Semantic Segmentation of Brain Tumors Using a Local–Global Attention Model
Abstract
1. Introduction
- (1)
- We propose semantic-oriented masked attention, a novel mechanism that applies global attention while integrating semantic supervision to enhance the precision of feature extraction in the decoder.
- (2)
- We propose network-in-network blocks to replace the residual blocks in the feature fusion component of the original Swin UNETR architecture, with the goal of capturing inter-dependencies between feature channels.
- (3)
- Experimental results show that our model achieves higher segmentation accuracy than several recent leading models on two public brain tumor datasets.The source code with our proposed model are available at: https://github.com/laizhui/LG-UNETR (accessed on 26 March 2025)
2. Related Works
2.1. CNN-Based Segmentation Models
2.2. Transformer-Based Segmentation Models
2.3. Hybrid Models
3. Methods
3.1. The Overall Architecture of Swin UNETR
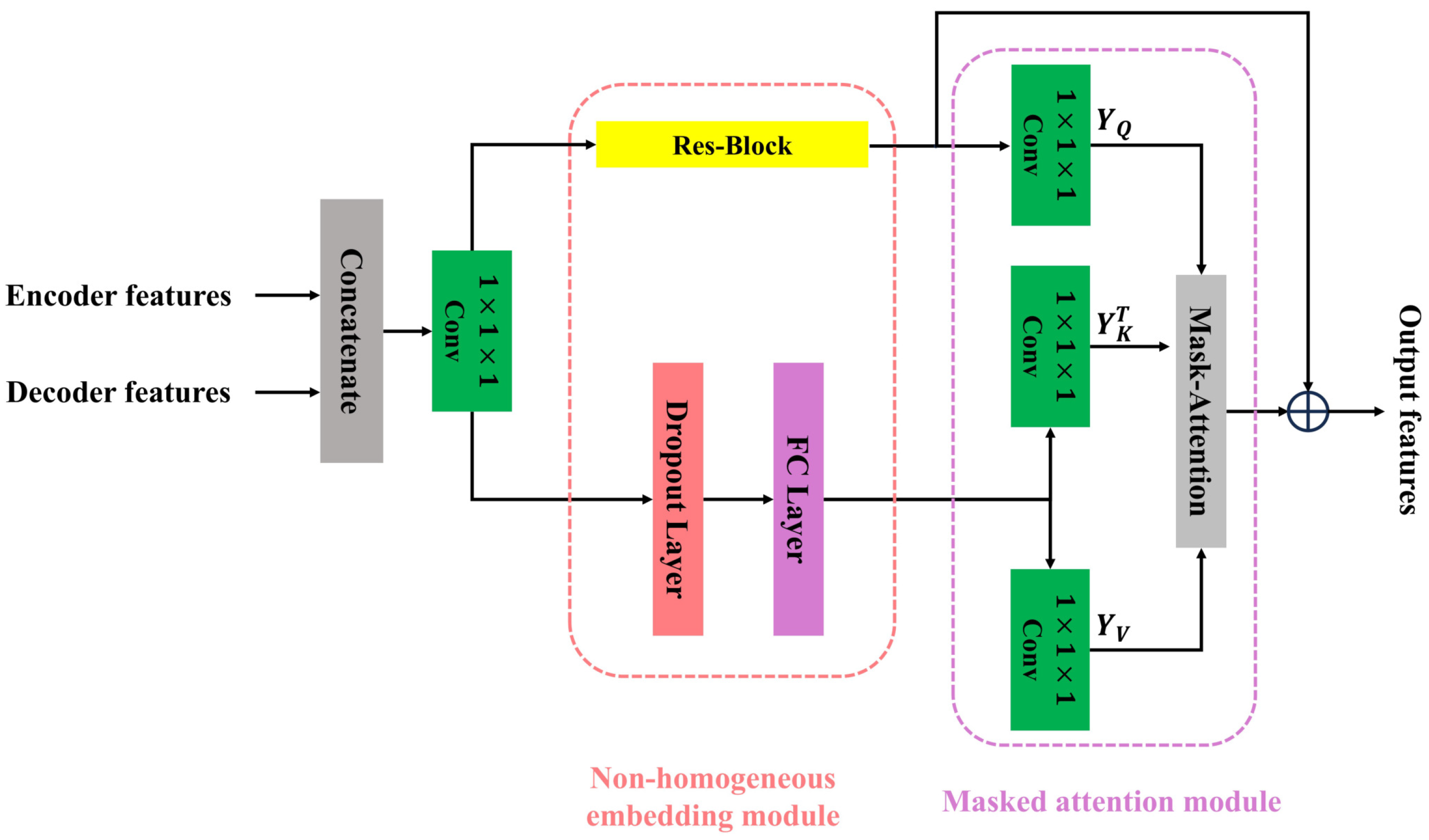
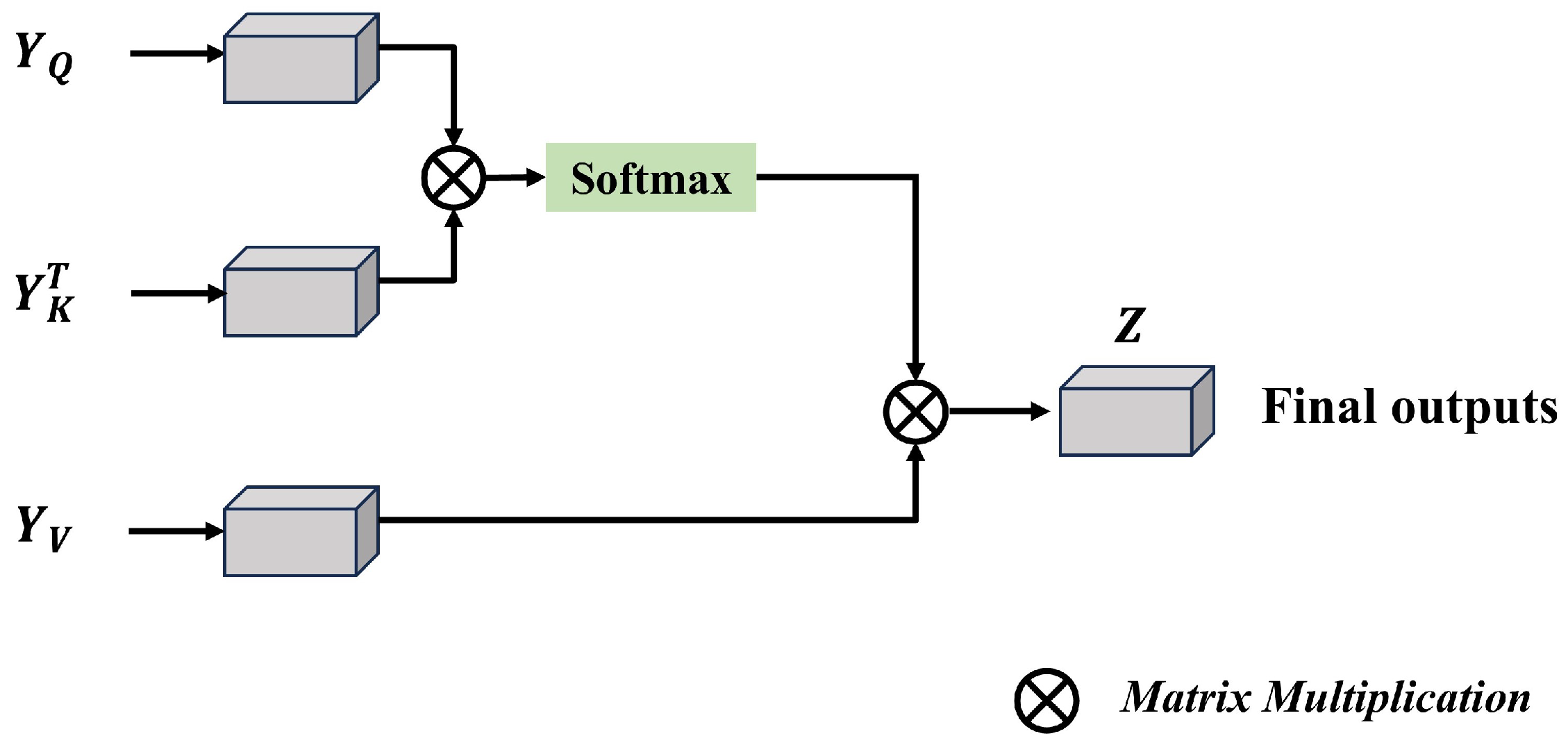
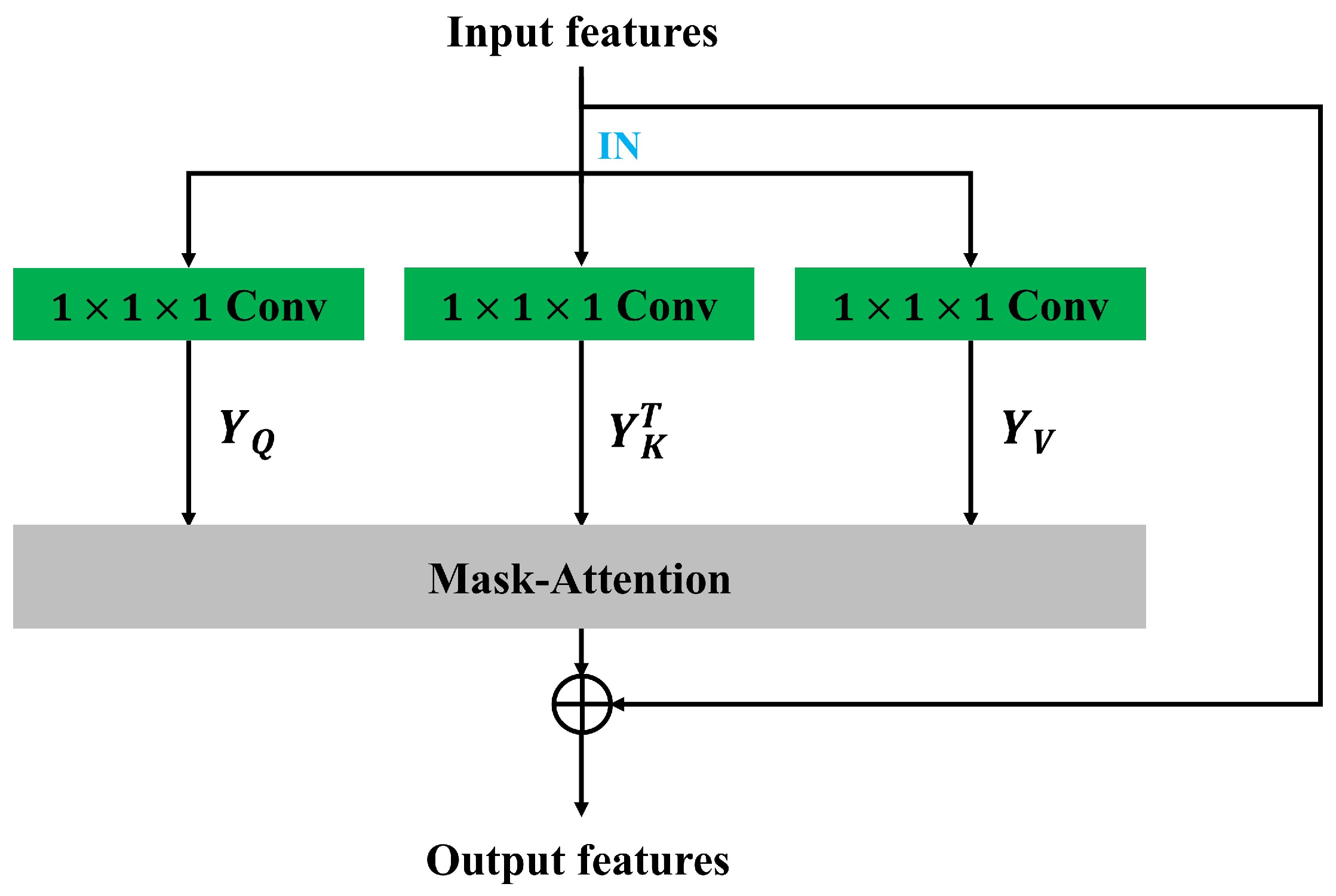
3.2. Semantic-Oriented Masked Attention
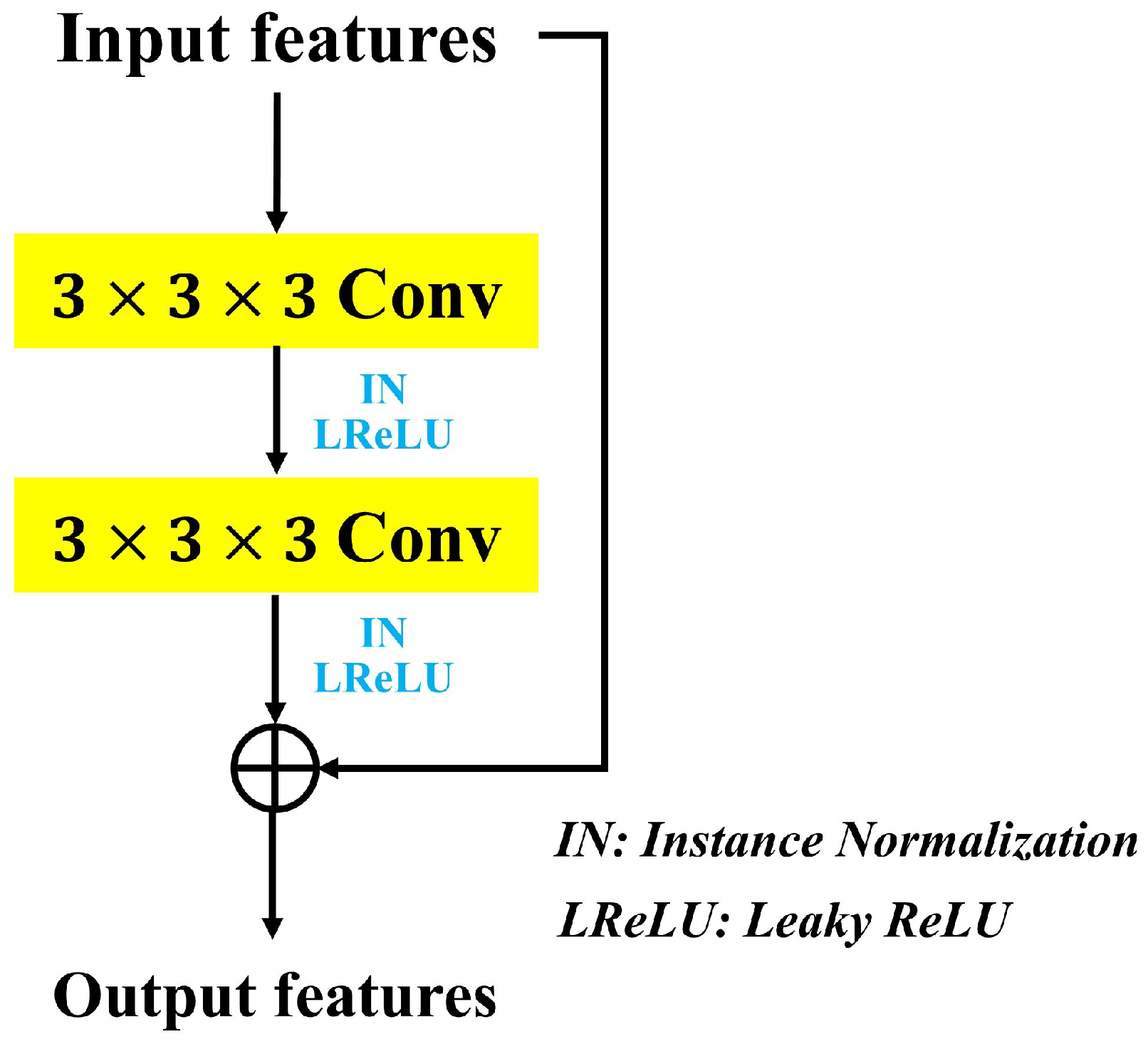
3.3. Network-in-Network Blocks
3.4. Loss Function
4. Experiments and Results
4.1. Datasets
4.1.1. BraTS2023-GLI Dataset
4.1.2. BraTS2024-GLI Dataset
4.1.3. Comparison Between the Two Datasets
4.2. Evaluation Metrics
4.3. Implementation Details
4.4. Comparison with State-of-the-Art Methods
4.4.1. Comparison on the BraTS2024-GLI Dataset
4.4.2. Comparison on the BraTS2023-GLI Dataset
4.5. Ablation Studies
- (1)
- Evaluating the contribution of each proposed block to the overall performance of our model.
- (2)
- Examining the influence of the number and location of SMA blocks on segmentation accuracy.
- (3)
- Investigating the effect of model width on model performance.
- (4)
- Exploring the impact of hyperparameter P on model performance.
4.5.1. The Effectiveness of Proposed Blocks
4.5.2. The Effects of the Number and Location of SMA Blocks on Performance
4.5.3. The Effects of Model Width on Performance
4.5.4. The Effects of Hyperparameter P on Performance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, C.; Wang, R.; Liu, Z.; Wang, M.; Tan, H.; Wu, Y.; Liu, X.; Sun, H.; Yang, R.; et al. Annotation-efficient deep learning for automatic medical image segmentation. Nat. Commun. 2020, 12, 5915. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, N.F.; Miller, G.; Moen, E.; Kong, A.; Kagel, A.; Dougherty, T.; Fullaway, C.C.; McIntosh, B.J.; Leow, K.S.; Schwartz, M.; et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 2021, 40, 555–565. [Google Scholar] [CrossRef]
- Ma, J.; He, Y.; Li, F.; Han, L.J.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2023, 15, 654. [Google Scholar] [CrossRef]
- Cinarer, G.; Emiroglu, B.G. Classificatin of Brain Tumors by Machine Learning Algorithms. In Proceedings of the 2019 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), Ankara, Turkey, 11–13 October 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Padlia, M.; Sharma, J. Fractional Sobel Filter Based Brain Tumor Detection and Segmentation Using Statistical Features and SVM. In Nanoelectronics, Circuits and Communication Systems; Springer: Singapore, 2018. [Google Scholar] [CrossRef]
- Virupakshappa; Amarapur, B. An Automated Approach for Brain Tumor Identification using ANN Classifier. In Proceedings of the 2017 International Conference on Current Trends in Computer, Electrical, Electronics and Communication (CTCEEC), Mysore, India, 8–9 September 2017; pp. 1011–1016. [Google Scholar] [CrossRef]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.C.; Bengio, Y.; Pal, C.J.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with Deep Neural Networks. Med Image Anal. 2016, 35, 18–31. [Google Scholar] [CrossRef]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Nyúl, L.G.; Udupa, J.K.; Zhang, X. New variants of a method of MRI scale standardization. IEEE Trans. Med. Imaging 2000, 19, 143–150. [Google Scholar] [CrossRef]
- Syazwany, N.S.; Nam, J.H.; Chul Lee, S. MM-BiFPN: Multi-Modality Fusion Network With Bi-FPN for MRI Brain Tumor Segmentation. IEEE Access 2021, 9, 160708–160720. [Google Scholar] [CrossRef]
- Sagar, A. ViTBIS: Vision Transformer for Biomedical Image Segmentation. arXiv 2022, arXiv:2201.05920. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Wu, Y.; Liao, K.Y.; Chen, J.; Wang, J.; Chen, D.Z.; Gao, H.; Wu, J. D-former: A U-shaped Dilated Transformer for 3D medical image segmentation. Neural Comput. Appl. 2022, 35, 1931–1944. [Google Scholar] [CrossRef]
- Wei, C.; Ren, S.; Guo, K.; Hu, H.; Liang, J. High-Resolution Swin Transformer for Automatic Medical Image Segmentation. Sensors 2023, 23, 3420. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Xiao, B.; Liu, D.; Wang, J. Deep High-Resolution Representation Learning for Human Pose Estimation. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 5686–5696. [Google Scholar] [CrossRef]
- Wang, L.; Dong, X.; Wang, Y.; Liu, L.; An, W.; Guo, Y.K. Learnable Lookup Table for Neural Network Quantization. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 12413–12423. [Google Scholar] [CrossRef]
- Liang, J.; Yang, C.; Zeng, M.; Wang, X. TransConver: Transformer and convolution parallel network for developing automatic brain tumor segmentation in MRI images. Quant. Imaging Med. Surg. 2021, 12, 2397–2415. [Google Scholar] [CrossRef]
- Wang, W.; Chen, C.; Ding, M.; Li, J.; Yu, H.; Zha, S. TransBTS: Multimodal Brain Tumor Segmentation Using Transformer. arXiv 2021, arXiv:2103.04430. [Google Scholar]
- Hatamizadeh, A.; Nath, V.; Tang, Y.; Yang, D.; Roth, H.R.; Xu, D. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. arXiv 2022, arXiv:2201.01266. [Google Scholar]
- Karargyris, A.; Umeton, R.; Sheller, M.J.; Aristizabal, A.; George, J.; Bala, S.; Beutel, D.J.; Bittorf, V.; Chaudhari, A.; Chowdhury, A.; et al. Federated benchmarking of medical artificial intelligence with MedPerf. Nat. Mach. Intell. 2021, 5, 799–810. [Google Scholar] [CrossRef]
- de Verdier, M.C.; Saluja, R.; Gagnon, L.; Labella, D.; Baid, U.; Tahon, N.E.H.M.; Foltyn-Dumitru, M.; Zhang, J.; Alafif, M.M.; Baig, S.; et al. The 2024 Brain Tumor Segmentation (BraTS) Challenge: Glioma Segmentation on Post-treatment MRI. arXiv 2024, arXiv:2405.18368. [Google Scholar]
- Baid, U.; Ghodasara, S.; Bilello, M.; Mohan, S.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S.; et al. The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification. arXiv 2021, arXiv:2107.02314. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.S.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Xiao, X.; Lian, S.; Luo, Z.; Li, S. Weighted Res-UNet for High-Quality Retina Vessel Segmentation. In Proceedings of the 2018 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, China, 19–21 October 2018; pp. 327–331. [Google Scholar] [CrossRef]
- Guan, S.; Khan, A.A.; Sikdar, S.; Chitnis, P.V. Fully Dense UNet for 2D Sparse Photoacoustic Tomography Artifact Removal. IEEE J. Biomed. Health Inform. 2018, 24, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. arXiv 2018, arXiv:1807.10165. [Google Scholar]
- Milletarì, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.J.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2016, 36, 61–78. [Google Scholar] [CrossRef]
- Chen, C.; Liu, X.; Ding, M.; Zheng, J.; Li, J. 3D Dilated Multi-Fiber Network for Real-time Brain Tumor Segmentation in MRI. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2020, 18, 203–211. [Google Scholar] [CrossRef]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.; Wu, J. UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation. In Proceedings of the ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–9 May 2020; pp. 1055–1059. [Google Scholar] [CrossRef]
- Peiris, H.; Hayat, M.; Chen, Z.; Egan, G.F.; Harandi, M. A Robust Volumetric Transformer for Accurate 3D Tumor Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021. [Google Scholar] [CrossRef]
- Xing, Z.; Yu, L.; Wan, L.; Han, T.; Zhu, L. NestedFormer: Nested Modality-Aware Transformer for Brain Tumor Segmentation. arXiv 2022, arXiv:2208.14876. [Google Scholar]
- Pinaya, W.H.L.; Tudosiu, P.D.; Gray, R.J.; Rees, G.; Nachev, P.; Ourselin, S.; Cardoso, M.J. Unsupervised brain imaging 3D anomaly detection and segmentation with transformers. Med. Image Anal. 2022, 79, 102475. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021; pp. 9992–10002. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like Pure Transformer for Medical Image Segmentation. arXiv 2021, arXiv:2105.05537. [Google Scholar]
- Liang, J.; Yang, C.; Zhong, J.; Ye, X. BTSwin-Unet: 3D U-shaped Symmetrical Swin Transformer-based Network for Brain Tumor Segmentation with Self-supervised Pre-training. Neural Process. Lett. 2022, 55, 3695–3713. [Google Scholar] [CrossRef]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. arXiv 2018, arXiv:1807.10165. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Multi-Atlas Labeling Beyond the Cranial Vault. 2015. Available online: https://www.synapse.org/Synapse:syn3193805/wiki/ (accessed on 25 March 2025).
- Hatamizadeh, A.; Yang, D.; Roth, H.R.; Xu, D. UNETR: Transformers for 3D Medical Image Segmentation. In Proceedings of the 2022 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 3–8 January 2022; pp. 1748–1758. [Google Scholar] [CrossRef]
- Roy, S.; Koehler, G.; Ulrich, C.; Baumgartner, M.; Petersen, J.; Isensee, F.; Jaeger, P.F.; Maier-Hein, K.H. MedNeXt: Transformer-driven Scaling of ConvNets for Medical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2023. [Google Scholar] [CrossRef]
- Shaker, A.M.; Maaz, M.; Rasheed, H.A.; Khan, S.H.; Yang, M.; Khan, F.S. UNETR++: Delving into Efficient and Accurate 3D Medical Image Segmentation. IEEE Trans. Med. Imaging 2024, 43, 3377–3390. [Google Scholar] [CrossRef]
- Xie, Q.; Chen, Y.; Liu, S.; Lu, X. SSCFormer: Revisiting ConvNet-Transformer Hybrid Framework From Scale-Wise and Spatial-Channel-Aware Perspectives for Volumetric Medical Image Segmentation. IEEE J. Biomed. Health Inform. 2024, 28, 4830–4841. [Google Scholar] [CrossRef]
- Lin, C.W.; Chen, Z. MM-UNet: A novel cross-attention mechanism between modules and scales for brain tumor segmentation. Eng. Appl. Artif. Intell. 2024, 133, 108591. [Google Scholar] [CrossRef]
- Yu, F.; Cao, J.; Liu, L.; Jiang, M. SuperLightNet: Lightweight Parameter Aggregation Network for Multimodal Brain Tumor Segmentation. In Proceedings of the 2025 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 19–25 October 2025. [Google Scholar]
- Yang, Y.; Wang, Y.; Qin, C. Pancreas segmentation with multi-channel convolution and combined deep supervision. J. Biomed. Eng. 2025, 42, 140–147. [Google Scholar]
- Lee, H.H.; Bao, S.; Huo, Y.; Landman, B.A. 3D UX-Net: A Large Kernel Volumetric ConvNet Modernizing Hierarchical Transformer for Medical Image Segmentation. arXiv 2022, arXiv:2209.15076. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. arXiv 2019, arXiv:1912.01703. [Google Scholar]
- Cardoso, M.J.; Li, W.; Brown, R.; Ma, N.; Kerfoot, E.; Wang, Y.; Murrey, B.; Myronenko, A.; Zhao, C.; Yang, D.; et al. MONAI: An open-source framework for deep learning in healthcare. arXiv 2022, arXiv:2211.02701. [Google Scholar]
- Loshchilov, I.; Hutter, F. Decoupled Weight Decay Regularization. arXiv 2017, arXiv:1711.05101. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 11966–11976. [Google Scholar] [CrossRef]




| Dataset | Total No. of Images | No. of Images with Red Labels | No. of Images with Green Labels | No. of Images with Blue Labels | No. of Images with At Least One Missing Label |
|---|---|---|---|---|---|
| BraTS2023-GLI | 1251 | 1208 | 1250 | 1218 | 71 |
| BraTS2024-GLI | 1350 | 565 | 1350 | 990 | 791 |
| Model | Params (M) (↓) | GFLOPs (↓) | Dice (%) (↑) | (mm) (↓) | |||
|---|---|---|---|---|---|---|---|
| ET | TC | WT | Avg. | ||||
| MedNeXt [47] | 61.7 | 1079.9 | 74.61 | 71.49 | 85.45 | 77.19 | 12.24 |
| UNETR [46] | 173.6 | 2095.4 | 78.92 | 75.44 | 86.16 | 80.17 | 8.42 |
| UNETR++ [48] | 280.7 | 1912.6 | 79.22 | 75.77 | 87.19 | 80.72 | 8.45 |
| Swin UNETR [21] | 97.1 | 1234.5 | 79.14 | 76.36 | 87.39 | 80.96 | 8.42 |
| 3D UX-NET [53] | 82.9 | 2362.4 | 80.18 | 76.96 | 87.22 | 81.45 | 8.40 |
| LG UNETR (ours) | 83.9 | 1150.9 | 80.80 | 78.25 | 88.50 | 82.51 | 8.02 |
| Model | Params (M) (↓) | GFLOPs (↓) | Dice (%) (↑) | (mm) (↓) | |||
|---|---|---|---|---|---|---|---|
| ET | TC | WT | Avg. | ||||
| MedNeXt [47] | 61.7 | 1079.9 | 84.80 | 86.25 | 90.84 | 87.30 | 5.11 |
| UNETR [46] | 173.6 | 2095.4 | 84.91 | 86.86 | 90.79 | 87.52 | 5.59 |
| UNETR++ [48] | 280.7 | 1912.6 | 85.58 | 86.85 | 91.91 | 88.12 | 5.08 |
| Swin UNETR [21] | 97.1 | 1234.5 | 85.77 | 87.95 | 92.10 | 88.61 | 5.06 |
| 3D UX-NET [53] | 82.9 | 2362.4 | 86.21 | 88.79 | 92.51 | 89.17 | 4.90 |
| LG UNETR (ours) | 83.9 | 1150.9 | 86.41 | 88.28 | 92.69 | 89.12 | 4.81 |
| Model | Blocks | Params (M) (↓) | GFLOPs (↓) | Dice (%) (↑) | ||||
|---|---|---|---|---|---|---|---|---|
| SMA | NiN | ET | TC | WT | Avg. | |||
| Model 1 | 97.1 | 1234.5 | 79.14 | 76.36 | 87.39 | 80.96 | ||
| Model 2 | ✓ | 99.8 | 1242.9 | 80.54 | 77.95 | 88.13 | 82.21 | |
| Model 3 | ✓ | 72.0 | 1199.5 | 80.24 | 77.39 | 87.95 | 81.86 | |
| Ours | ✓ | ✓ | 83.9 | 1150.9 | 80.80 | 78.25 | 88.50 | 82.51 |
| Setting | Stage 1 | Stage 2 | Stage 3 | Stage 4 | Bottom | Params (M) (↓) | GFLOPs (↓) | Dice (%) (↑) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ET | TC | WT | Avg. | ||||||||
| Setting 1 | ✓ | ✓ | ✓ | 46.6 | 1186.4 | 80.40 | 77.70 | 88.00 | 82.04 | ||
| Setting 2 | ✓ | ✓ | ✓ | 91.3 | 1198.5 | 80.42 | 77.68 | 88.25 | 82.12 | ||
| Setting 3 | ✓ | ✓ | ✓ | ✓ | 83.0 | 1150.8 | 79.78 | 76.30 | 88.11 | 81.40 | |
| Setting 4 | ✓ | ✓ | ✓ | ✓ | ✓ | 83.9 | 1150.9 | 80.80 | 78.25 | 88.50 | 82.51 |
| Feature Size | Params (M) (↓) | GFLOPs (↓) | Dice (%) (↑) | |||
|---|---|---|---|---|---|---|
| ET | TC | WT | Avg. | |||
| [36,72,144,288] 1 | 57.9 | 438.8 | 79.83 | 77.11 | 88.53 | 81.82 |
| [48,96,192,384] | 72.9 | 771.8 | 80.42 | 77.74 | 88.14 | 82.10 |
| [60,120,240,480] | 83.9 | 1150.9 | 80.80 | 78.25 | 88.50 | 82.51 |
| P | Params (M) (↓) | GFLOPs (↓) | Dice (%) (↑) | |||
|---|---|---|---|---|---|---|
| ET | TC | WT | Avg. | |||
| 64 | 64.7 | 1149.4 | 80.30 | 77.13 | 87.92 | 81.78 |
| 128 | 83.9 | 1150.9 | 80.80 | 78.25 | 88.50 | 82.51 |
| 256 | 122.2 | 1153.7 | 80.42 | 77.52 | 88.05 | 82.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, S.; Lai, Z.; Zhu, J.; He, W.; Mao, G. Semantic Segmentation of Brain Tumors Using a Local–Global Attention Model. Appl. Sci. 2025, 15, 5981. https://doi.org/10.3390/app15115981
Xing S, Lai Z, Zhu J, He W, Mao G. Semantic Segmentation of Brain Tumors Using a Local–Global Attention Model. Applied Sciences. 2025; 15(11):5981. https://doi.org/10.3390/app15115981
Chicago/Turabian StyleXing, Shuli, Zhenwei Lai, Junxiong Zhu, Wenwu He, and Guojun Mao. 2025. "Semantic Segmentation of Brain Tumors Using a Local–Global Attention Model" Applied Sciences 15, no. 11: 5981. https://doi.org/10.3390/app15115981
APA StyleXing, S., Lai, Z., Zhu, J., He, W., & Mao, G. (2025). Semantic Segmentation of Brain Tumors Using a Local–Global Attention Model. Applied Sciences, 15(11), 5981. https://doi.org/10.3390/app15115981






