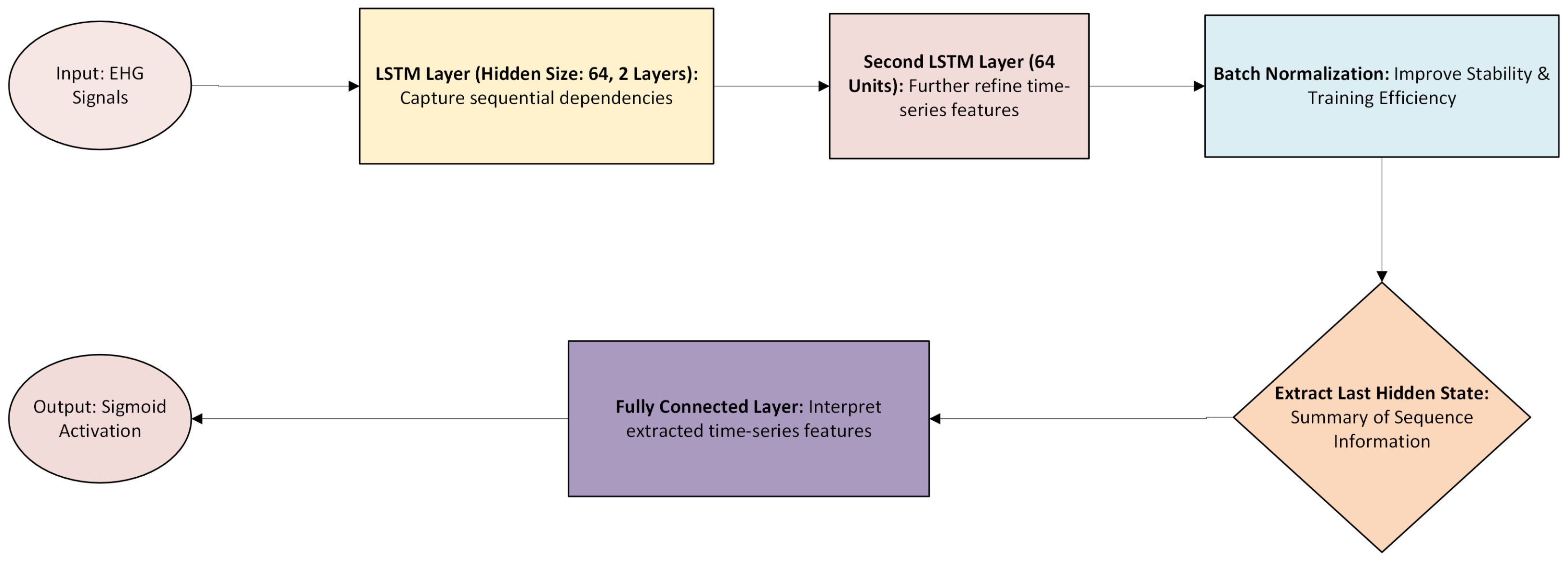
To predict pregnancy complications, convolutional neural networks (CNNs) are designed for ultrasound images, long short-term memory (LSTM) networks for electrohysterogram (EHG) time series, and multi-layer perceptron (MLP) networks for patient health data are used. The overall architecture is shown in
Figure 11.
Data Preprocessing and Balancing: Fetal ultrasound images were resized to 224 × 224 pixels, normalized, and augmented for CNN training. Electrohysterogram (EHG) signals were resampled, min–max-scaled, and class-balanced using SMOTE. Tabular data underwent imputation, feature normalization, and LASSO-based selection to ensure seamless integration across modalities.
Model Development: The multimodal model consists of three key components:
Convolutional Neural Networks (CNNs): Used for analyzing ultrasound images and detecting fetal anomalies.
(Convolution + ReLU activation: implemented as
nn.Conv2d followed by
nn.ReLU())
(Pooling operation:
nn.MaxPool2d)
(Final [-25]feature vector via global pooling and flattening: nn.AdaptiveAvgPool2d + nn.Flatten)
Long Short-Term Memory (LSTM) Networks: Used for processing EHG time-series data and detecting uterine contraction patterns.
(Forget gate computation) (Output gate computation) (Final LSTM output: implemented using nn.LSTM, taking last hidden state)
Multi-Layer Perceptron (MLP): Used for structured tabular data, capturing demographic and clinical factors.
(First dense layer with ReLU activation:nn.Linear → ReLU) (Second dense layer, producing final tabular embedding)
Outputs from MLP (
Figure 12), CNN (
Figure 13), and LSTM (
Figure 14) models are fused in a shared integration layer before making final predictions. This fusion mechanism ensures that tabular, spatial, and temporal information contributes to more accurate predictions.
Training and Evaluation: The model was trained using the Adam optimizer (learning rate = 0.0005) and a batch size of 16/32. Early stopping (patience = 3 epochs) prevented overfitting, while the ReduceLROnPlateau scheduler dynamically adjusted the learning rate upon performance plateau. A 5-fold stratified cross-validation approach was applied to ensure model robustness across different data distributions.
Performance Metrics and Explainable AI: SHAP was used for interpreting feature contributions in tabular and time-series inputs, while Grad-CAM visualizations provided heatmaps highlighting critical regions in ultrasound images, ensuring model transparency.
3.9. Ethical Considerations
This study utilized the publicly accessible and fully anonymized NFHS-5 dataset, provided by the Ministry of Health and Family Welfare, Government of India. As a secondary dataset, it contains no personally identifiable information, and no direct interaction with human subjects occurred. All preprocessing and analysis steps were conducted in accordance with data privacy regulations (
Figure 15).
4. Results
To guarantee the generalization and robustness of the constructed deep learning models, ResRNN-Net, MultiScaleFusion-Net, and the multimodal model underwent a stringent cross-validation scheme. The multimodal model was evaluated separately in two experimental settings.
Training solely on tabular data (NFHS-5 dataset);
Training on a fusion of tabular, image, and time-series data.
4.1. Cross-Validation Strategy
The dataset was divided employing 5-fold stratified cross-validation such that every fold had an equal ratio of positive and negative pregnancy complications. This approach enabled deep learning models to be trained and tested on several data splits, minimizing variance and providing better estimation of model performance. For ResRNN and MultiScaleFusion-Net, in every fold, the data were split into 80% training and 20% validation subsets. Due to the heterogeneous nature of the datasets, special measures were taken for the images, the data were augmented to prevent overfitting, and, for the time series, the data were segmented and processed with padding to maintain sequence length consistency, and then a combination strategy was adopted where each fold contained a balanced subset of all three data types.
4.1.1. Model Training and Evaluation Using Cross-Validation
In both the folds, ResRNN-Net and MultiScaleFusion-Net were trained for 10 epochs with a batch size of 32. The models were validated on the validation set during training using the following key performance metrics: accuracy (overall accuracy of predictions), precision (proportion of true positive predictions out of all positive predictions), recall (capacity to spot true positive cases), F1-score (harmonic mean of precision and recall), and ROC–AUC (robustness over different thresholds). The performance metrics were estimated from the validation set using the evaluation function and contrasted against true labels, with each performance measure being computed. The performance metrics for the final model were obtained by averaging the results from all five folds. ResRNN-Net had an F1-score of 0.82, an ROC–AUC of 0.79, and an accuracy of 79%, which is indicative of excellent predictive power. Likewise, MultiScaleFusion-Net achieved an F1-score of 0.82, an ROC–AUC of 0.80, and an accuracy of 80%, which is very close to the performance of ResRNN-Net. A detailed comparison of the cross-validation results for both models is presented in
Table 3.
For the multimodal model, two experiments, as shown in
Table 4, were observed and both were trained with 10 epochs, 32 as the batch size, BCE-Loss, and an Adam optimizer with a learning rate of 0.001. The tabular-only model achieved 80% accuracy with balanced performance across all the metrics, while the integrated multimodal model had higher recall (100%), indicating that it detected all the positive cases but at the cost of lower precision (55%).
4.1.2. Advanced Model Training and Evaluation
For increased model refinement, an advanced evaluation and training scheme was adopted involving addressing class imbalance, efficient pipelines, early termination, and maximized threshold learning. The measures ensured stable performance while avoiding significant overfitting. The data were shown to have extreme class imbalance, with complications more common than non-complications. To counter this, SMOTE was used only on the training set, creating synthetic samples for the minority class. Class weights were also calculated to provide greater significance to under-represented samples, encouraging balanced learning. An efficient data pipeline was produced based on TensorFlow’s tf.data API. This pipeline enabled effective batch processing, shuffling, and prefetching, minimizing training bottlenecks. A batch size of 128 was used to trade off computational efficiency against memory usage. Both ResRNN-Net and MultiScaleFusion-Net were trained with the Adam optimizer (learning rate 0.0005) and binary cross-entropy loss. Early stopping (patience of two epochs) avoided overfitting, and ReduceLROnPlateau reduced the learning rate by half upon performance plateau. In order to optimize the F1-score, predictions were tested over thresholds from 0.3 to 0.7. The optimal threshold that yielded the highest F1-score was chosen for ultimate testing. To compare the deep learning models, XGBoost and a voting ensemble (random forest + logistic regression) were trained under the same conditions. XGBoost utilized early stopping, whereas the ensemble utilized soft voting for stable predictions.
Table 5 illustrates the performance metrics of all the models, wherein ResRNN-Net and MultiScaleFusion-Net stood out as the best. The outcome proved that both deep learning models—ResRNN-Net and MultiScaleFusion-Net—performed better than the conventional models, with higher F1-scores, recall, and ROC–AUC. MultiScaleFusion-Net showed slightly higher recall, while ResRNN-Net maintained better balance across all the measures.
4.1.3. Hyperparameter Tuning for ResRNN-Net and MultiScale Fusion Net
To maximize model performance and generalizability, hyperparameter tuning was performed for the ResRNN-Net and MultiScaleFusion-Net architectures. This was conducted to determine the optimal learning rate, batch size, and dropout rate configurations, which would maximize prediction accuracy and model efficiency. The following hyperparameters were tuned:
Learning Rate: [0.0003, 0.0005]—Controls gradient update step size.
Batch Size: Evaluated at discrete values of 32, 64, and 128—Number of samples processed before model weight updates.
Dropout Rate: [0.3, 0.5]—Prevents overfitting by randomly deactivating neurons.
A random search approach was used to explore hyperparameter combinations for both models. For each trial, the models were initialized with the Adam optimizer and binary cross-entropy loss using early stopping (patience: one epoch). The models were evaluated using the F1-score, balancing precision and recall. ResRNN-Net was tested with ten trials and MultiScaleFusion-Net with five trials due to early convergence.
As shown in
Table 6, Both models achieved F1-scores above 0.81, showcasing strong prediction performance with balanced precision and recall. The learning rate of 0.0005 ensured smooth convergence, while a batch size of 64 optimized computational efficiency. A 50% dropout rate effectively prevented overfitting without compromising learning capacity.
To determine whether the observed performance difference between ResRNN-Net and MultiScaleFusion-Net was statistically significant, a paired t-test was conducted on the F1-scores across cross-validation folds. The paired t-test yielded a t-statistic of 5.9761 with a p-value of 0.0039, indicating a statistically significant difference at the 0.01 level. These results suggest that, while both models performed competitively, ResRNN-Net demonstrates statistically superior performance under the t-test criterion.
4.1.4. Hyperparameter Tuning for Multimodal Model
To optimize the performance of the multimodal deep learning model, hyperparameter tuning was conducted as shown in
Table 7 to identify the best configurations for the learning rate, batch size, and optimizer selection. The search aimed to enhance the model stability and predictive power across the three modalities: tabular, imaging, and time-series data. The following key hyperparameters were considered for tuning:
Learning Rate: [0.001, 0.0005].
Batch Size: [16, 32].
Optimizer: [Adam, AdamW].
A grid search approach was applied across eight hyperparameter configurations, where the model was trained and evaluated on validation accuracy, F1-score, precision, recall, and ROC–AUC. We can obtain better F1-score (83%), accuracy (78.9%), and recall (100%) values by choosing the optimal hyperparameters, which are learning rate = 0.0005, batch size = 16, and Adam as the optimizer.
By selecting optimized hyperparameters for cross-validation, the performance of the multimodal model improved. The accuracy increased from 60% to 63%, as shown in
Table 8. This highlights the critical impact of hyperparameter tuning in enhancing model robustness and predictive power.
Discussion on Multimodal Integration: The multimodal model represents an important first step toward integrating structured tabular data, medical imaging, and temporal signals for more comprehensive risk prediction. Initially, the model achieved perfect recall, but fine-tuning the hyperparameters was crucial to improving the balance across the other performance metrics, as shown in
Table 8. Currently, we use early fusion to combine the learned embeddings from each modality. However, the drop in precision and only moderate accuracy suggest that this approach may not fully harness the power of cross-modal interactions. To address this, future work will explore alternative fusion strategies, such as late fusion, attention-based modality weighting, and hierarchical feature aggregation. Additionally, we plan to conduct ablation studies using combinations of two modalities to better understand each data type’s contribution and refine our integration strategy further.
Table 9 presents a consolidated overview of the key hyperparameters used across the ResRNN-Net, MultiScaleFusion-Net, and multimodal models. It details the architectural components, such as recurrent layers, convolutional blocks, and fusion strategies, along with their associated configurations, like unit sizes, activation functions, and optimizers.
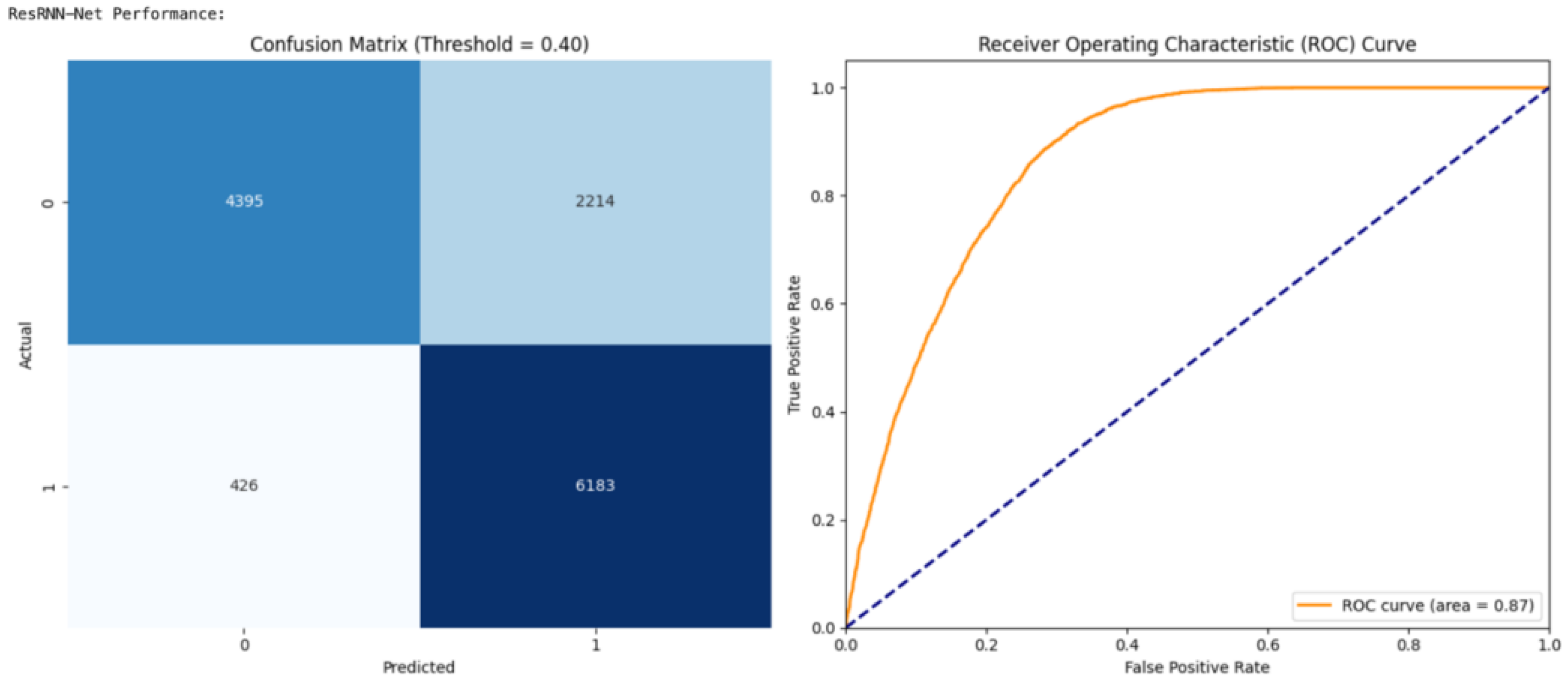
4.2. Performance Interpretation for ResRNN and MSF Net
The confusion matrix and ROC curve analyses present interesting observations of the efficacy and reliability of ResRNN-Net as well as MultiScaleFusion-Net to predict pregnancy complications. The first important observation is the high recall by both the models, guaranteeing that as many actual complication cases as possible were labeled correctly. The high sensitivity in this context is essential in real-world clinical uses, where the failure to recognize a potential complication would have a negative consequence.
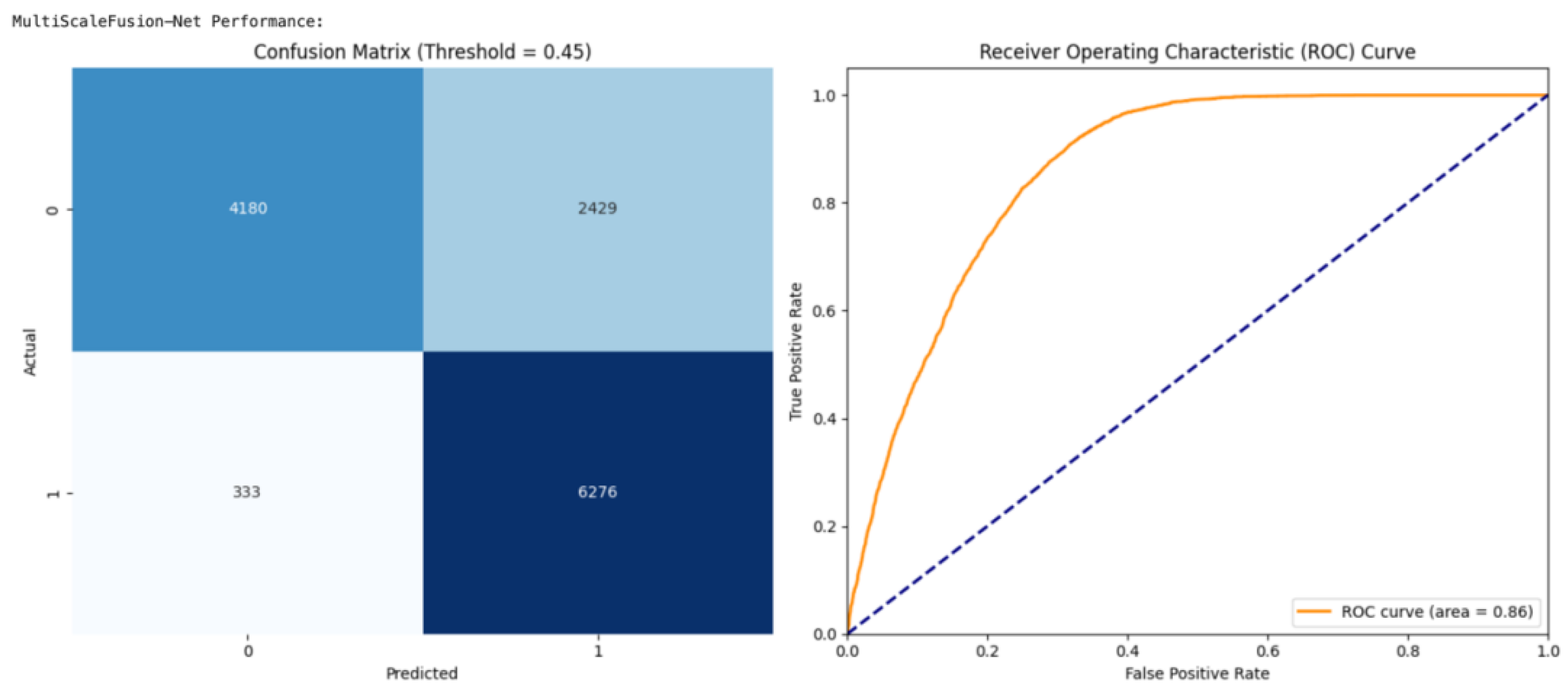
Figure 16 illustrates that the optimal performance for ResRNN-Net was at a threshold of 0.40, whereas the best results for MultiScaleFusion-Net, as shown in
Figure 17, were obtained at a marginally increased threshold of 0.45. This variation indicates the need for threshold optimization to realize clinically meaningful predictions. Finally, the collective evidence from the confusion matrix and ROC curve analyses corroborated the models’ performance, proving their feasibility for real-world application in maternal health tracking and early complication identification.
4.3. Multimodal Model Performance
The multimodal model was evaluated in two configurations: tabular-only and integrated (tabular + image + EHG). The tabular-only model achieved 80% accuracy with balanced precision and recall, making it a strong baseline. However, it lacked deeper insights from imaging and physiological signals. The integrated model improved recall (90%), ensuring fewer missed complications, but at the cost of lower precision (55%), leading to more false positives. The integrated model leveraged diverse data for richer predictions. Its increased training complexity and computational demands require further optimization for real-world deployment.
4.4. Discussion and Comparative Analysis
Although competitive performance was shown by all the models, ResRNN-Net and MultiScaleFusion-Net were remarkable due to their higher recall and F1-scores. TabNet was a very interpretable choice, and the MLP was computationally efficient, with excellent accuracy. The model selection was based on the deployment context—ResRNN-Net for clinical sensitivity, TabNet for interpretability, and an MLP for real-time resource-scarce scenarios. The integrated multimodal model, despite its complexity, reinforced the value of combining tabular, image, and time-series data for robust pregnancy complication prediction.
Table 10 provides a critical evaluation of all implemented models, comparing key metrics including accuracy, precision, recall, F1-score, and ROC–AUC.
Statistical and Deployment Oriented Insights
To support the experimental findings with practical insights, we examined computational efficiency and deployment feasibility. Both ResRNN-Net and MultiScaleFusion-Net were trained with the best hyperparameters obtained from a random search (learning rate = 0.0005; batch size = 64).
4.5. Explainable AI and Feature Importance
For transparency and trust in the pregnancy complication prediction models, explainable AI (XAI) methods were utilized, with SHapley Additive exPlanation (SHAP) being the main focus. This method provided both global feature importance insights and local explanations for each prediction, increasing the interpretability of the ResRNN-Net and MultiScaleFusion-Net models.
To provide deeper interpretability of the SHAP visualizations, it is essential to understand what each feature index represents.
Figure 18 presents the complete mapping of the SHAP feature indices to their corresponding clinical, demographic, and lifestyle variables in the NFHS-5 dataset. This mapping aids clinicians and researchers in translating model insights into actionable factors for maternal health evaluation.
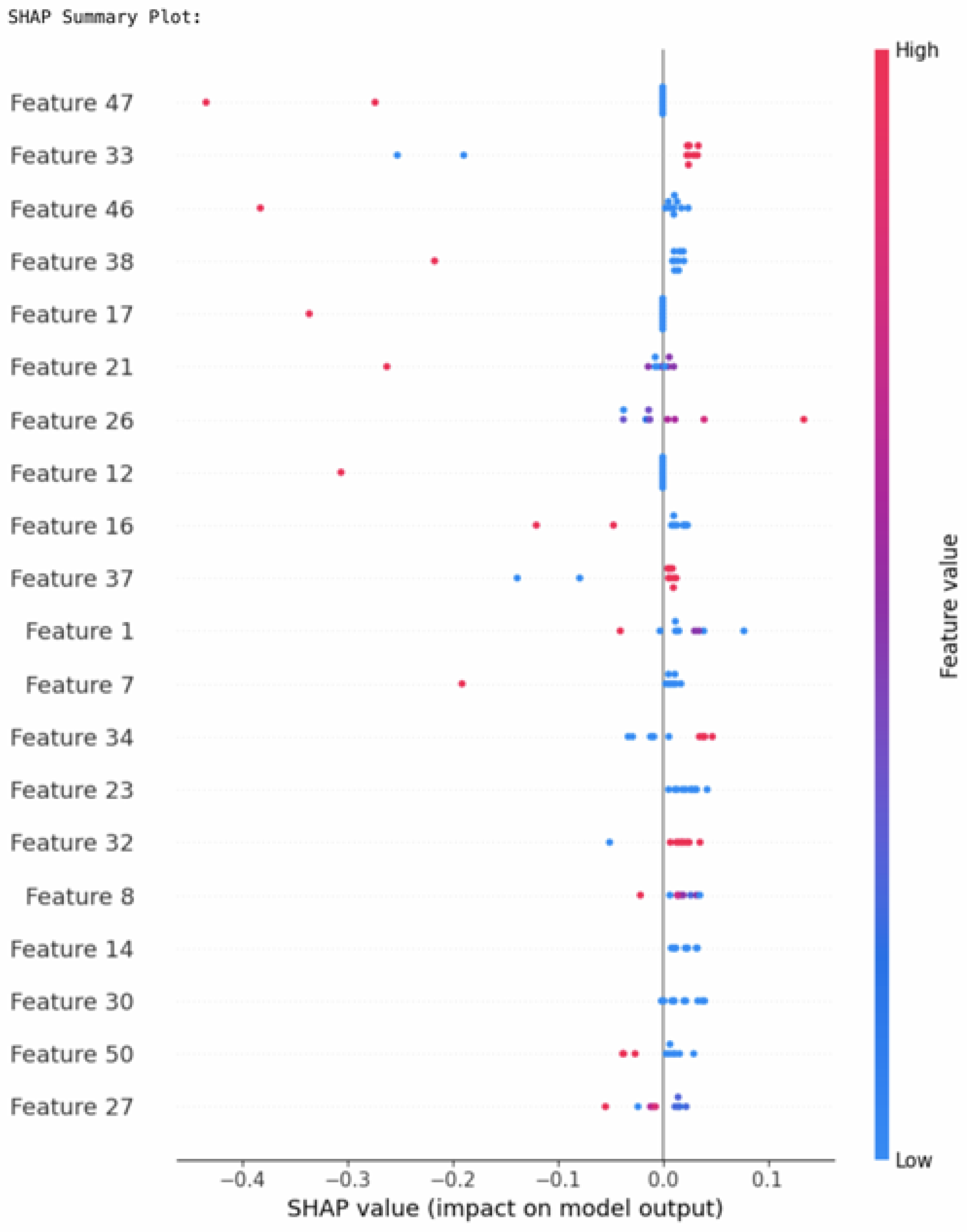
The SHAP summary plot in
Figure 19 depicts the contribution of every feature regarding model predictions. For ResRNN-Net (including 10 samples), the major features were
Feature 47 (Water-Source-Other),
Feature 33 (Postnatal-check),
Feature 46 (Water-Source-Natural-Source), and
Feature 38 (Smoke), as presented by the summary plot. Positive values of SHAP represented features contributing to increased pregnancy complication prediction probabilities, whereas negative values decreased the prediction probability.
Feature 17 (Child-putToBrst) and
Feature 21 (Child-Brst) had significant contributions, which coincided with the clinical expectations.
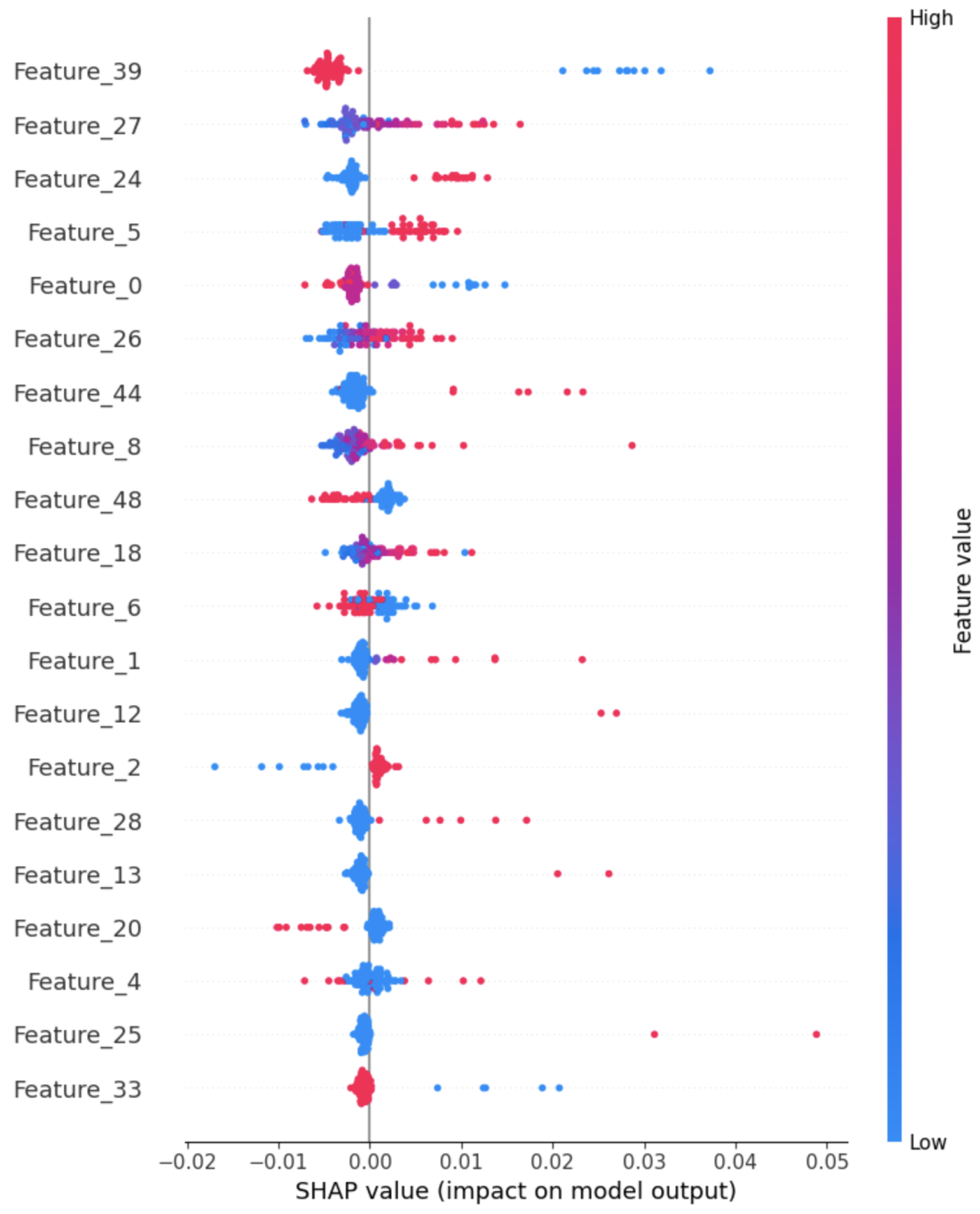
Figure 20 presents the Tabular-SHAP analysis for the multimodal model, which reveals that Feature 39 (Prenatal_Care) proves to be a prominent feature, although Feature 27 (Hg_child_adjust) and Feature 24 (Preg_intParaDrug) contribute notably.
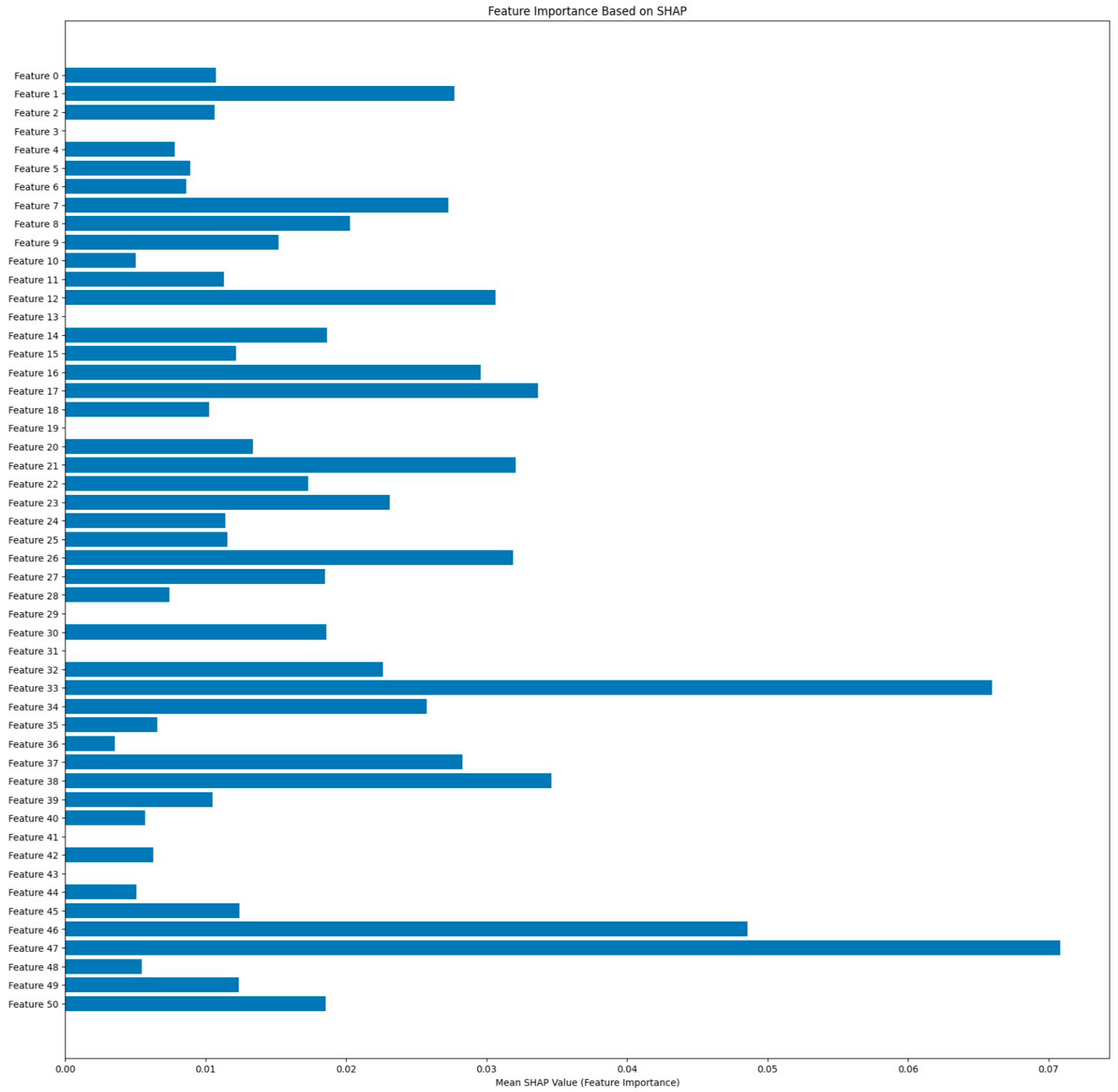
The SHAP feature importance bar plot as shown in
Figure 21 also calculates the mean contribution of every feature regarding the model predictions. Features such as
47,
33, and
46 recorded the highest mean SHAP values, justifying their critical role in classification. The visualization shows a more significant ordering of feature impact in the direction of decision interpretability by ResRNN-Net. These findings validate the model’s adherence to the clinical expectations and its trustworthiness in practice.
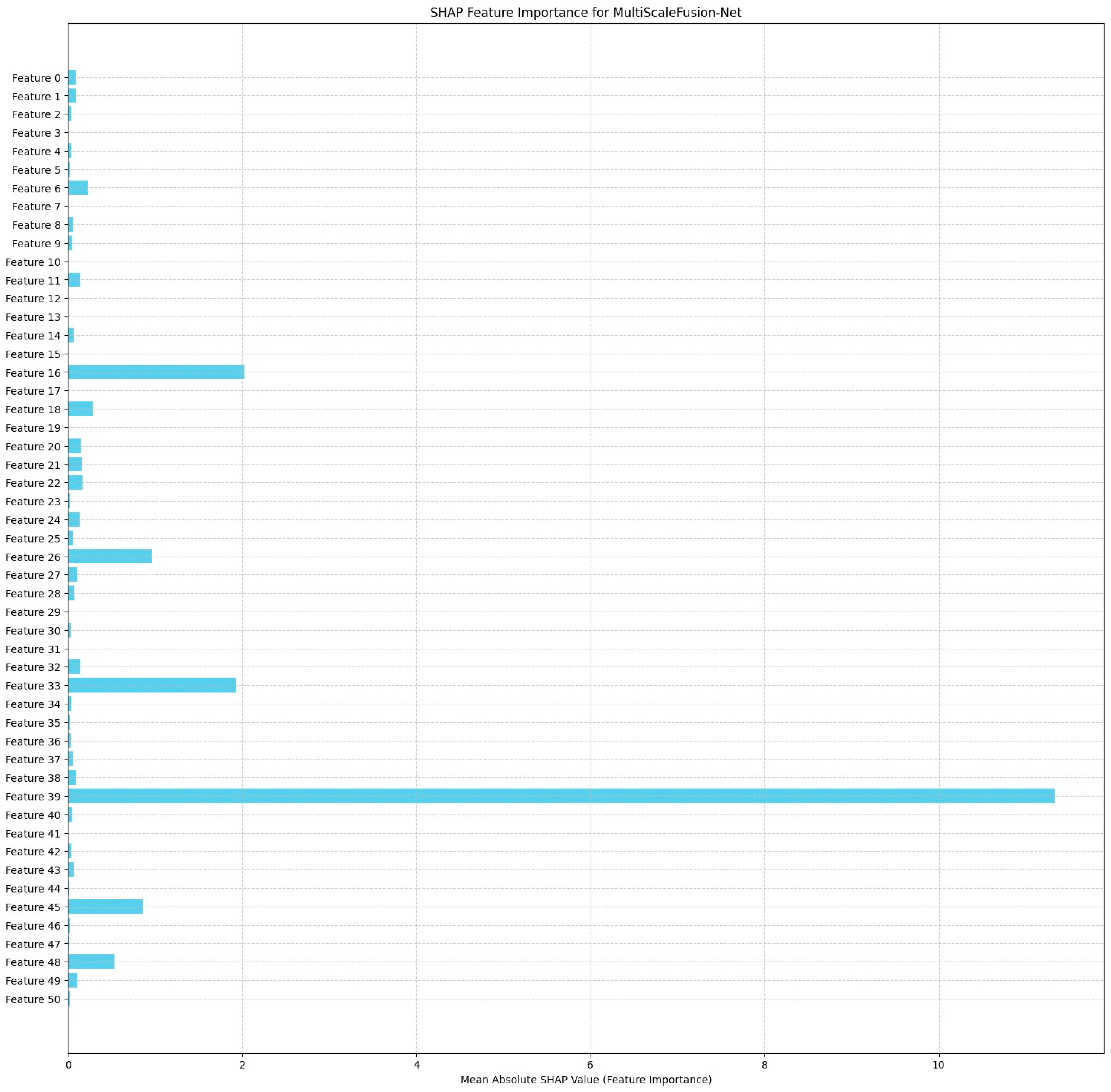
The MultiScaleFusion-Net SHAP feature importance plot in
Figure 22 reveals Feature
39 (Prenatal_Care) to be the most influential on the model predictions, with a much greater mean SHAP value than the others. Features
16 (ChildFood_bottle),
26 (ChildAge_mnths), and
33 (PostnatalChk) also make strong contributions, confirming their contribution to pregnancy complication classification. The distribution shows that MultiScaleFusion-Net is founded on a more specific collection of predictive variables than ResRNN-Net.
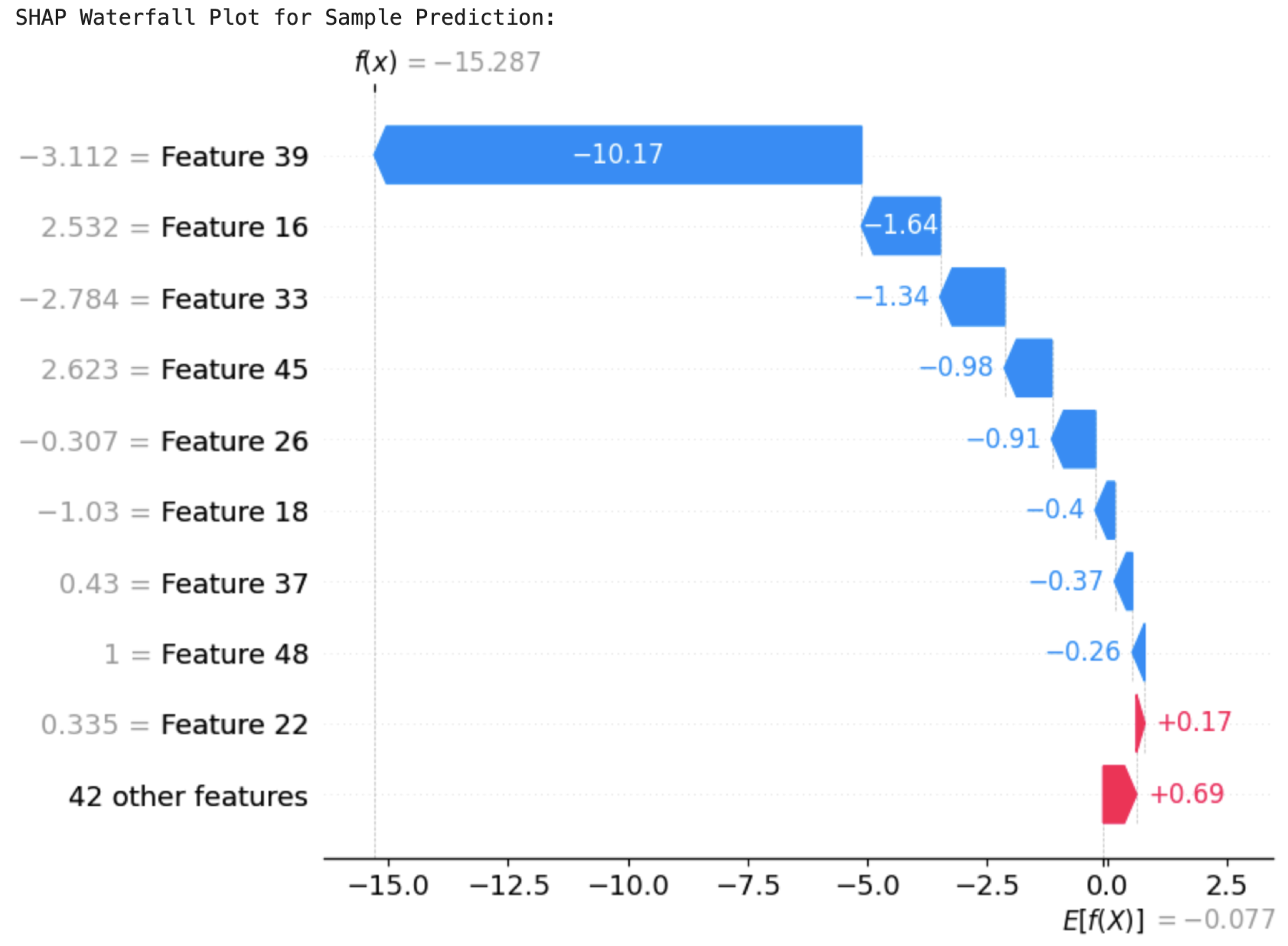
In order to better understand the individual predictions, SHAP waterfall plots were created for example predictions of the two models. These plots decomposed the contributions of each feature, illustrating how the model arrived at its final prediction. For ResRNN-Net,
Figure 23 identified
Feature 39 (Prenatal_Care) as the most contributory factor in reducing the prediction probability greatly. On the other hand,
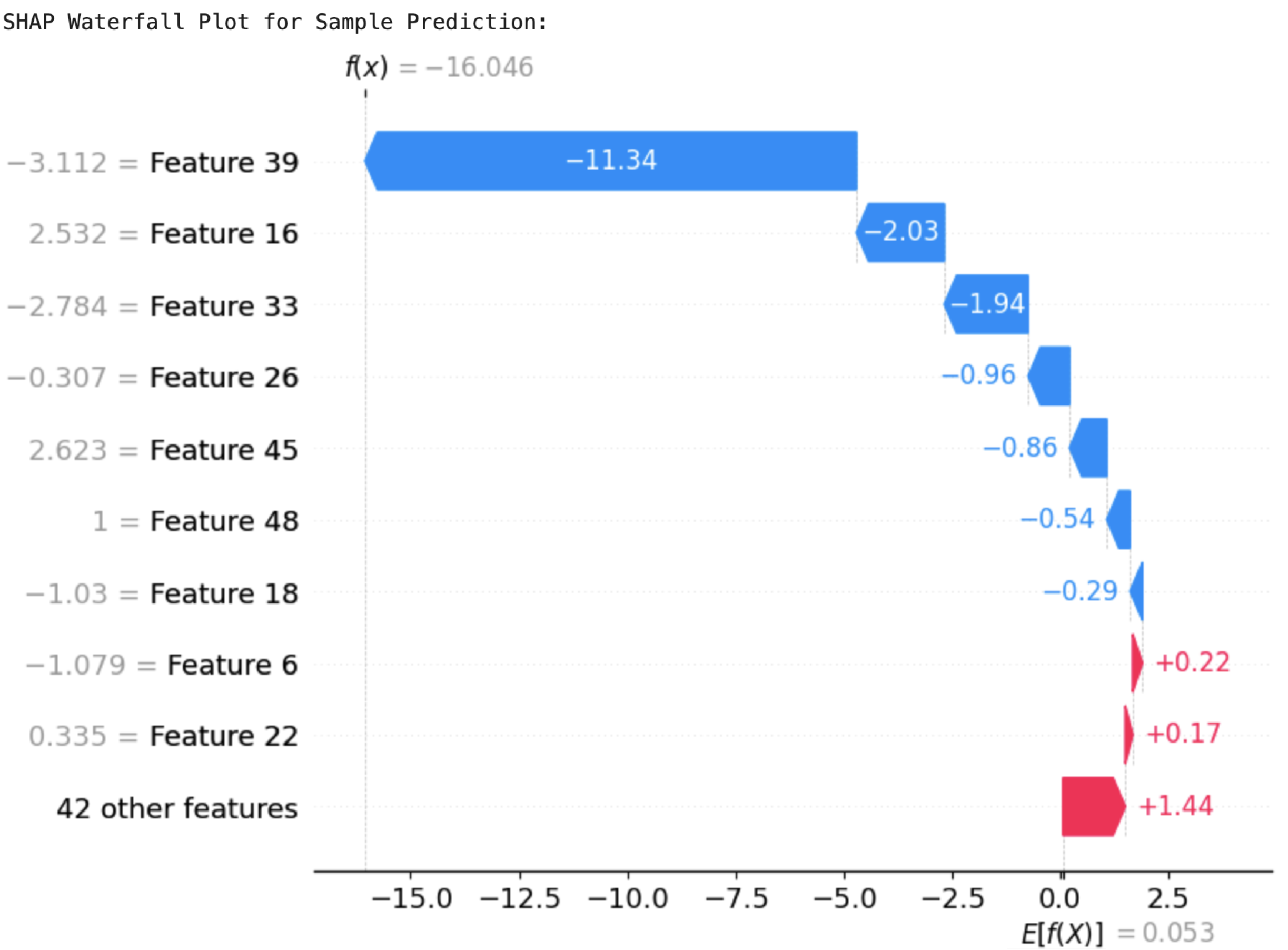
Feature 22 (Preg_iron) and others improved the likelihood of a positive prediction. The SHAP values cumulatively accounted for the last output, supporting the decision-making process of the model. Likewise, the waterfall plot of MultiScaleFusion-Net, shown in
Figure 24, identified
Feature 39 (Prenatal_Care) and
Feature 33 (PostnatalChk) as major contributors to the prediction results.
Feature 39 (Prenatal_Care) drove the prediction towards the negative class, while
Feature 22 (Preg_iron) influenced it positively. The plot emphasizes the model’s reliance on clinically significant features, justifying its conformity to domain expertise.
4.6. Grad-CAM Visualization
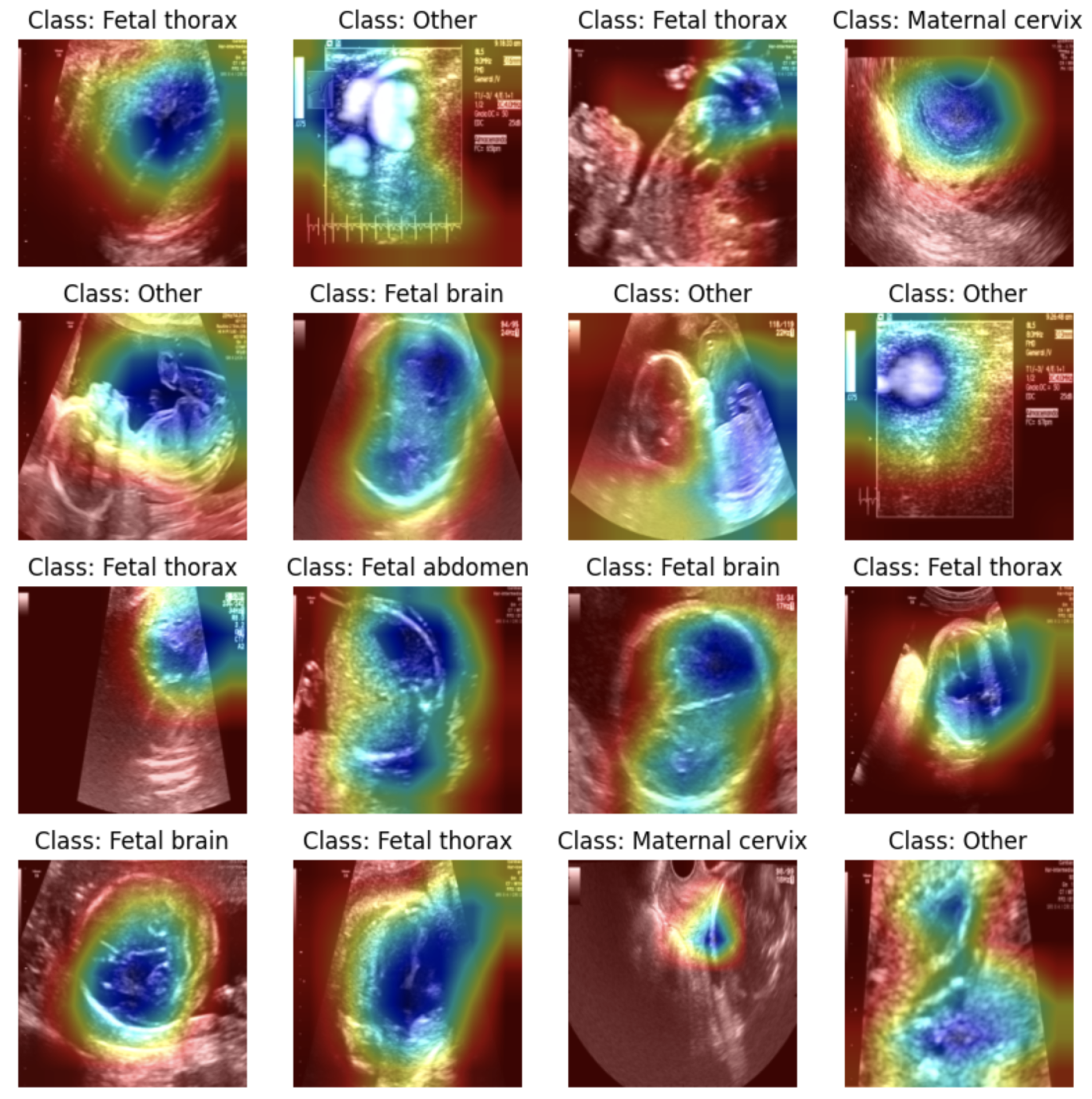
To effectively demonstrate the Grad-CAM approach, an alternative image dataset was employed: the Fetal Planes Ultrasound Dataset [
41]. This dataset includes ultrasound images of fetal structures, categorized into different planes, such as fetal thorax, fetal brain, fetal abdomen, and maternal cervix. A ResNet-18 model was fine-tuned on this dataset, achieving high accuracy in classifying the fetal planes. The Grad-CAM technique was then applied to visualize the regions that influenced the model’s predictions for each image. As shown in all the images in
Figure 25, the heatmaps effectively highlight the critical regions within the ultrasound scans, such as the thoracic cavity in the fetal thorax images or the brain structures in the fetal brain images. This visualization confirmed that the model was making predictions based on medically relevant areas rather than irrelevant background features.
These results demonstrate the potential of Grad-CAM for enhancing interpretability in medical imaging tasks. While it was not directly applicable to the tabular pregnancy dataset, the image-based evaluation provided a clear understanding of how Grad-CAM highlights important features in deep learning models.
5. Conclusions
Worldwide healthcare initiatives consider maternal health to be their foundational building block while affecting public health in addition to social and economic development. The implementation of technology, especially artificial intelligence (AI), in maternal healthcare presents a complete solution for the existing challenges. In this study, we have constructed a stable deep learning framework to predict pregnancy complications based on the NFHS-5 dataset. By adopting a systematic pipeline with data preprocessing, feature selection, model building, and evaluation, the project achieved identifying high-risk pregnancies with great accuracy and explainability.
ResRNN-Net and MultiScaleFusion-Net proved to be the top-performing architectures, utilizing Bi-LSTM, CNNs, and attention techniques to identify subtle relationships in the dataset. ResRNN-Net obtained the best F1-score (82%), balancing precision and recall, while MultiScaleFusion-Net provided similar performance (81%) with quicker inference. Conventional machine learning algorithms such as XGBoost and ensemble classifier lagged behind the deep learning strategies, highlighting the significance of sophisticated architectures for high-complexity clinical predictions. The multimodal model further enhanced predictive power by integrating tabular, image, and time-series data, ensuring higher recall (100%), making it ideal for risk-sensitive applications. The incorporation of explainable AI (XAI) methods, including SHAP and Grad-CAM, additionally improved the model explainability, ensuring that the essential features influencing the predictions were interpretable and clinically meaningful. The deep learning models not only achieved better prediction accuracy but also provided interpretable results, so the system is quite suitable for deployment in maternal health monitoring in the real world.
Fostering the implementation of predictive models in maternal healthcare requires negotiating numerous important directions. Broadening datasets to incorporate more divergent populations—across geographic regions, age groups, and volatile risk factors—will complement the adaptability and tenacity of these models. Refining data quality through state-of-the-art techniques, such as generative models for attributing missing values and the merging of additional data modalities (e.g., genetic, environmental, and behavioral), is predicted to significantly elevate predictive performance. Coherent integration with clinical systems, specifically electronic health records (EHRs), would empower real-time predictions and actionable alerts, thus promoting more reactive clinical decision-making. Enriching model comprehensibility through explainable AI (XAI) techniques and developing intuitive interfaces for healthcare professionals will be crucial for effective implementation. Moreover, sequential studies that track maternal health over time will be fundamental for evaluating the long-term influence and clinical relevance of these predictive frameworks.