1. Introduction
Breast cancer remains one of the most prevalent cancers worldwide. In 2022, it accounted for over 2.3 million new cases, ranking second among all cancer types globally. Moreover, with about 666,000 deaths, breast cancer ranks as the fourth most frequent contributor to cancer mortality worldwide. A timely and precise diagnosis of breast cancer can prolong the lives of patients or increase their chance of survival [
1]. However, the accuracy of pathological diagnosis relies not only on image texture quality but also on subjective elements like the pathologist’s focus and level of experience. Consequently, the risk of misdiagnosis remains, which may negatively influence patient prognosis and clinical outcomes. In order to achieve more accurate and efficient breast cancer pathology assessments, leveraging advanced computational techniques to assist pathologists in pathological tissue analysis holds significant clinical value [
2].
Convolutional neural networks (CNNs) have demonstrated outstanding performance in medical image processing tasks [
3,
4,
5,
6,
7]. CNN-related breast cancer histopathological image (BCHI) classification methods have also made considerable progress [
8,
9,
10,
11,
12,
13,
14]. Currently, CNN-based approaches for BCHI classification can be broadly divided into two categories.
Non-end-to-end training model can be used to describe the first category. Prior to using machine learning classifiers to categorize samples based on the extracted features, researchers first extract features from input images using conventional or newly developed CNNs. In this process, researchers typically pre-train CNN models using the ImageNet dataset [
15]. For example, Kumar et al. [
16] used the VGG-16 model pre-trained on ImageNet to extract deep features from BCHI and evaluate classification performance by combining support vector machines and random forest methods. The second stream of classification for BCHI use end-to-end training of the deep neural network model. This stream prevents the multi-module network model’s discontinuity between the stage of feature extraction and the classifier, which is further divided into two subcategories. The first subtype is task-specific CNN approaches, which involve fine-tuning traditional pre-trained networks for certain tasks [
10,
11]. For example, in the work of Shaalu and Mehra [
10], commonly adopted pre-trained models including VGG16, VGG19, and ResNet50 were evaluated against fully trained counterparts on BCHI. Zhang et al. [
11] incorporated global and local characteristics on six standard VGGNet and ResNet models, and examined the models’ ability to classify BCHI systematically. ResNet50 outperforms the other six traditional network models, according to experimental findings. Benhammou et al. [
17] performed BCHI classification by fine-tuning Inception-V3 convolutional neural network pre-trained on ImageNet. Said et al. [
18] utilized a ResNet18 model pre-trained on ImageNet to fine-tune its terminal residual modules. They also combine global contrast normalization and data augmentation, achieving significant performance advantages on the BreakHis dataset.
Another subclass is to add representative convolutional neural network units to the classical network or the newly constructed network. Bardou et al. [
19] proposed a seven-layer BCHI classification network, with two fully connected layers placed after five convolutional stages. Through data enhancement technology, the accuracy of the two models on the BreakHis dataset was between 96.15% and 98.33%. In order to extract picture features and address the multi-classification problem with fewer computational resources, Murtaza et al. [
20] suggested a tree-based breast tumor model. Studies have revealed that a single network with a straightforward structure is also capable of achieving great classification accuracy for this medical purpose. Since the attention mechanism can swiftly choose the focus point and provide a more distinct feature representation, more and more researchers have started incorporating it into CNN models. Jiang et al. [
14] proposed a lightweight CNN that combines residual structures with a squeeze-and-excitation mechanism, achieving superior performance in BCHI classification with fewer parameters. In the classification of BCHI, it achieved higher performance with fewer parameters. Then, Budak et al. [
21] used FCN and BiLSTM to detect BCHI, and achieved significant results in the BreakHis dataset. Toğaçar et al. [
22] combined the residual module and the convolutional block attention module, and utilized HyperColumn technology to better perform fine-grained positioning of BreakHis images. The performance of BCHI classification has significantly improved with the continuous development of deep neural network models.
In addition, Transformer-based [
23] and attention-driven models for medical image classification continue to emerge, demonstrating strong capabilities in feature extraction and representation [
24]. For example, ST-Double-Net [
25] integrates the Swin Transformer (SwinT) [
26] with a weakly supervised object localization strategy, effectively combining global and local features without requiring bounding box annotations, and significantly improves the recognition of ambiguous lesion regions. SupCon-ViT [
27] combines a pre-trained vision Transformer with supervised contrastive learning, enhancing feature discriminability and improving generalization. Additionally, DecT [
28] enhances color feature representation by jointly processing RGB and HED images using color deconvolution and a self-attention mechanism. Zhuang et al. [
29] further combined ResNet101 with the SwinT and incorporated the CBAM attention mechanism, along with focal loss, to increase sensitivity to diverse lesion types. Although these Transformer-based models achieve notable improvements in classification performance, most still face limitations, such as high computational demands and increased training costs.
Although notable advancements have been achieved in medical image analysis, several challenges remain unaddressed. For instance, in BCHI classification, most existing methods rely solely on first-order statistics while ignoring high-order statistical features, which limits the expressive capacity of deep features. In fact, CNN models with high-order statistics recently impressed with their promising efficacy on a variety of visual tasks. Included among the representative deep high-order models are
P, Bilinear CNN, MPN-COV, and GSoP [
30,
31,
32,
33]. In particular, Li et al. [
32] developed a novel trainable second-order network called MPN-COV that successfully applies matrix power normalization to capture the global covariance statistical information of deep features. In our previous work [
34], we conducted a preliminary investigation into the role of deep second-order statistics in BCHI classification. Experimental results demonstrate that it gains average performance improvement of 4.92% over its baseline. Meanwhile, dilated convolution, which perceives global features by expanding the perception field, has been widely employed for building more discriminant deep models in recent years. Among them, Chen et al. [
35] introduced hole convolution, allowing the network to extract denser features and improve the reliability of segmentation results. Receptive field block network (RFBNet) [
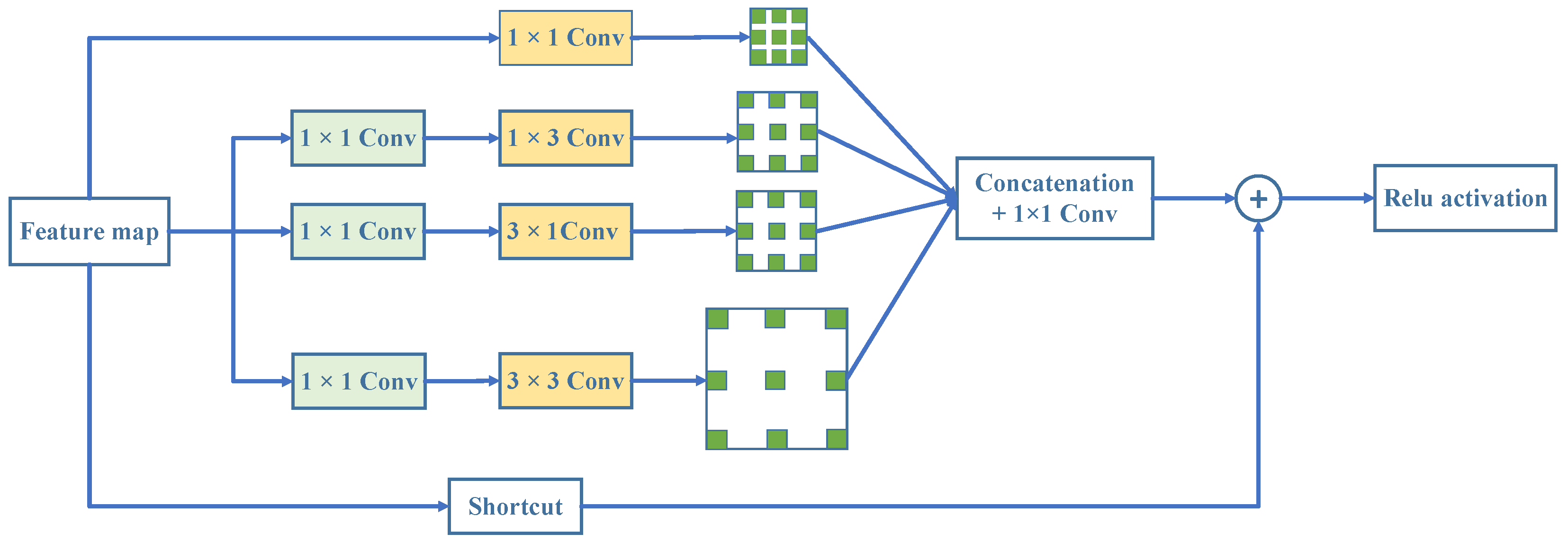
36] optimizes receptive field design by accounting for spatial eccentricity, which enhances feature extraction in lightweight networks and has a significant effect on target detection. Nevertheless, the challenge of classifying medical images has not yet been effectively solved by hole convolution migration.
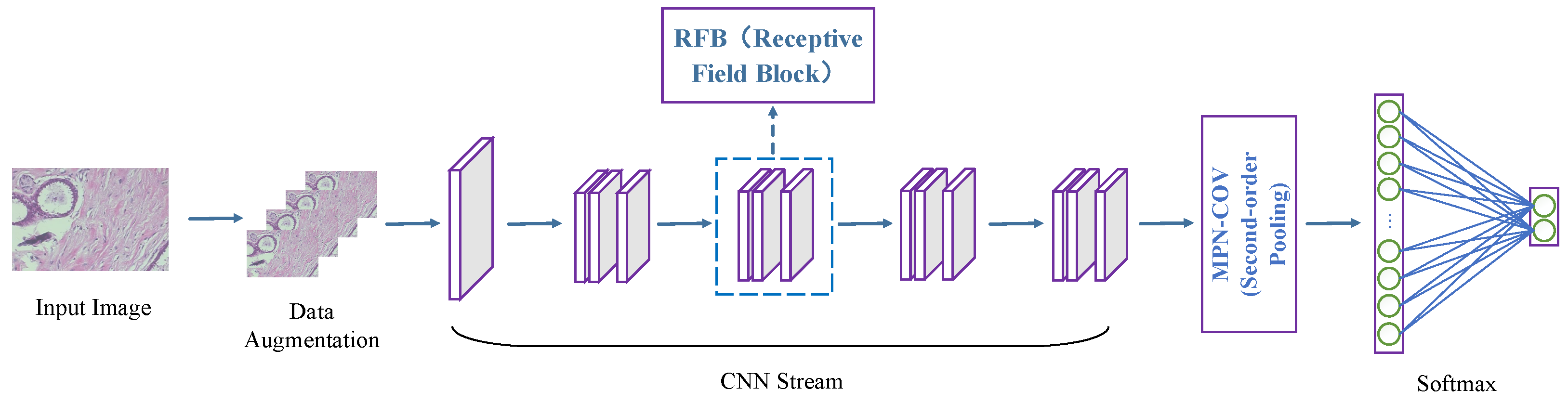
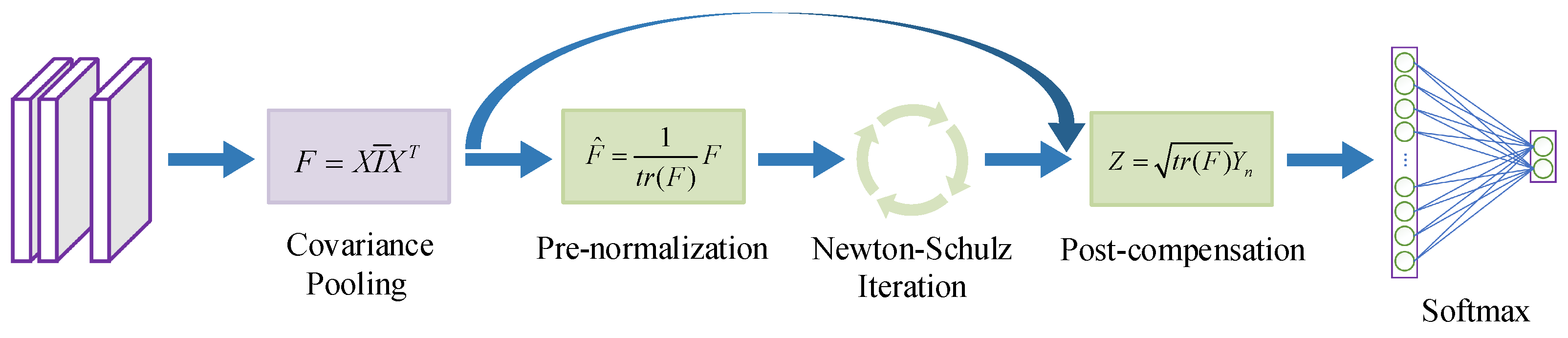
This study proposes an innovative high-order receptive field network (HoRFNet) focused on the efficient classification of BCHI. The core design concept of HoRFNet lies in integrating multi-branch receptive field expansion with high-order statistical modeling streams, thereby overcoming the limitations of traditional CNNs in spatial dependency modeling and feature representation. HoRFNet introduces multi-branch convolutions and dilated convolutional layers to replace the square-kernel convolutional layers in traditional backbone networks. This design breaks the limitations of fixed receptive fields in traditional CNNs by employing a multi-scale perception strategy, capturing local details while integrating global contextual information, thereby achieving a more precise understanding of complex breast cancer tumor morphologies. Moreover, the introduction of dilated convolutions effectively avoids information loss caused by downsampling, preserves high-resolution feature representation, and significantly enhances the network’s discriminative power and detail depiction capabilities. To capture the global interactions between convolutional features, HoRFNet introduces covariance pooling based on matrix power normalization (MPN) after the final convolutional layer. This design overcomes the limitations of traditional CNNs that rely solely on first-order feature aggregation, by explicitly modeling high-order statistical information to more accurately represent the structural characteristics of pathological tissues. This enables histopathological images to obtain more informative global feature representations.
Figure 1 depicts the overall structure of HoRFNet, and the three areas listed below sum up as the key contributions of this study.
To categorize breast cancer histopathological image, a high-order receptive field network (HoRFNet) from end to end is proposed. To the best of our knowledge, this is the first instance in which the medical image classification problem simultaneously incorporates second-order statistical characteristics and receptive field block (RFB).
HoRFNet replaces the third layer with a multi-branch convolution layer using various convolution kernels, followed by an expansion convolution layer. This successfully enhances receptive fields and the discriminality of the deep model. To further enhance performance, covariance statistics of deep features computed using matrix power normalization (MPN) are used to collect more discriminant information from breast cancer histopathological image.
HoRFNet demonstrates superior performance on the open-source BreakHis dataset, achieving 99.50% and 99.23% classification accuracy at the image level and patient level, respectively, and surpassing existing approaches.
3. Experiments
This section presents the experimental validation of the proposed method and is organized into four main parts. First, we introduce the BreakHis dataset and its key characteristics. Next, we describe the evaluation metrics used to assess model performance. Then, we detail the experimental setup, including data pre-processing, model training, and parameter configuration. Finally, we report the experimental results and provide a systematic analysis and comparison of model performance, thereby validating the effectiveness and feasibility of the proposed approach.
3.1. Datasets
3.1.1. BreakHis Dataset
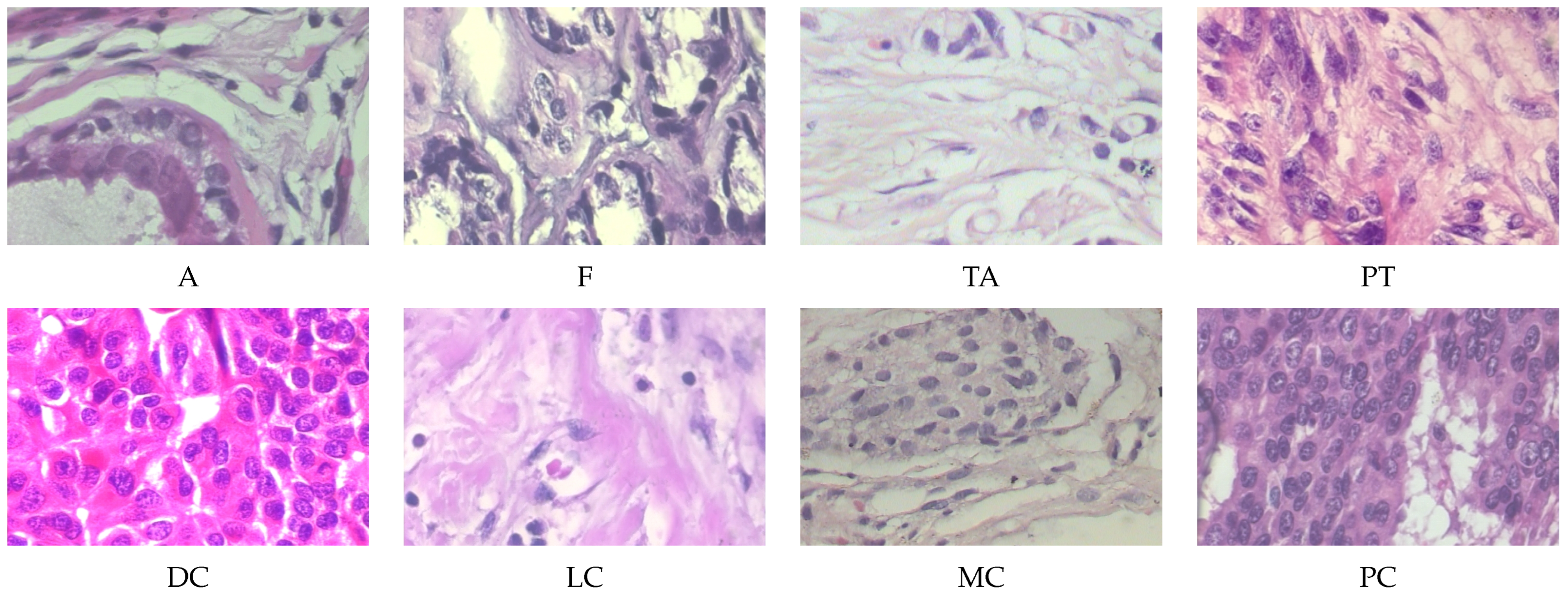
The BreakHis dataset is a publicly available dataset widely used for breast cancer histopathological image analysis. It contains 7909 breast cancer tissue slice images from 82 patients, with images captured at four magnification levels including
,
,
, and
. All images are divided into two main categories, benign and malignant, with further subdivisions into specific subtypes. The benign tumors include adenosis (A), fibroadenoma (F), tubular adenoma (TA), and phyllodes tumor (PT). The malignant tumors include ductal carcinoma (DC), lobular carcinoma (LC), mucinous carcinoma (MC), and papillary carcinoma (PC). Some typical histopathological images in breakHis at 400× magnification are given in
Figure 4.
3.1.2. BACH Dataset
The BACH dataset, introduced by ICIAR 2018 (International Conference on Image Analysis and Recognition), is designed for breast cancer histopathological image classification. It consists of 400 high-resolution (2048 × 1536 pixels) H&E-stained images. The images are equally distributed across four tissue categories: Normal, Benign, In situ carcinoma, and Invasive carcinoma, with 100 images per class, all annotated by expert pathologists. To accommodate different levels of classification tasks, this study establishes both four-class and two-class classification settings. In the two-class classification task, the Normal and Benign categories are merged into the Benign class, while In situ carcinoma and Invasive carcinoma are combined into the Malignant class, simulating a typical clinical scenario of benign versus malignant decision making in breast cancer screening.
3.2. Evaluation Metrics
To comprehensively evaluate the classification performance of HoRFNet on the BreakHis dataset, this study introduces six commonly used evaluation metrics including image level accuracy, patient level accuracy, precision, recall, F1-score, and confusion matrix. These metrics provide a systematic assessment of model performance from multiple perspectives, including recognition capability at the image and patient levels, as well as classification stability and generalization ability.
Image level accuracy (ILA) measures the proportion of correctly classified images to the total number of images and is used to evaluate the model’s recognition capability at the individual image level:
where
represents the number of correctly classified images and
denotes the total number of images.
Patient level accuracy (PLA) is computed on a per-patient basis by aggregating the classification results of all associated images using a majority voting strategy. This metric reflects the model’s consistency and stability in overall patient level classification:
where
represents the number of correctly classified patients and
denotes the total number of patients.
A confusion matrix is an intuitive tool for evaluating the performance of a classification model. It summarizes the relationship between the model’s predictions and the actual labels, illustrating how the model performs across different classes. It includes four fundamental cases: a true positive (TP) is a sample that is actually positive and correctly predicted as positive; a false positive (FP) is a sample that is actually negative but incorrectly predicted as positive; a true negative (TN) is a sample that is actually negative and correctly predicted as negative; and a false negative (FN) is a sample that is actually positive but incorrectly predicted as negative.
Precision measures the proportion of true positive samples among all samples predicted as positive, reflecting the model’s ability to reduce false positives. Recall measures the proportion of true positive samples among all actual positive samples, evaluating the model’s ability to capture positive cases. The F1-score is the harmonic mean of precision and recall, used to comprehensively evaluate the model’s ability to identify positive samples in classification tasks.
3.3. Experimental Setup
In the experiments, the original dataset is randomly divided into training set and test set in the ratio of 7:3. Among them, 25% of the training set is used for cross-validation. The data augmentation strategy is used during the training process to improve the generalization ability of the model. The optimizer is chosen to be stochastic gradient descent (SGD) with the momentum factor set to 0.9. The initial learning rate is 0.001 and is dynamically adjusted by applying the inverse decay strategy in eight rounds of training. The experimental environment consists of SitOnHoly dual NVIDIA 2080Ti GPUs (each with 11 GB of VRAM), 64 GB of RAM, and uses the PyTorch 2.1.0 deep learning framework.
3.4. Ablation Study
3.4.1. The Impact of Newton–Schulz Iteration Count
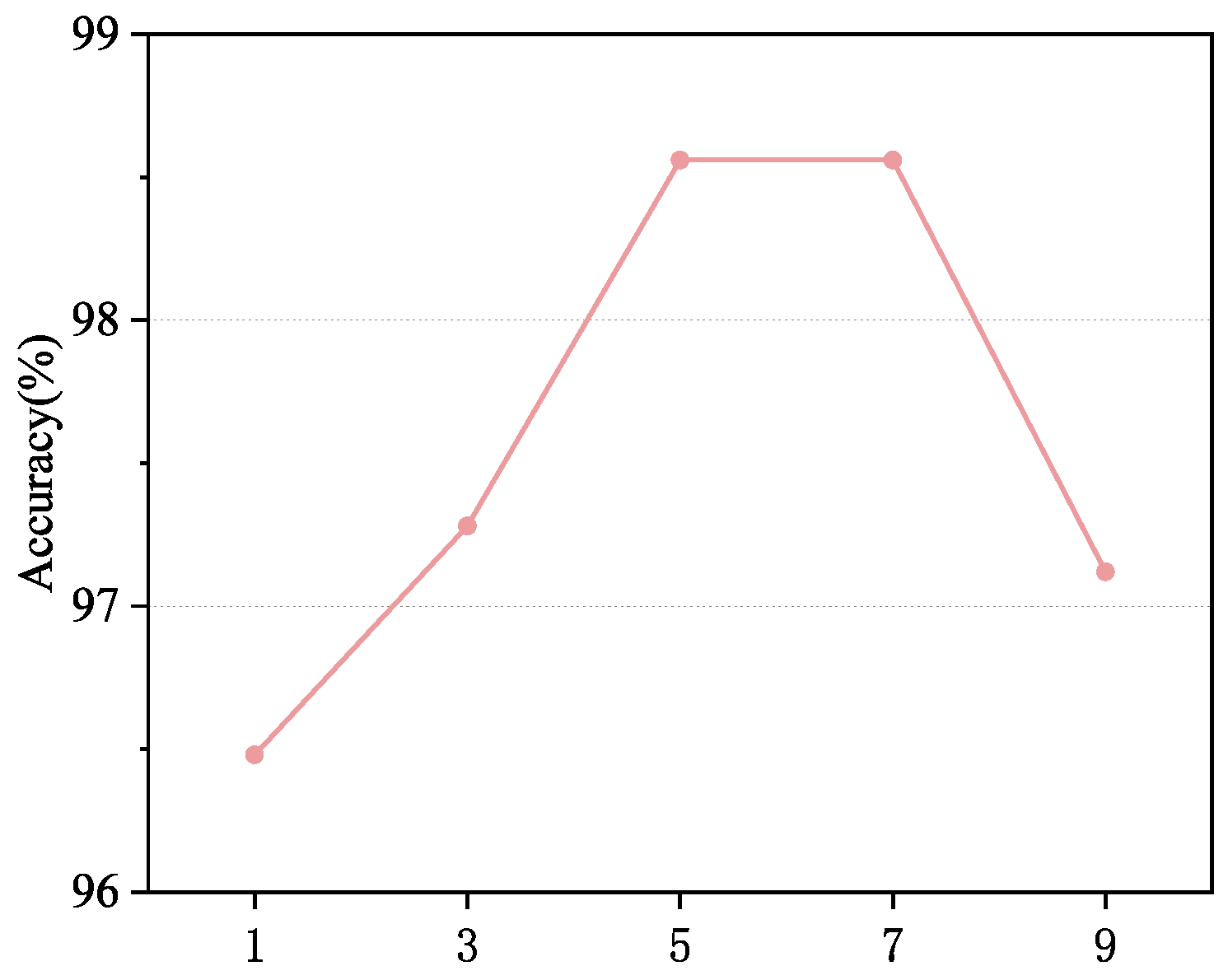
In this experiment, we investigate the impact of the number of Newton–Schulz iterations on the classification model’s accuracy. This experiment is evaluated on histopathological images from the BreakHis dataset at 100× magnification. As shown in
Figure 5, increasing the number of iterations significantly enhances the model’s ability to represent high-order features. As the number of iterations increases from 1 to 5, classification accuracy steadily improves, reaching a peak of 98.56% at 5 iterations. This indicates that a moderate number of iterations better captures covariance information and enhances feature representation quality. However, when the number of iterations further increases to seven, although the model’s accuracy remains at a high level, the improvement tends to saturate. Beyond seven iterations, the model’s performance starts to decline. This phenomenon is likely due to the accumulation of numerical instability caused by excessive iterations, which weakens the feature’s discriminative ability. Meanwhile, a higher number of iterations significantly increases computational cost and reduces inference efficiency. Considering both performance and computational complexity, we ultimately choose five iterations as the optimal setting, ensuring high classification accuracy while effectively controlling computational costs, achieving an optimal balance between performance and efficiency.
3.4.2. The Impact of the Structural Configuration of the RFB Module
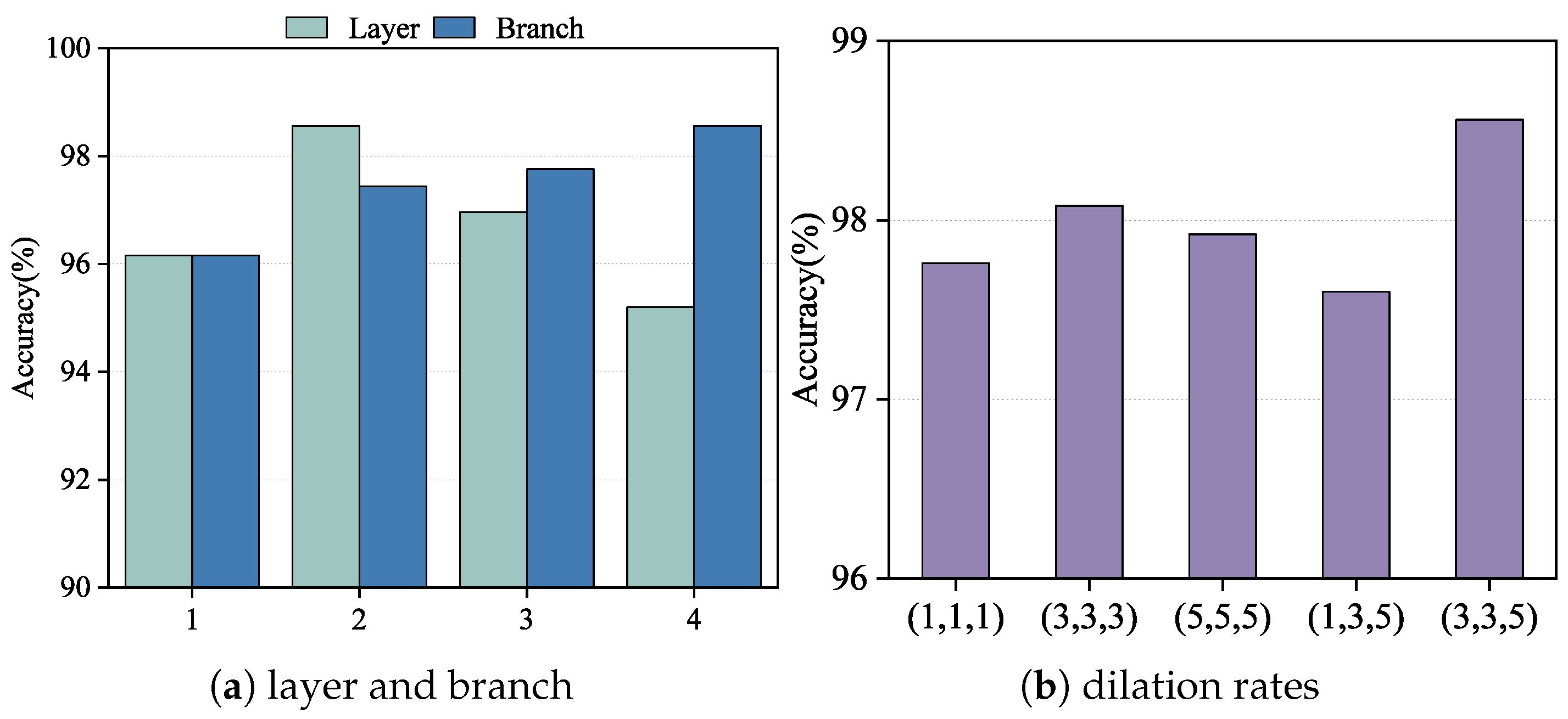
We conducted an ablation study on the BreakHis dataset at 100× magnification to evaluate the effect of replacing different convolutional layers within HoRFNet with the receptive field block (RFB) module. The results are shown in
Figure 6a. Specifically, we replaced the second, third, fourth, and fifth convolutional layers with RFB modules, corresponding to configurations numbered 1 through 4. The experimental results show that replacing the third convolutional layer yields the highest classification accuracy, with improvements of 2.40%, 1.60%, and 3.54% compared to replacing the second, fourth, and fifth layers, respectively. This suggests that the third convolutional layer plays a critical role in feature extraction, as it retains relatively high spatial resolution while also capturing semantic information. Integrating the multi-branch RFB module at this stage significantly enhances the model’s feature representation capability. In contrast, replacing the fifth convolutional layer results in the poorest performance, likely because the RFB module is less effective at this deeper stage due to limited benefits from receptive field expansion. Based on this analysis, we select the third convolutional layer as the optimal insertion point for the RFB module in subsequent experiments.
Building on this, we further investigate the impact of varying the number of branches within the RFB module on model performance, as shown in
Figure 6a. We incrementally increase the number of branches: “1” represents using only the first branch, “2” includes the first and second branches, and so on, up to all four branches. The results show a consistent increase in classification accuracy as the number of branches increases, with the highest performance achieved when all four branches are included. Specifically, using four branches improves accuracy by 2.40%, 1.12%, and 0.80% compared to using only one, two, and three branches, respectively. This demonstrates that the multi-branch structure effectively simulates receptive fields at multiple scales, enhancing the model’s ability to capture complex structures and diverse textures. The combination of different kernel sizes and dilation rates in each branch facilitates the extraction of key features across scales, improving the model’s discriminative capacity. Therefore, we adopt the configuration with all four branches as the optimal RFB module setting in our model design.
To further investigate the impact of dilation rate configuration on model performance, we conduct ablation experiments using five different sets of dilation rate combinations within the RFB module. The experimental results are shown in
Figure 6b. The tested configurations include (1, 1, 1), (3, 3, 3), (5, 5, 5), (1, 3, 5), and (3, 3, 5), with each triplet corresponding to the dilation rates of the three branches in the RFB module. The results show that the (3, 3, 5) configuration achieves the highest classification accuracy among all settings, outperforming (1,1,1), (3,3,3), (5, 5, 5), and (1, 3, 5) by 0.80%, 0.48%, 1.04%, and 0.96%, respectively. These findings demonstrate that a progressively increasing dilation structure is more effective for modeling multi-scale receptive fields, thereby enhancing the model’s ability to capture both global and local contextual information. In contrast, fixed dilation configurations such as (3, 3, 3) and (5, 5, 5), while capable of enlarging the receptive field to some extent, are limited in feature diversity and information integration. Additionally, while the asymmetric design of (1, 3, 5) offers advantages in deeper receptive field coverage, it lacks adequate receptive field in the shallow branches, thereby compromising overall feature extraction. Based on this analysis, we select (3, 3, 5) as the optimal dilation rate configuration for the RFB module in subsequent experiments, aiming to maximize the representation and discrimination of structural information across multiple scales.
3.4.3. The Impact of the Second-Order Feature and Receptive Field Modules
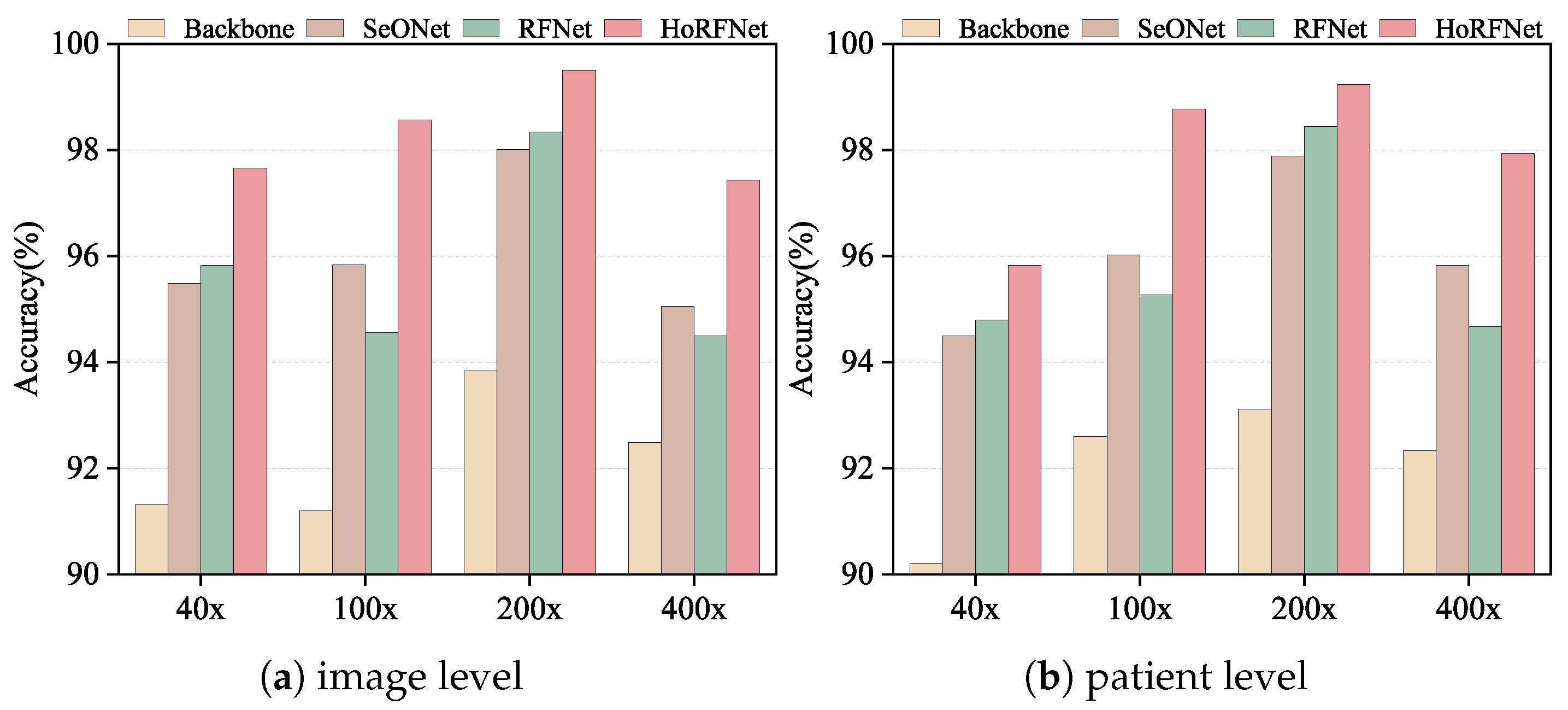
In this section, we conduct ablation studies at both image level and patient level to verify the effectiveness of each module in HoRFNet under four different magnifications. In
Figure 7, we use the backbone network as the baseline and compare the performance changes after introducing the second-order feature module (SeONet), the receptive field module (RFNet), and their combination (HoRFNet) to verify each module’s contribution to performance improvement.
In
Figure 7a, HoRFNet achieves image level recognition accuracies of 97.66%, 98.56%, 99.50%, and 97.43% at 40×, 100×, 200×, and 400× magnifications, respectively, demonstrating clear advantages over the other three models. In particular, compared with the Backbone model at the image level, the HoRFNet model has an accuracy increase of 6.35%, 7.36%, 5.63%, and 4.94% at the four magnifications, respectively.
On the BreakHis dataset, we conduct an in-depth analysis of the performance of the Backbone, SeONet, and RFNet models, and also compares the performance improvement of high-order and receptive field block units on the Backbone model. In the image level evaluation results presented in
Figure 7a, both the second-order feature module (SeONet) and the receptive field module (RFNet) effectively improve classification accuracy across all magnifications, demonstrating clear improvements over the backbone baseline. For example, at 200× magnification, the backbone achieves an accuracy of 93.87%, while SeONet and RFNet improve it to 98.01% and 98.34%, respectively, demonstrating the positive contribution of both modules to image level recognition performance. The second-order features more effectively model the high-order relationships among local textures, while the receptive field module enhances the model’s ability to perceive multiscale contextual information. However, at 100× and 400× magnifications, RFNet performs slightly lower than SeONet. For example, at 400×, RFNet achieves 94.50% compared to 95.05% for SeONet, which may be attributed to the distribution of fine-grained details in the images. At these magnifications, structural boundaries and local contrast become more prominent, giving second-order features an advantage in modeling local relationships. Nevertheless, RFNet still outperforms the backbone across all magnifications, indicating its effectiveness in enhancing overall feature representation. However, by combining the second-order statistical features and receptive field block, the HoRFNet model can obtain additional gain of 1.84%, 4.00%, 1.16%, and 2.93% higher than the RFNet model, and 2.17%, 2.72%, 1.49%, and 2.38% higher than the SeONet model.
In the patient level evaluation presented in
Figure 7b, all models exhibit stability across different magnifications, similar to the image level evaluation. For example, at 200× magnification, the backbone achieves an accuracy of 93.11%, while SeONet and RFNet increase it to 97.89% and 98.44%, respectively. HoRFNet further improves the accuracy to 99.23%. This trend is consistent across most magnifications. Notably, similar to the image level results, at 100× and 400× magnifications, RFNet slightly underperforms SeONet but still significantly outperforms the backbone. These results suggest that the receptive field module is advantageous for extracting global structural information, whereas at 100× and 400× magnifications, second-order features may play a more critical role in ensuring consistency in patient level predictions. Overall, HoRFNet achieves the best performance by combining the strengths of SeONet and RFNet. At 40×, 100×, 200×, and 400× magnifications, it achieves patient level recognition accuracies of 95.82%, 98.77%, 99.23%, and 97.94%, respectively. Therefore, the ablation study results further confirm the effectiveness of second-order statistical features, the receptive field module, and their integration strategy in BCHI classification.
3.4.4. Statistical Significance Analysis
To further validate the statistical significance of the performance improvements achieved by the proposed HoRFNet model in BCHI classification, this study conducts a
t-test comparing SeONet, RFNet, and HoRFNet against the Backbone model across four magnification levels (40×, 100×, 200×, and 400×). As shown in
Table 2, HoRFNet consistently achieves the highest mean improvement (Mean) at each magnification, with values of 7.515, 6.560, 5.710, and 4.395, respectively. The corresponding
p-values are 0.044, 0.010, 0.005, and 0.011, all of which are significantly below the 0.05 threshold, indicating that the performance gains of HoRFNet over the Backbone model are statistically significant. In contrast, although SeONet and RFNet show some performance improvements at certain magnifications, most of their
p-values exceed 0.05. Notably, RFNet yields a
p-value of 0.836 at 400× magnification, failing the significance test. These results demonstrate that HoRFNet not only outperforms the Backbone in classification accuracy but also exhibits statistically more significant and robust advantages, validating the effectiveness of its architectural design.
3.5. Experimental Results
3.5.1. Experimental Results Under Other Metrics
To more comprehensively evaluate the classification performance of HoRFNet under various magnifications, this study introduces precision, recall, and F1-score as supplementary evaluation metrics. The corresponding experimental results are presented in
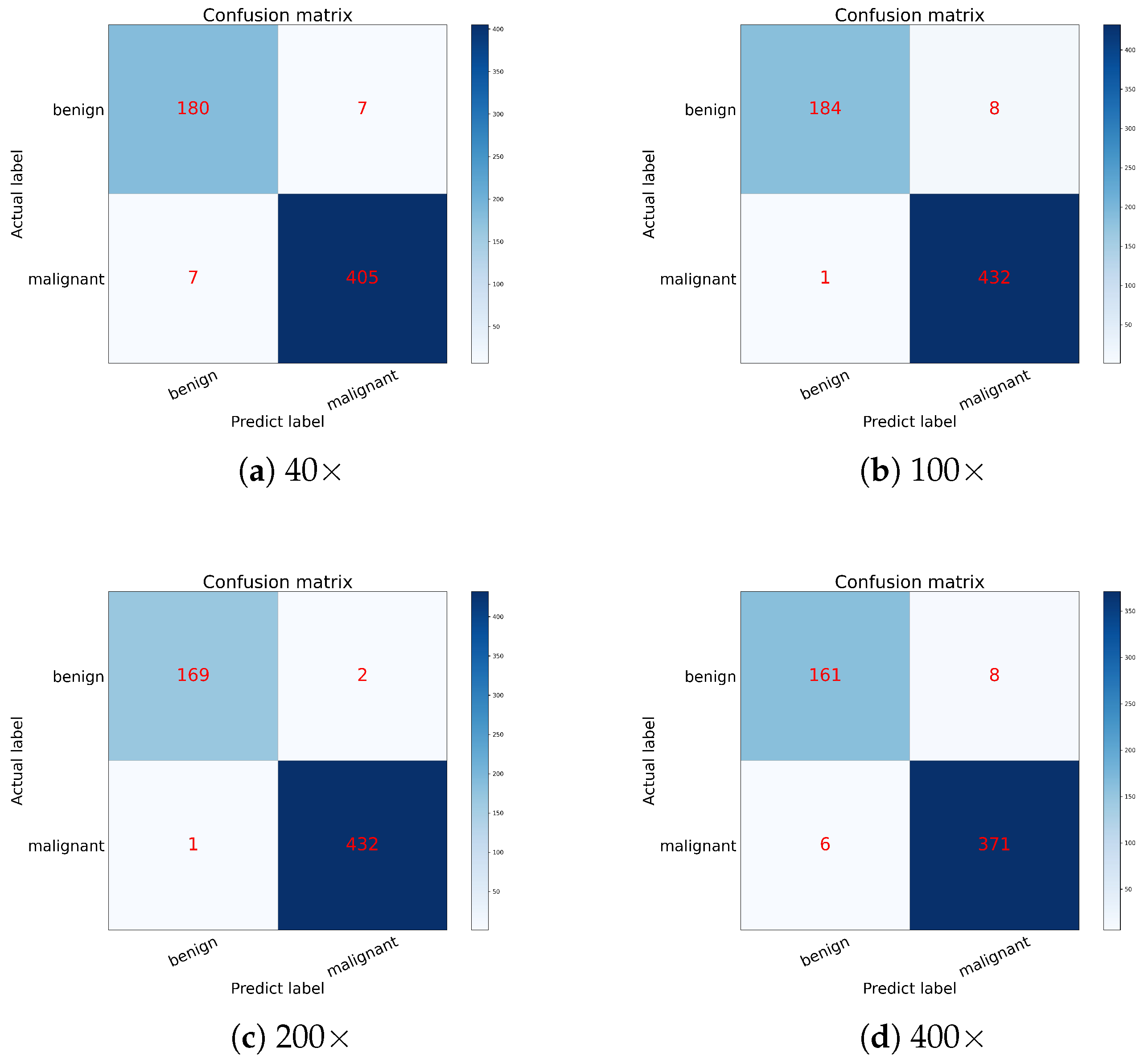
Table 3, and the confusion matrices for each magnification are shown in
Figure 8.
As shown in
Table 3, HoRFNet achieves high classification performance across all magnification levels. The average precision reaches 98.47%, the average recall is 99.05%, and the average F1-score is 98.88%. Among the four magnifications, the model achieves the best performance at 200×, with precision and F1-score reaching 99.53% and 99.64%, respectively. At 100× and 200×, recall reaches 99.76%, indicating that the model demonstrates strong sensitivity in detecting malignant samples.
Figure 8 further illustrates the model’s classification performance under different magnification conditions. For example, on the 40× dataset (
Figure 8a), the model correctly classifies 180 benign and 405 malignant samples, with only 14 misclassifications. On the 100× dataset (
Figure 8b), only one malignant sample and eight benign samples are misclassified. In the 200× dataset (
Figure 8c), only three misclassifications occur, representing the lowest error rate among all magnifications. In contrast, the classification performance at 400× (
Figure 8d) is slightly lower, with 14 misclassifications primarily occurring in the prediction of malignant cases. The results indicate that HoRFNet maintains stable and strong classification performance across different magnifications, with particularly high accuracy at 100× and 200×, further confirming the model’s reliability in BCHI classification.
3.5.2. Comparison with Other Methods
To further evaluate the classification performance of HoRFNet on multi-magnification images, this study conducts a systematic comparison with representative CNN methods from recent years. The comparison includes accuracy at both the image level and the patient level, as shown in
Table 4. In particular, SeONet and RFNet are included as reference models to further assess the improvements brought by HoRFNet.
Based on
Table 4, we conclude that HoRFNet demonstrates a significant competitive advantage over previously proposed models. Specifically, on the 200× dataset, HoRFNet achieves an optimal image level accuracy reaching 99.50%, while the performance at the patient level attains 99.23%. Compared with studies [
43,
44,
45,
46,
52,
54], HoRFNet improves accuracy at the image level by 8.50%, 14.60%, 10.40%, 11.02%, 7.05%, and 5.70%, respectively, and at the patient level by 8.23%, 15.13%, 9.43%, 10.03%, 6.95%, and 4.93%, respectively. Additionally, SeONet yields 98.01% accuracy for image level classification and 97.89% for patient-level prediction on the 200× dataset, while RFNet achieves 98.34% and 98.44%, respectively. Both models significantly outperform those in studies [
43,
44,
45,
46,
49,
52,
53]. However, HoRFNet reaches 97.66% accuracy on the 40× dataset in terms of image-level performance, which is slightly 0.33% lower than BreastNet proposed by Toğaçar et al. [
22]. Nevertheless, at other magnifications, the proposed model outperforms BreastNet in recognition accuracy. Compared with the models in recent studies [
53,
54,
55], HoRFNet shows consistently better results under all magnification settings of the BreakHis dataset. For instance, Xiao et al. [
54] employ Inception-V3 combined with image segmentation and fusion strategies to enhance BCHI classification accuracy. However, its average performance is approximately 5.70% and 4.93% lower than that of HoRFNet on image level and patient level evaluations, respectively. According to the above analysis results, compared with the network model in recent years, our HoRFNet has considerable advantages.
In addition, to comprehensively evaluate the performance of HoRFNet, we compare it with recent models based on the Transformer architecture. As shown in
Table 4, at the image level, Zhuang et al. [
29] achieved accuracies of 97.50%, 96.60%, 96.30%, and 96.10% across different magnifications. ST-Double-Net [
25] achieves corresponding accuracies of 97.47%, 96.86%, 97.25%, and 95.05%. HoRFNet consistently outperforms both Transformer-based methods at all magnification levels. In particular, at 100× and 200×, HoRFNet improves upon ResNet101+SwinT by 1.96% and 3.20%, respectively, and surpasses ST-Double-Net by 1.70% and 2.25%, respectively. Therefore, although Transformer-based architectures have recently shown impressive performance in vision tasks, HoRFNet demonstrates superior discriminative power in the BCHI classification.
3.5.3. Evaluation on the BACH Dataset
The above experiments demonstrate that HoRFNet achieves high classification accuracy on the BreakHis dataset. To further validate the model’s generalization ability and robustness, we conduct additional experiments on the BACH dataset, involving both two-class and four-class classification tasks. As shown in
Table 5, HoRFNet consistently outperforms the Backbone model on the BACH dataset. In the two-class classification task, HoRFNet achieves an accuracy of 88.75%, reflecting an 8.75% improvement over the Backbone’s 80.00%. In the four-class classification task, HoRFNet attains an accuracy of 82.50%, significantly outperforming the Backbone’s 76.25%, with a margin of 6.25%. These results indicate that HoRFNet maintains stable recognition performance across different data distributions and tissue types, further validating its generalizability and robustness in BCHI classification.
3.5.4. Model Efficiency Comparison
To comprehensively evaluate the proposed model in terms of computational efficiency, resource consumption, and classification performance, we conduct a systematic comparison at 100× magnification between HoRFNet and several baseline models, including the Backbone, SeONet, RFNet, and the lightweight EfficientNet-lite. The experimental results are presented in
Table 6. Compared to the Backbone model, HoRFNet shows significant advantages across multiple dimensions. First, in terms of classification performance, HoRFNet achieves the highest accuracy of 98.56%, significantly outperforming the Backbone and other comparison models. This result demonstrates that HoRFNet possesses stronger feature representation and discriminative capabilities. In terms of training efficiency, HoRFNet completes training in just 41 min, clearly outperforming the Backbone’s 64 min. This indicates that its optimized architecture and enhanced feature extraction capability effectively reduce training time. During inference, HoRFNet achieves an average inference time of 7.80 s, which is higher than the Backbone’s 3.28 s. However, this difference primarily results from the incorporation of high-order statistical modeling and multi-branch receptive field modules. Specifically, the matrix power normalization in covariance pooling and the use of dilated convolutions enhance feature representation and global perception, but also increase the complexity of the inference path. Despite this, HoRFNet still performs much faster than EfficientNet-lite, which takes 26.40 s, highlighting its strong deployment responsiveness in practical settings.
In terms of resource consumption, HoRFNet has 101.72 M parameters, which is only about one-fourth the size of the Backbone’s 396.98 M. This enhances the model’s compactness and deployment flexibility. Regarding computational complexity, HoRFNet requires 3.57 G FLOPs, which is comparable to the Backbone’s 3.45 G FLOPs. This demonstrates that HoRFNet substantially enhances representational capacity without introducing a significant computational burden. In contrast, although EfficientNet-lite has fewer parameters (81.64 M), its computational complexity reaches 6.80G FLOPs, which results in a disadvantage in actual computational efficiency. Overall, HoRFNet maintains excellent classification performance while achieving a favorable balance between computational efficiency and model effectiveness.
3.5.5. Visualization Results
To more clearly illustrate the advantages of HoRFNet in BCHI classification, this study presents a set of representative examples: images that are misclassified by the backbone model but correctly identified by HoRFNet. These examples are shown in
Figure 9. Upon inspection, these histopathological slide images often contain highly complex and subtly varying texture patterns, which challenge the feature extraction capability of the backbone model and may result in misclassification. In contrast, HoRFNet improves the ability to capture fine-grained information by introducing second-order statistical features and a receptive field enhancement module, enabling stronger discriminative power when processing complex images. This structural optimization significantly enhances recognition accuracy, further demonstrating the effectiveness of HoRFNet in BCHI classification.
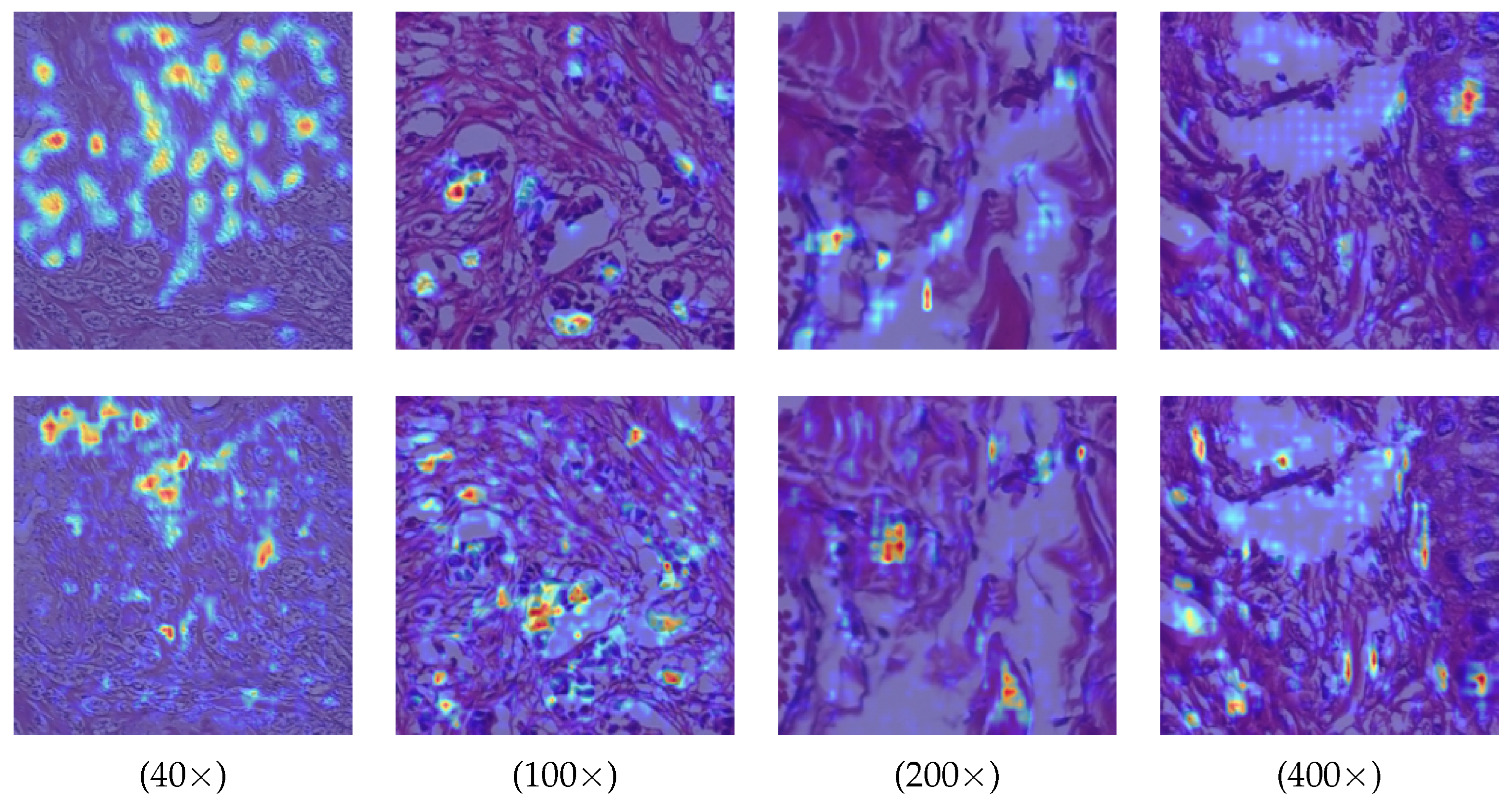
In addition, we provide visualizations of model attention regions under different magnification levels to further evaluate the discriminative capability of HoRFNet. As shown in
Figure 10, HoRFNet (second row) consistently exhibits more concentrated high-response regions across all magnifications, clearly outperforming the dispersed activation maps produced by the Backbone (first row). At 40× and 100×, HoRFNet effectively focuses on key tissue areas with pathological significance, whereas the Backbone shows substantial background interference and lacks precise region localization. At 200× and 400×, HoRFNet demonstrates superior sensitivity to fine-grained structures, accurately capturing edges and detailed regions, while the Backbone displays blurred boundaries and unstable attention patterns. These results indicate that HoRFNet has stronger multi-scale feature extraction capabilities, maintaining stable and accurate discriminative performance across varying resolutions.
4. Discussion
This work proposes the HoRFNet model, which integrates high-order modeling with a multi-branch receptive field mechanism and demonstrates notable advantages in BCHI classification. Experimental results show that the model achieves leading image-level and patient-level recognition performance across all four magnification levels in the BreakHis dataset, confirming its strong generalization and discriminative capabilities. It is important to note that BreakHis, as a widely used public dataset, provides strong representativeness and comparability, serving as a standardized evaluation platform for various histopathological image analysis methods. However, histopathological image of breast cancer may exhibit structural differences across countries, regions, climates, and population backgrounds, such as variations in tissue morphology, cell density, or staining conditions.
Looking ahead, we expand the scope of our study by exploring the model’s adaptability on multicenter datasets collected from different countries and regions. We also introduce data with diverse sources, staining styles, and preparation protocols to systematically evaluate the model’s cross-domain robustness and generalization performance. In addition, we consider incorporating other publicly available BCHI datasets to comprehensively assess the model’s performance across varying data environments and to further advance the practical deployment of HoRFNet in real-world clinical applications.