D4Care: A Deep Dynamic Memory-Driven Cross-Modal Feature Representation Network for Clinical Outcome Prediction
Abstract
1. Introduction
2. Methods
2.1. Time Series Model
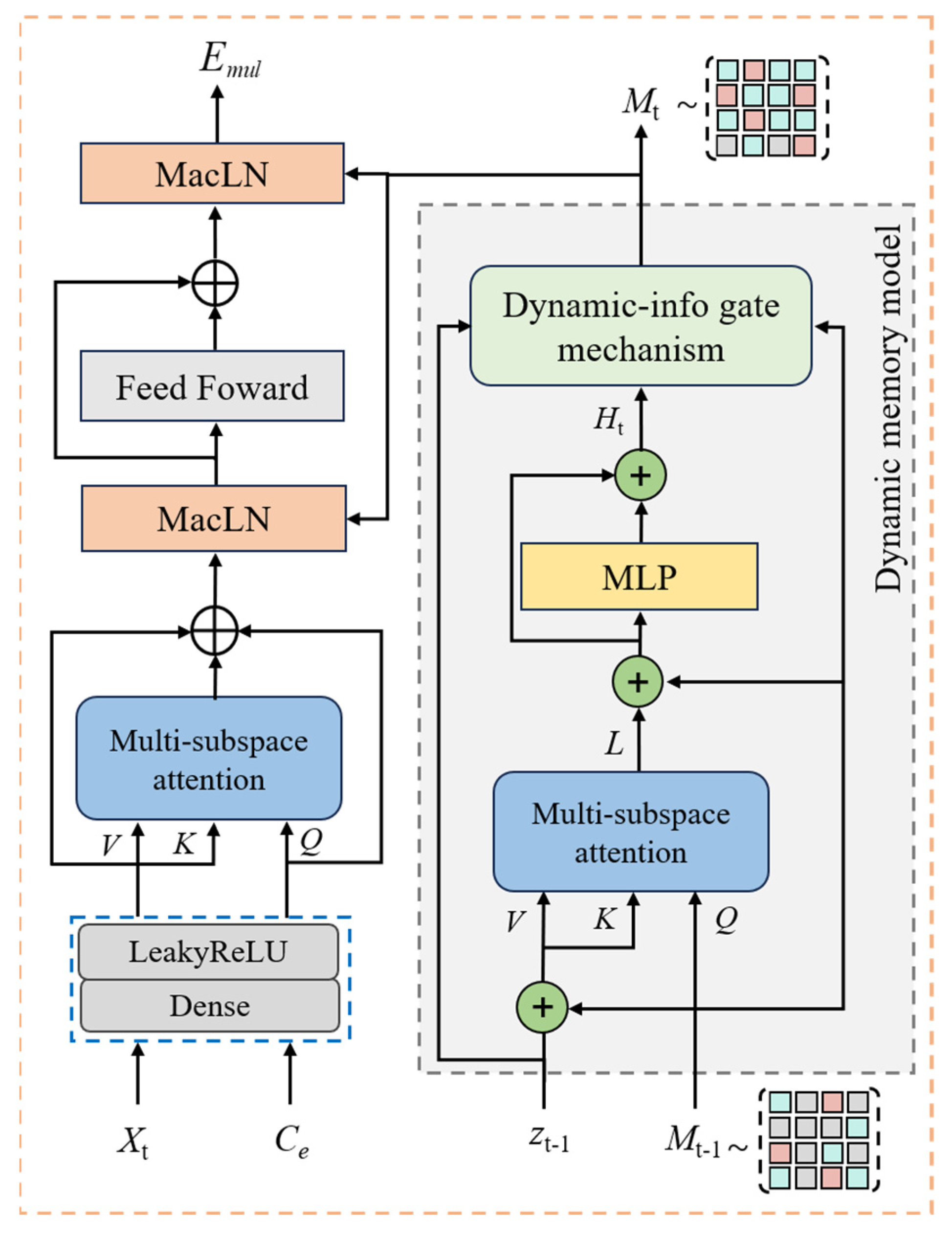
2.2. Dual-View Feature Encoding Model
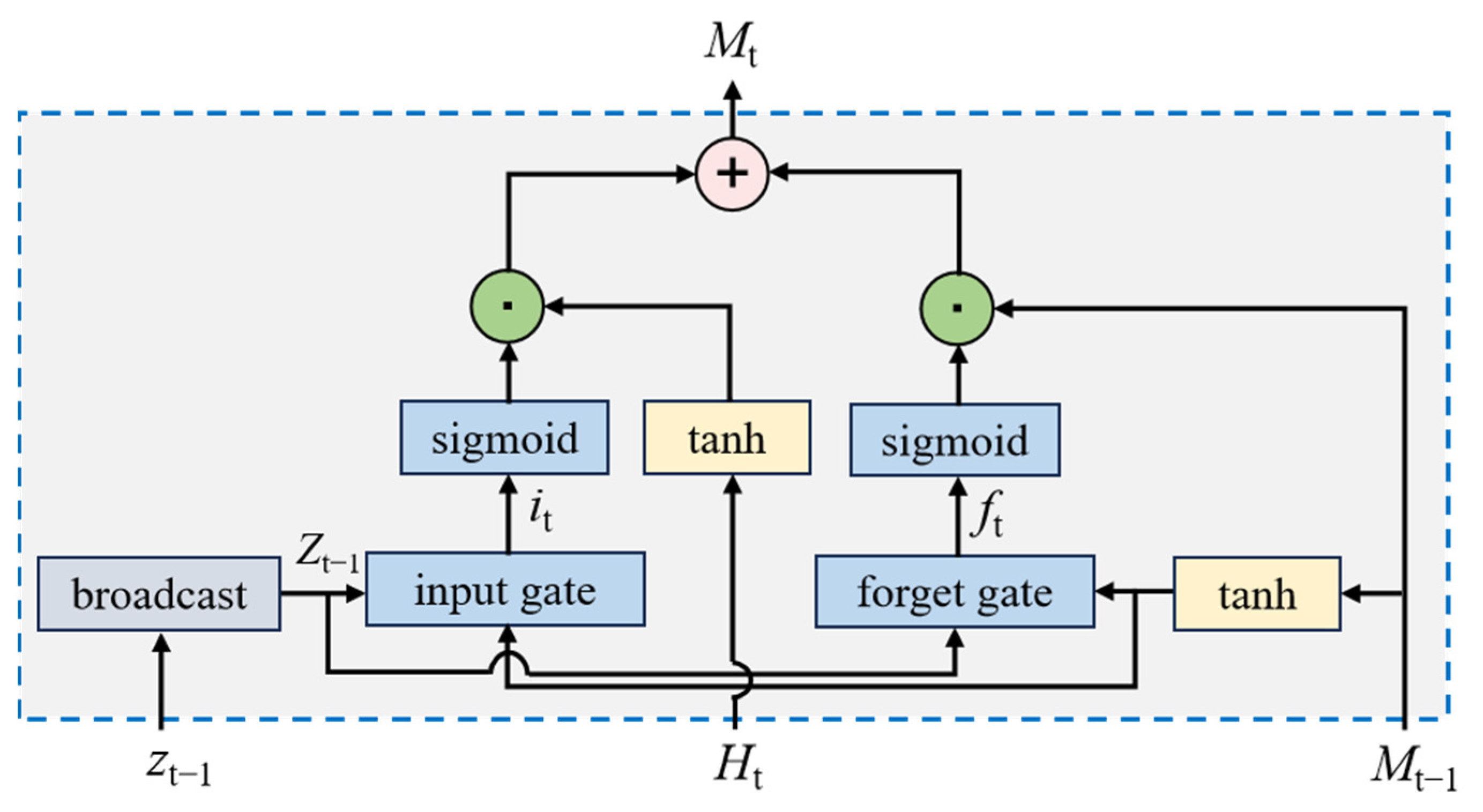
2.3. Memory-Driven Cross-Modal Attention Model
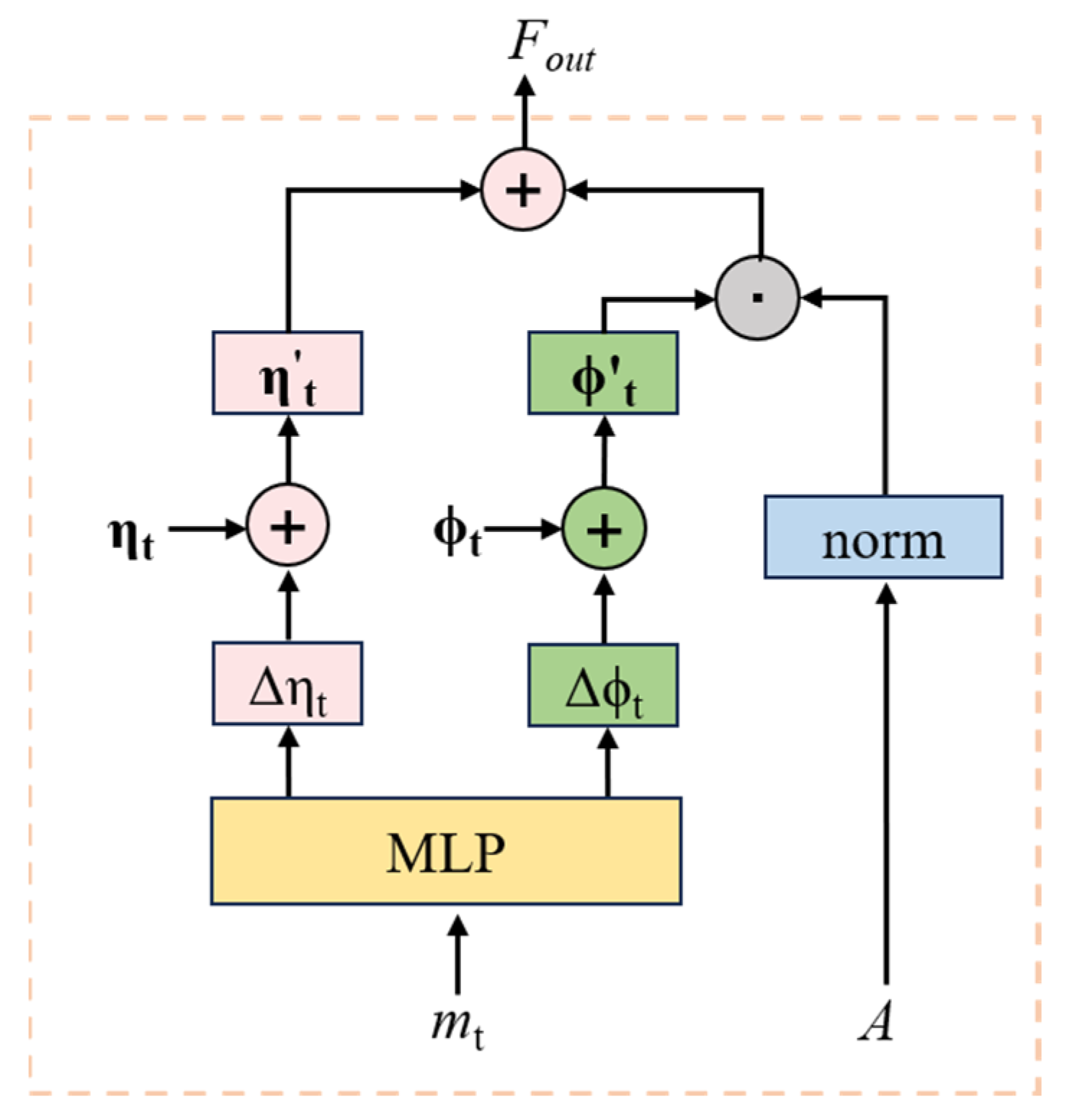
2.4. Clinical Prediction
3. Experiment and Discussion
3.1. Data Description
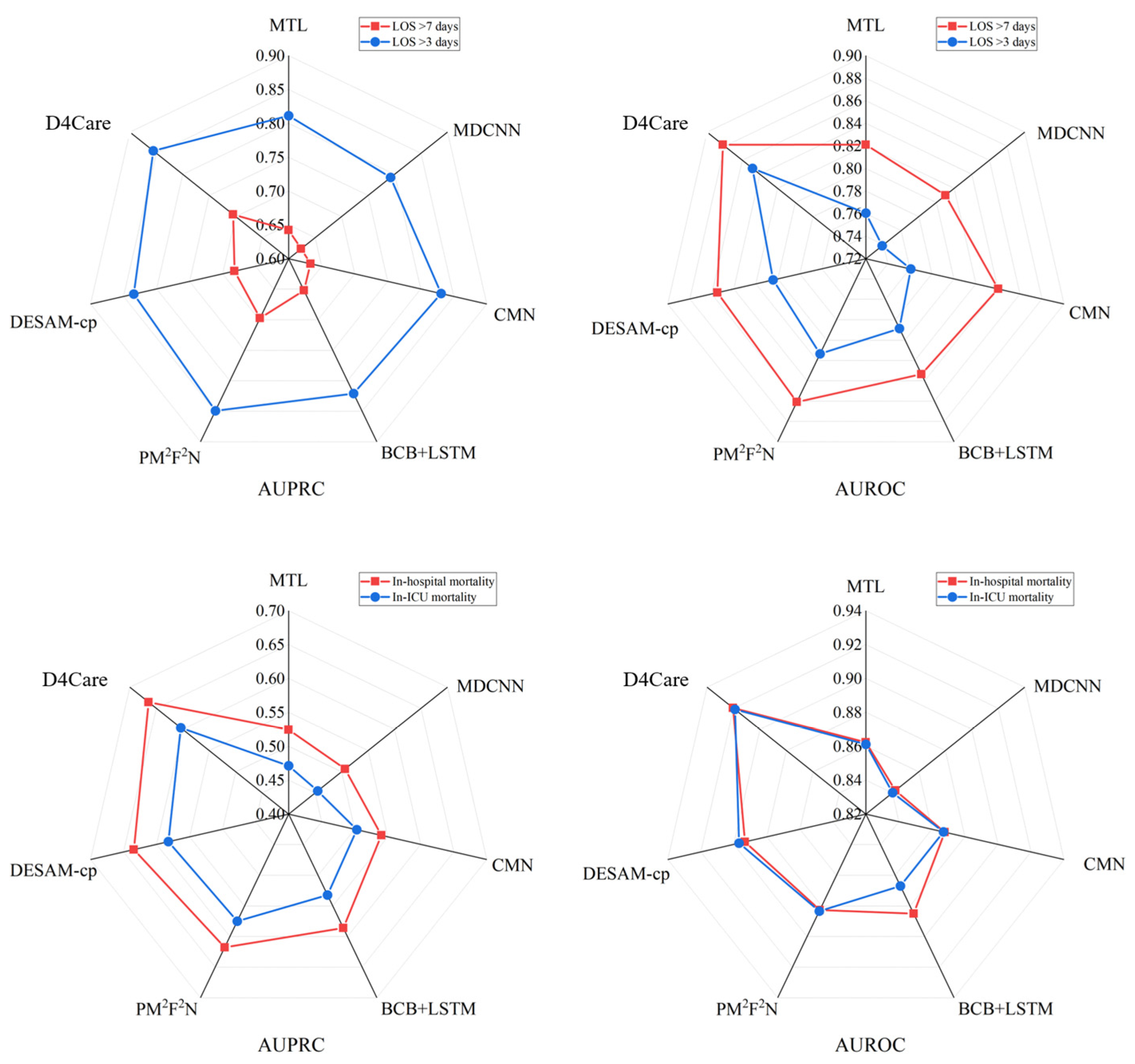
3.2. Results and Analysis
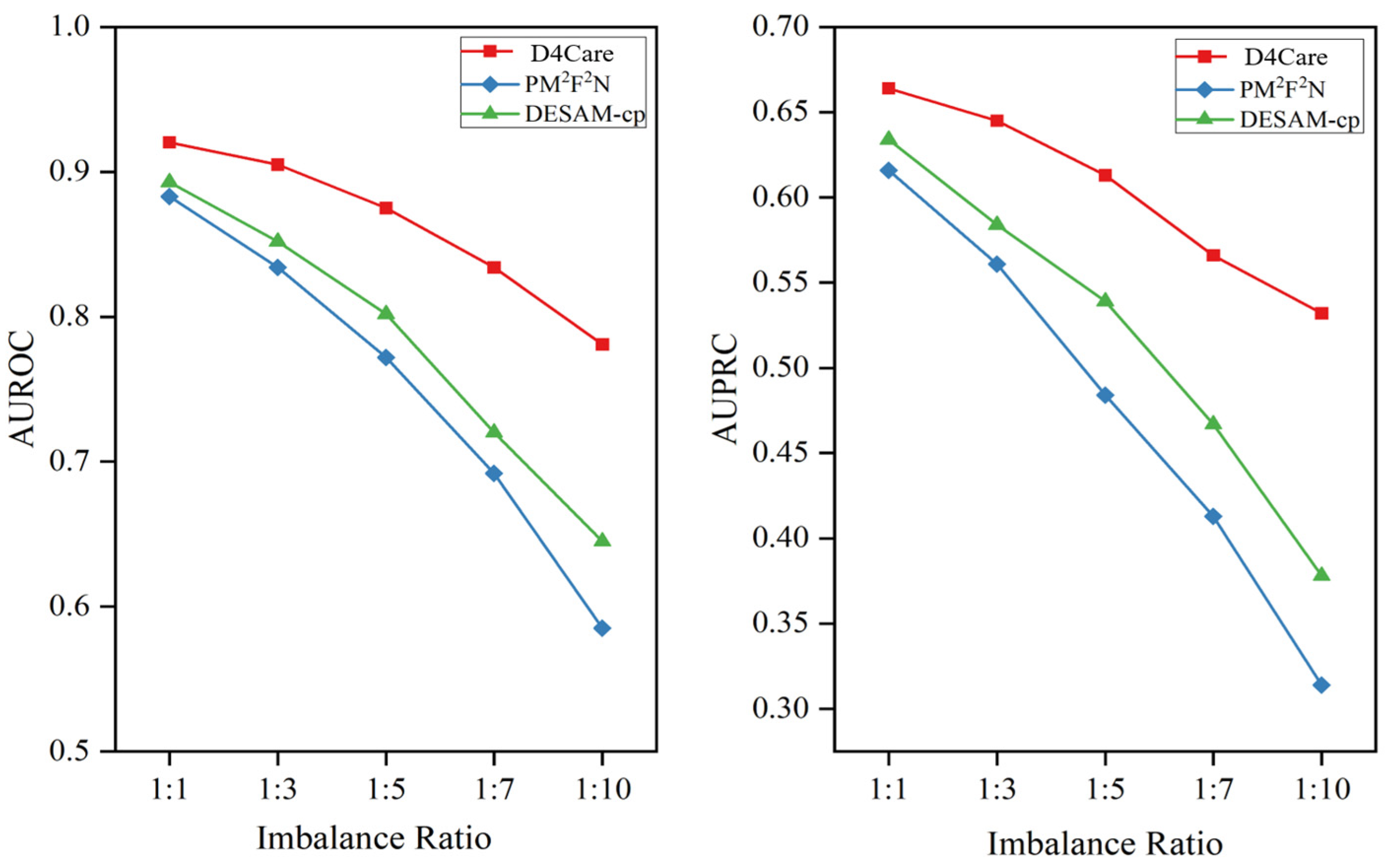
3.3. Performance on Imbalanced Data
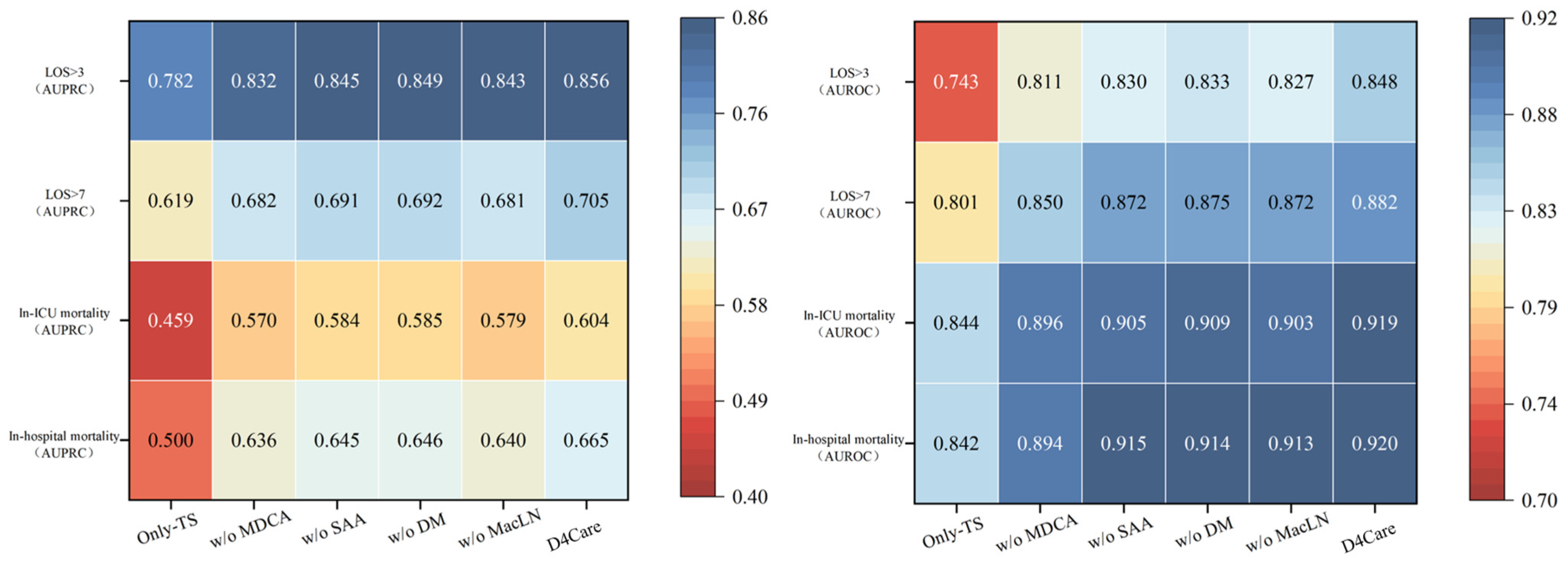
3.4. Ablation Study
3.5. Interpretability Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| BiGRU | Bi-directional Gated Recurrent Unit |
| BCB | Bio + ClinicalBERT |
| EHRs | Electronic health records |
| DM | Dynamic memory model |
| DGM | Dynamic-info gate mechanism |
| DVFE | Dual-view feature encoding model |
| MacLN | Memory-aware constrained layer normalization |
| MDCA | Memory-driven cross-modal attention model |
| NER | Named entity recognition |
| SAA | Sentence-aware attention model |
| TS | Time series model |
References
- Yang, C.; Kors, J.A.; Ioannou, S.; John, L.H.; Markus, A.F.; Rekkas, A.; Ridder, M.A.J.d.; Seinen, T.M.; Williams, R.D.; Rijnbeek, P.R. Trends in the conduct and reporting of clinical prediction model development and validation: A systematic review. J. Am. Med. Inform. Assoc. 2022, 29, 983–989. [Google Scholar] [CrossRef]
- Awad, A.; Bader-El-Den, M.; Mcnicholas, J.; Briggs, J. Early Hospital Mortality Prediction of Intensive Care Unit Patients Using an Ensemble Learning Approach. Int. J. Med. Inform. 2017, 108, 185–195. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, B.; Liu, G. Artificial intelligence can dynamically adjust strategies for auxiliary diagnosing respiratory diseases and analyzing potential pathological relationships. J. Breath Res. 2023, 17, 046007. [Google Scholar] [CrossRef]
- Chaudhry, B.; Wang, J.; Wu, S.; Maglione, M.; Mojica, W.; Roth, E.; Morton, S.C.; Shekelle, P.G. Systematic Review: Impact of Health Information Technology on Quality, Efficiency, and Costs of Medical Care. Ann. Intern. Med. 2006, 144, 742–752. [Google Scholar] [CrossRef]
- Burger, M.; Rätsch, G.; Kuznetsova, R. Multi-modal Graph Learning over UMLS Knowledge Graphs. In Proceedings of the Machine Learning for Health (ML4H), New Orleans, LA, USA, 10 December 2023; PMLR: Birmingham, UK, 2023; pp. 52–81. [Google Scholar]
- Gomes, B.; Pilz, M.; Reich, C.; Leuschner, F.; Konstandin, M.; Katus, H.A.; Meder, B. Machine learning-based risk prediction of intrahospital clinical outcomes in patients undergoing TAVI. Clin. Res. Cardiol. 2020, 110 (Suppl. S1), 343–356. [Google Scholar] [CrossRef]
- Zhang, X.; Qian, B.; Li, Y.; Liu, Y.; Chen, X.; Guan, C.; Li, C. Learning robust patient representations from multi-modal electronic health records: A supervised deep learning approach. In Proceedings of the 2021 SIAM International Conference on Data Mining (SDM), Alexandria, Egypt, 29 April–1 May 2021; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2021; pp. 585–593. [Google Scholar]
- Pham, T.; Tran, T.; Phung, D.; Venkatesh, S. DeepCare: A Deep Dynamic Memory Model for Predictive Medicine; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Huang, S.C.; Pareek, A.; Seyyedi, S.; Banerjee, I.; Lungren, M.P. Fusion of medical imaging and electronic health records using deep learning: A systematic review and implementation guidelines. npj Digit. Med. 2020, 3, 136. [Google Scholar] [CrossRef]
- Soenksen, L.R.; Ma, Y.; Zeng, C.; Boussioux, L.; Carballo, K.V.; Na, L.; Wiberg, H.M.; Li, M.L.; Fuentes, I.; Bertsimas, D. Integrated multimodal artificial intelligence framework for healthcare applications. npj Digit. Med. 2022, 5, 149. [Google Scholar] [CrossRef]
- Yang, B.; Wu, L. How to Leverage Multimodal EHR Data for Better Medical Predictions? arXiv 2021. [CrossRef]
- Ma, M.; Ren, J.; Zhao, L.; Tulyakov, S.; Wu, C.; Peng, X. SMIL: Multimodal Learning with Severely Missing Modality. arXiv 2021. [Google Scholar] [CrossRef]
- Msosa, Y.J.; Grauslys, A.; Zhou, Y.; Wang, T.; Buchan, I.; Langan, P.; Foster, S.; Walker, M.; Pearson, M.; Folarin, A.; et al. Trustworthy Data and AI Environments for Clinical Prediction: Application to Crisis-Risk in People With Depression. J. Biomed. Health Inform. (J-BHI) 2023, 27, 11. [Google Scholar] [CrossRef]
- Yang, Z.; Mitra, A.; Liu, W.; Berlowitz, D.; Yu, H. TransformEHR: Transformer-based encoder-decoder generative model to enhance prediction of disease outcomes using electronic health records. Nat. Commun. 2023, 14, 7857. [Google Scholar] [CrossRef]
- Van Aken, B.; Papaioannou, J.M.; Mayrdorfer, M.; Budde, K.; Gers, F.; Loeser, A. Clinical outcome prediction from admission notes using self-supervised knowledge integration. arXiv 2021, arXiv:2102.04110. [Google Scholar]
- Chandak, P.; Huang, K.; Zitnik, M. Building a knowledge graph to enable precision medicine. Sci. Data 2023, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Sauer, C.M.; Chen, L.C.; Hyland, S.L.; Girbes, A.; Elbers, P.; Celi, L.A. Leveraging electronic health records for data science: Common pitfalls and how to avoid them. Lancet Digit. Health 2022, 4, e893–e898. [Google Scholar] [CrossRef]
- Jiang, P.; Xiao, C.; Cross, A.; Sun, J. GraphCare: Enhancing Healthcare Predictions with Personalized Knowledge Graphs. arXiv 2023, arXiv:2305.12788. [Google Scholar]
- Zhao, Y.; Hong, Q.; Zhang, X.; Deng, Y.; Wang, Y.; Petzold, L. Bertsurv: Bert-based survival models for predicting outcomes of trauma patients. arXiv 2021, arXiv:2103.10928. [Google Scholar]
- Miotto, R.; Li, L.; Kidd, B.A.; Dudley, J.T. Deep Patient: An Unsupervised Representation to Predict the Future of Patients from the Electronic Health Records. Sci. Rep. 2016, 6, 26094. [Google Scholar] [CrossRef]
- Ye, X.; Wu, J.; Mou, C.; Dai, W. Medlens: Improve mortality prediction via medical signs selecting and regression. In Proceedings of the 2023 IEEE 3rd International Conference on Computer Communication and Artificial Intelligence (CCAI), Taiyuan, China, 26–28 May 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 169–175. [Google Scholar]
- Jain, S.; Burger, M.; Rätsch, G.; Kuznetsova, R. Knowledge Graph Representations to enhance Intensive Care Time-Series Predictions. arXiv 2023, arXiv:2311.07180. [Google Scholar]
- Zhang, K.; Niu, K.; Zhou, Y.; Tai, W.; Lu, G. MedCT-BERT: Multimodal Mortality Prediction using Medical ConvTransformer-BERT Model. In Proceedings of the 2023 IEEE 35th International Conference on Tools with Artificial Intelligence (ICTAI), Atlanta, GA, USA, 6–8 November 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 700–707. [Google Scholar]
- Thilak, V.; Huang, C.; Saremi, O.; Dinh, L.; Goh, H.; Nakkiran, P.; Susskind, J.M.; Littwin, E. LiDAR: Sensing Linear Probing Performance in Joint Embedding SSL Architectures. arXiv 2023, arXiv:2312.04000. [Google Scholar]
- Niu, K.; Zhang, K.; Peng, X.; Xiao, N. Deep multi-modal intermediate fusion of clinical record and time series data in mortality prediction. Front. Mol. Biosci. 2023, 10, 1136071. [Google Scholar] [CrossRef]
- Chen, Z.; Song, Y.; Chang, T.H.; Wan, X. Generating radiology reports via memory-driven transformer. arXiv 2020, arXiv:2010.16056. [Google Scholar]
- Lyu, W.; Dong, X.; Wong, R.; Zheng, S.; Abell-Hart, K.; Wang, F.; Chen, C. A multimodal transformer: Fusing clinical notes with structured EHR data for interpretable in-hospital mortality prediction. In Proceedings of the AMIA Annual Symposium Proceedings, Washington, DC, USA, 5–9 November 2022; American Medical Informatics Association: Bethesda, MA, USA, 2022; Volume 2022, p. 719. [Google Scholar]
- An, Y.; Li, R.; Chen, X. Merge: A multi-graph attentive representation learning framework integrating group information from similar patients. Comput. Biol. Med. 2022, 151, 106245. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Chen, D.; Jin, X.; Xu, G.; Tang, C.; Guo, X.; Tang, Z.; Bao, Y.; Wang, F.; Shen, R. Association between acute kidney injury and prognoses of cardiac surgery patients: Analysis of the MIMIC-III database. Front. Surg. 2023, 9, 1044937. [Google Scholar] [CrossRef]
- Park, Y.; Ho, J.C. Califorest: Calibrated random forest for health data. In Proceedings of the ACM Conference on Health, Inference, and Learning, New York, NY, USA, 2–4 April 2020; pp. 40–50. [Google Scholar]
- Xia, Z.; Xu, P.; Xiong, Y.; Lai, Y.; Huang, Z. Survival Prediction in Patients with Hypertensive Chronic Kidney Disease in Intensive Care Unit: A Retrospective Analysis Based on the MIMIC-III Database. J. Immunol. Res. 2022, 2022, 3377030. [Google Scholar] [CrossRef]
- An, Y.; Cai, G.; Chen, X.; Guo, L. PARSE: A personalized clinical time-series representation learning framework via abnormal offsets analysis. Comput. Methods Programs Biomed. 2023, 242, 107838. [Google Scholar] [CrossRef]
- Liu, R.; Gutiérrez, R.; Mather, R.V.; Stone, T.A.D.; Mercado, L.A.S.C.; Bharadwaj, K.; Johnson, J.; Das, P.; Balanza, G.; Uwanaka, E.; et al. Development and prospective validation of postoperative pain prediction from preoperative EHR data using attention-based set embeddings. npj Digit. Med. 2023, 6, 209. [Google Scholar] [CrossRef]
- Ni, P.; Li, Y.; Zhu, J.; Peng, J.; Dai, Z.; Li, G.; Bai, X. Disease diagnosis prediction of emr based on BiGRU-ATT-capsnetwork model. In Proceedings of the 2019 IEEE International Conference on Big Data (Big Data), Los Angeles, CA, USA, 9–12 December 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 6166–6168. [Google Scholar]
- Huang, K.; Altosaar, J.; Ranganath, R. ClinicalBERT: Modeling Clinical Notes and Predicting Hospital Readmission. arXiv 2019. [Google Scholar] [CrossRef]
- Kormilitzin, A.; Vaci, N.; Liu, Q.; Nevado-Holgado, A. Med7: A transferable clinical natural language processing model for electronic health records. Artif. Intell. Med. 2021, 118, 102086. [Google Scholar] [CrossRef] [PubMed]
- Alsentzer, E.; Murphy, J.R.; Boag, W.; Weng, W.H.; Jin, D.; Naumann, T.; McDermott, M. Publicly available clinical BERT embeddings. arXiv 2019, arXiv:1904.03323. [Google Scholar]
- Wang, S.; Mcdermott, M.B.A.; Chauhan, G.; Ghassemi, M.; Hughes, M.C.; Naumann, T. MIMIC-Extract: A Data Extraction, Preprocessing, and Representation Pipeline for MIMIC-III. In Proceedings of the ACM Conference on Health, Inference, and Learning, Toronto, ON, Canada, 2–4 April 2020. [Google Scholar] [CrossRef]
- Si, Y.; Roberts, K. Deep patient representation of clinical notes via multi-task learning for mortality prediction. AMIA Summits Transl. Sci. Proc. 2019, 2019, 779. [Google Scholar]
- Khadanga, S.; Aggarwal, K.; Joty, S.; Srivastava, J. Using clinical notes with time series data for icu management. arXiv 2019, arXiv:1909.09702. [Google Scholar]
- Bardak, B.; Tan, M. Improving clinical outcome predictions using convolution over medical entities with multimodal learning. Artif. Intell. Med. 2021, 117, 102112. [Google Scholar] [CrossRef] [PubMed]
- Deznabi, I.; Iyyer, M.; Fiterau, M. Predicting in-hospital mortality by combining clinical notes with time-series data. In Proceedings of the Findings of the Association for Computational Linguistics: ACL-IJCNLP, Online Event, 1–6 August 2021; Volume 2021, pp. 4026–4031. [Google Scholar]
- Zhang, Y.; Zhou, B.; Song, K.; Sui, X.; Zhao, G.; Jiang, N.; Yuan, X. PM2F2N: Patient multi-view multi-modal feature fusion networks for clinical outcome prediction. In Proceedings of the Findings of the Association for Computational Linguistics: EMNLP, Abu Dhabi, United Arab Emirates, 7–11 December 2022; Volume 2022, pp. 1985–1994. [Google Scholar]
- Lee, S.; Jang, G.; Kim, C.; Park, S.; Yoo, K.; Kim, J.; Kim, S.; Kang, J. Enhancing Clinical Outcome Predictions through Auxiliary Loss and Sentence-Level Self-Attention. In Proceedings of the 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Istanbul, Türkiye, 5–8 December 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1210–1217. [Google Scholar]
- Sundararajan, M.; Taly, A.; Yan, Q. Axiomatic attribution for deep networks. In Proceedings of the International conference on machine learning, Sydney, NSW, Australia, 6–11 August 2017; PMLR: Birmingham, UK, 2017; pp. 3319–3328. [Google Scholar]

| DATA | # Patient | # Hospital | # ICU |
|---|---|---|---|
| MIMIC-III (>15 years old) | 38,597 | 49,785 | 53,423 |
| MIMIC-Extract | 34,472 | 34,472 | 34,472 |
| MIMIC-Extract (at least 24 + 6 (gap) hours patient) | 23,937 | 23,937 | 23,937 |
| Final cohort | 21,080 | 21,080 | 21,080 |
| Type | Mortality | Length of Stay (LOS) | ||
|---|---|---|---|---|
| In-Hospital Mortality | In-ICU Mortality | LOS > 7 | LOS > 3 | |
| ratio | 89.5%:10.5% | 93%:7% | 56.8%:43.2% | 92.1%:7.9% |
| Entity Type | Total Entity | Unique Entity | Example |
|---|---|---|---|
| Drug | 742,231 | 18,204 | Magnesium |
| Strength | 152,234 | 10,680 | 400 mg/5 mL |
| Route | 207,876 | 1192 | PO |
| Dosage | 126,756 | 7230 | 30 mL |
| Form | 40,885 | 597 | suspension |
| Frequency | 71,285 | 3279 | bid |
| Duration | 5830 | 1185 | next 5 days |
| Model | In-Hospital Mortality | In-ICU Mortality | LOS > 7 | LOS > 3 | ||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | |
| MTL | 0.8623 (±0.0143) | 0.5243 (±0.0140) | 0.8611 (±0.0123) | 0.4712 (±0.0088) | 0.8211 (±0.0012) | 0.6423 (±0.0123) | 0.7605 (±0.0043) | 0.8113 (±0.0034) |
| MDCNN | 0.8423 (±0.0042) | 0.5067 (±0.0051) | 0.8402 (±0.0044) | 0.4548 (±0.0062) | 0.8102 (±0.0012) | 0.6236 (±0.0052) | 0.7385 (±0.0095) | 0.7925 (±0.0026) |
| CMN | 0.8678 (±0.0092) | 0.5403 (±0.0012) | 0.8670 (±0.0012) | 0.5032 (±0.0014) | 0.8402 (±0.0012) | 0.6332 (±0.0016) | 0.7608 (±0.0048) | 0.8308 (±0.0053) |
| BCB + LSTM | 0.8850 (±0.0021) | 0.5860 (±0.0048) | 0.8670 (±0.0012) | 0.5322 (±0.0036) | 0.8335 (±0.0019) | 0.6520 (±0.0085) | 0.7886 (±0.0023) | 0.8210 (±0.0019) |
| PM2F2N | 0.8827 (±0.0034) | 0.6178 (±0.0034) | 0.8834 (±0.0023) | 0.5750 (±0.0028) | 0.8609 (±0.0008) | 0.6974 (±0.0011) | 0.8135 (±0.0010) | 0.8492 (±0.0036) |
| DESAM-cp | 0.8933 (±0.0016) | 0.6344 (±0.0023) | 0.8968 (±0.0035) | 0.5822 (±0.0021) | 0.8549 (±0.0094) | 0.6820 (±0.0028) | 0.8043 (±0.0028) | 0.8345 (±0.0032) |
| D4Care | 0.9203 (±0.0028) | 0.6645 (±0.0014) | 0.9189 (±0.0045) | 0.6038 (±0.0033) | 0.8820 (±0.0056) | 0.7048 (±0.0012) | 0.8484 (±0.0011) | 0.8557 (±0.0022) |
| Model | In-Hospital Mortality | In-ICU Mortality | LOS > 7 | LOS > 3 | ||||
|---|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | AUROC | AUPRC | |
| Only-TS | 0.8420 (±0.0065) | 0.5002 (±0.0082) | 0.8436 (±0.0032) | 0.4587 (±0.0010) | 0.8010 (±0.0013) | 0.6189 (±0.0034) | 0.7432 (±0.0027) | 0.7824 (±0.0031) |
| w/o MDCA | 0.8940 (±0.0008) | 0.6358 (±0.0014) | 0.8964 (±0.0024) | 0.5702 (±0.0027) | 0.8501 (±0.0092) | 0.6822 (±0.0023) | 0.8110 (±0.0024) | 0.8322 (±0.0019) |
| w/o SAA | 0.9147 (±0.0011) | 0.6448 (±0.0027) | 0.9045 (±0.0031) | 0.5842 (±0.0018) | 0.8719 (±0.0010) | 0.6914 (±0.0011) | 0.8303 (±0.0015) | 0.8446 (±0.0011) |
| w/o DM | 0.9138 (±0.0017) | 0.6460 (±0.0028) | 0.9092 (±0.0012) | 0.5851 (±0.0008) | 0.8752 (±0.0016) | 0.6920 (±0.0044) | 0.8326 (±0.0023) | 0.8490 (±0.0017) |
| w/o MacLN | 0.9132 (±0.0082) | 0.6401 (±0.0056) | 0.9032 (±0.0019) | 0.5788 (±0.0021) | 0.8718 (±0.0032) | 0.6811 (±0.0024) | 0.8267 (±0.0020) | 0.8434 (±0.0024) |
| D4Care | 0.9203 (±0.0028) | 0.6645 (±0.0014) | 0.9189 (±0.0045) | 0.6038 (±0.0033) | 0.8820 (±0.0056) | 0.7048 (±0.0012) | 0.8484 (±0.0011) | 0.8557 (±0.0022) |
| IG Rank | Clinically Significant Words | Common Words | IG Rank | Clinically Significant Words | Common Words |
|---|---|---|---|---|---|
| 1 | pain | the | 10 | conditions | diseases |
| 2 | fever | with | 11 | failure | from |
| 3 | cough | medical | 12 | drug | history |
| 4 | respiratory | diagnosis | 13 | pulses | admitted |
| 5 | pneumonia | year | 14 | sob | end |
| 6 | heart | old | 15 | acute | let |
| 7 | brain | will | 16 | insulin | visit |
| 8 | clear | not | 17 | increasing | unspecified |
| 9 | mental | possible | 18 | seizure | status |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, B.; Liu, G. D4Care: A Deep Dynamic Memory-Driven Cross-Modal Feature Representation Network for Clinical Outcome Prediction. Appl. Sci. 2025, 15, 6054. https://doi.org/10.3390/app15116054
Chen B, Liu G. D4Care: A Deep Dynamic Memory-Driven Cross-Modal Feature Representation Network for Clinical Outcome Prediction. Applied Sciences. 2025; 15(11):6054. https://doi.org/10.3390/app15116054
Chicago/Turabian StyleChen, Binyue, and Guohua Liu. 2025. "D4Care: A Deep Dynamic Memory-Driven Cross-Modal Feature Representation Network for Clinical Outcome Prediction" Applied Sciences 15, no. 11: 6054. https://doi.org/10.3390/app15116054
APA StyleChen, B., & Liu, G. (2025). D4Care: A Deep Dynamic Memory-Driven Cross-Modal Feature Representation Network for Clinical Outcome Prediction. Applied Sciences, 15(11), 6054. https://doi.org/10.3390/app15116054






