Hybrid Transform-Based Feature Extraction for Skin Lesion Classification Using RGB and Grayscale Analysis
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Image Dataset
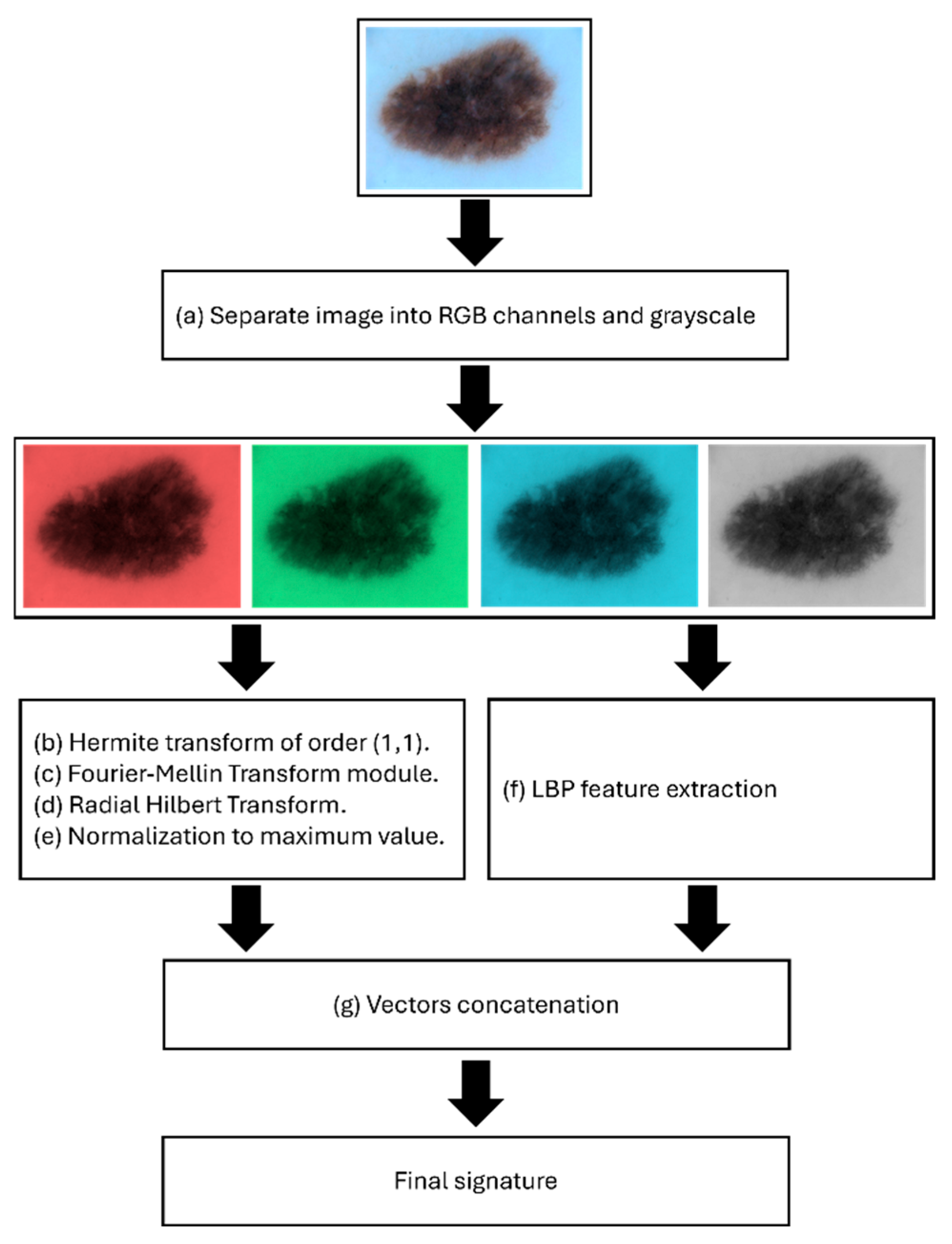
2.2. RGB Channel and Grayscale Separation
2.3. Hermite Transform
2.4. The Signatures
2.5. Radial Fourier–Mellin Signatures Through Hilbert Transform
2.6. Signatures Classification
2.7. Model Architecture
2.8. Model Compilation
2.9. Training Procedure
2.10. Model Performance
3. Results
- : true positives for class .
- : true negatives for class .
- : false positives for class .
- : false negatives for class .
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shetty, B.; Fernandes, R.; Rodrigues, A.P.; Chengoden, R.; Bhattacharya, S.; Lakshmanna, K. Skin lesion classification of dermoscopic images using machine learning and convolutional neural network. Sci. Rep. 2022, 12, 18134. [Google Scholar]
- Wu, Y.; Chen, B.; Zeng, A.; Pan, D.; Wang, R.; Zhao, S. Skin cancer classification with deep learning: A systematic review. Front. Oncol. 2022, 12, 893972. [Google Scholar] [CrossRef] [PubMed]
- Dubal, P.; Bhatt, S.; Joglekar, C.; Patil, S. Skin Cancer Detection and Classification. In Proceedings of the 2017 6th International Conference on Electrical Engineering and Informatics (ICEEI), Langkawi, Malaysia, 25–27 November 2017; pp. 1–6. [Google Scholar]
- Cullell-Dalmau, M.; Noé, S.; Otero-Viñas, M.; Meić, I.; Manzo, C. Convolutional neural network for skin lesion classification: Understanding the fundamentals through hands-on learning. Front. Med. 2021, 8, 644327. [Google Scholar] [CrossRef]
- Debelee, T.G. Skin lesion classification and detection using machine learning techniques: A systematic review. Diagnostics 2023, 13, 3147. [Google Scholar] [CrossRef]
- Sulthana, R.; Chamola, V.; Hussain, Z.; Albalwy, F.; Hussain, A. A novel end-to-end deep convolutional neural network based skin lesion classification framework. Expert Syst. Appl. 2024, 246, 123056. [Google Scholar]
- Nugroho, A.K.; Wardoyo, R.; Wibowo, M.E.; Soebono, H. Image dermoscopy skin lesion classification using deep learning method: Systematic literature review. Bull. Electr. Eng. Inform. 2024, 13, 1042–1049. [Google Scholar] [CrossRef]
- Hu, Z.; Mei, W.; Chen, H.; Hou, W. Multi-scale feature fusion and class weight loss for skin lesion classification. Comput. Biol. Med. 2024, 176, 108594. [Google Scholar] [CrossRef]
- Benjamin, J.R.; Jayasree, T. Improved medical image fusion based on cascaded PCA and shift invariant wavelet transforms. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 229–240. [Google Scholar] [CrossRef]
- Barajas-Garcia, C.; Solorza-Calderon, S.; Gutierrez-Lopez, E. Scale, translation and rotation invariant wavelet local feature descriptor. Appl. Math. Comput. 2019, 363, 124594. [Google Scholar] [CrossRef]
- Kim, H.K.; Kim, J.D. Region-based shape descriptor invariant to rotation, scale and translation. Signal Process. Image Commun. 2000, 16, 87–93. [Google Scholar] [CrossRef]
- Popa, C.A.; Cernăzanu-Glăvan, C. Fourier Transform-Based Image Classification Using Complex-Valued Convolutional Neural Networks. In Advances in Neural Networks–ISNN 2018, Proceedings of the 15th International Symposium on Neural Networks, Minsk, Belarus, 25–28 June 2018; Springer International Publishing: Cham, Switzerland, 2018; Volume 15, pp. 300–309. [Google Scholar]
- Uzun, I.S.; Amira, A.; Bouridane, A. FPGA implementations of fast Fourier transforms for real-time signal and image processing. IEE Proc. Vis. Image Signal Process. 2005, 152, 283–296. [Google Scholar] [CrossRef]
- Braccini, C.; Gambardella, G.; Grattarola, A. Digital Image Processing by Means of Generalized Scale-Invariant Filters. In Issues in Acoustic Signal—Image Processing and Recognition; Springer: Berlin/Heidelberg, Germany, 1983; pp. 315–329. [Google Scholar]
- De Sena, A.; Rocchesso, D. A fast Mellin and scale transform. EURASIP J. Adv. Signal Process. 2007, 2007, 089170. [Google Scholar] [CrossRef]
- Bigot, J.; Gamboa, F.; Vimond, M. Estimation of translation, rotation, and scaling between noisy images using the Fourier–Mellin transform. SIAM J. Imaging Sci. 2009, 2, 614–645. [Google Scholar] [CrossRef]
- Guerra-Rosas, E.; López-Ávila, L.F.; Garza-Flores, E.; Vidales-Basurto, C.A.; Álvarez-Borrego, J. Classification of skin lesion images using artificial intelligence methodologies through radial Fourier–Mellin and Hilbert transform signatures. Appl. Sci. 2023, 13, 11425. [Google Scholar] [CrossRef]
- Garza-Flores, E.; Guerra-Rosas, E.; Álvarez-Borrego, J. Spectral indexes obtained by implementation of the fractional Fourier and Hermite transform for the diagnosis of malignant melanoma. Biomed. Opt. Express 2019, 10, 6043–6056. [Google Scholar] [CrossRef]
- Martins, A.S.; Neves, L.A.; de Faria, P.R.; Tosta, T.A.A.; Longo, L.C.; Silva, A.B.; Roberto, G.F.; Nascimento, M.Z.D. A Hermite polynomial algorithm for detection of lesions in lymphoma images. Pattern Anal. Appl. 2021, 24, 523–535. [Google Scholar] [CrossRef]
- Barba-J, L.; Vargas-Quintero, L.; Calderón-Agudelo, J.A. Bone SPECT/CT image fusion based on the discrete Hermite transform and sparse representation. Biomed. Signal Process. Control 2022, 71, 103096. [Google Scholar] [CrossRef]
- Agrafioti, F.; Gao, J.; Hatzinakos, D.; Yang, J. Heart biometrics: Theory, methods and applications. Biometrics 2011, 3, 199–216. [Google Scholar]
- Damian, F.A.; Moldovanu, S.; Dey, N.; Ashour, A.S.; Moraru, L. Feature selection of non-dermoscopic skin lesion images for nevus and melanoma classification. Computation 2020, 8, 41. [Google Scholar] [CrossRef]
- McGuire, M. An image registration technique for recovering rotation, scale and translation parameters. NEC Res. Inst. Tech. Rep. 1998, TR-98-018, 1–29. [Google Scholar]
- Ngo, L.H.; Luong, M.; Sirakov, N.M.; Viennet, E.; Le-Tien, T.T. Skin lesion image classification using sparse representation in quaternion wavelet domain. Signal Image Video Process. 2022, 16, 1721–1729. [Google Scholar] [CrossRef]
- Was, L.; Milczarski, P.; Stawska, Z.; Wyczechowski, M.; Kot, M.; Wiak, S.; Wozniacka, A.; Pietrzak, L. Analysis of Dermatoses Using Segmentation and Color Hue in Reference to Skin Lesions. In Artificial Intelligence and Soft Computing, Proceedings of the 16th International Conference, ICAISC 2017, Zakopane, Poland, 11–15 June 2017; Springer International Publishing: Cham, Switzerland, 2017; Volume 16, pp. 677–689. [Google Scholar]
- Gerstenblith, M.R.; Shi, J.; Landi, M.T. Genome-wide association studies of pigmentation and skin cancer: A review and meta-analysis. Pigment. Cell Melanoma Res. 2010, 23, 587–606. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, R.P.; Hill, G.B.; Bajdik, C.D.; Coldman, A.J.; Fincham, S.; McLean, D.I.; Threlfall, W.J. Sunlight exposure, pigmentation factors, and risk of nonmelanocytic skin cancer: II. Squamous Cell carcinoma. Arch. Dermatol. 1995, 131, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Jiang, H.; Wang, H.; Alwageed, H.; Zhou, Y.; Sebdani, M.M.; Yao, Y.D. Modulation classification based on signal constellation diagrams and deep learning. IEEE Trans. Neural Netw. Learn. Syst. 2019, 30, 718–727. [Google Scholar] [CrossRef]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 318–327. [Google Scholar] [CrossRef]
- Malik, H.; Farooq, M.S.; Khelifi, A.; Abid, A.; Qureshi, J.N.; Hussain, M. A comparison of transfer learning performance versus health experts in disease diagnosis from medical imaging. IEEE Access 2020, 8, 139367–139386. [Google Scholar] [CrossRef]
- Mijwil, M.M. Skin cancer disease images classification using deep learning solutions. Multimed. Tools Appl. 2021, 80, 26255–26271. [Google Scholar] [CrossRef]
- Indraswari, R.; Rokhana, R.; Herulambang, W. Melanoma image classification based on MobileNetV2 network. Procedia Comput. Sci. 2022, 197, 198–207. [Google Scholar] [CrossRef]
- Zhi, S.; Li, Z.; Yang, X.; Sun, K.; Wang, J. A multiclassification model for skin diseases using dermatoscopy images with Inception-v2. Appl. Sci. 2024, 14, 10197. [Google Scholar] [CrossRef]
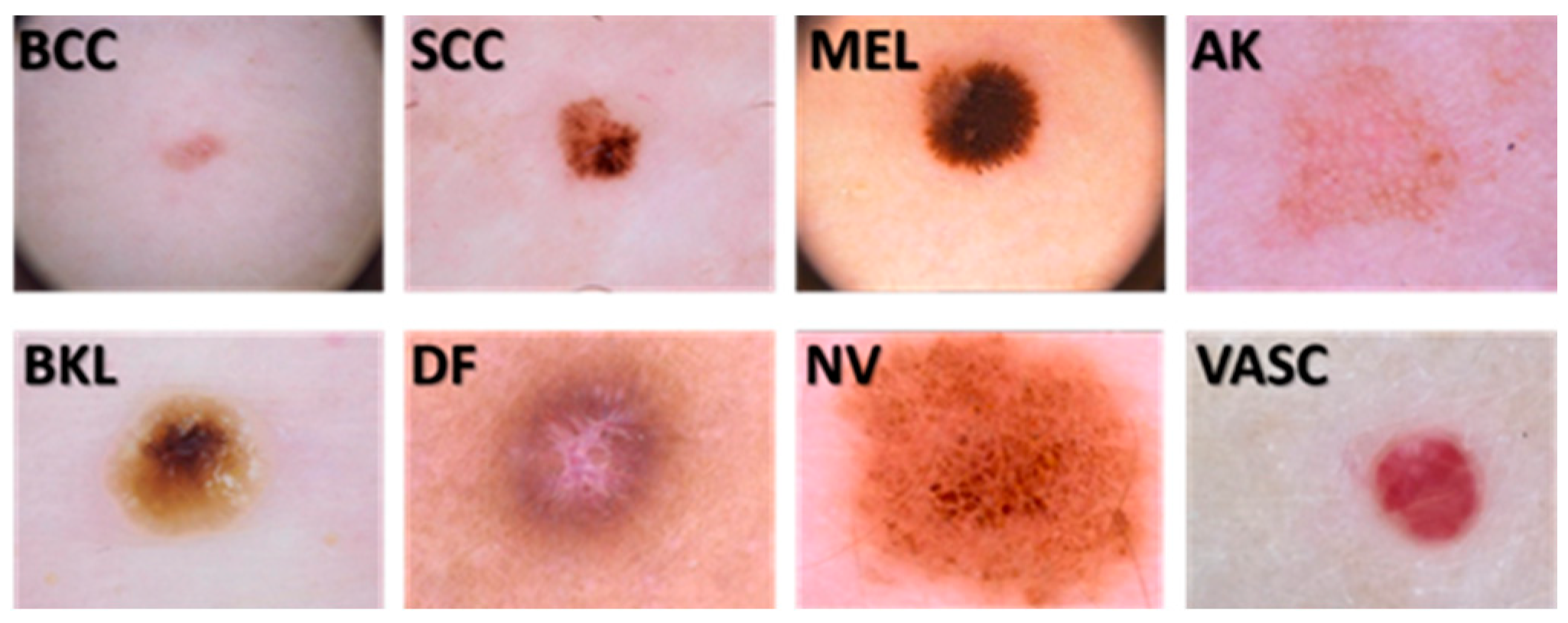





| Image | Actual Class | Predicted Class | Accurate Classified |
|---|---|---|---|
 | NV | NV | True |
 | MEL | MEL | True |
 | VASC | MEL | False |
 | BCC | BCC | True |
 | NV | BKL | False |
| Class | Recall | FP Rate | Specificity | Precision | Accuracy | F1 Score |
|---|---|---|---|---|---|---|
| NV | 55.85 ± 2.33 | 4.50 ± 0.33 | 95.50 ± 0.33 | 63.92 ± 1.43 | 90.60 ± 0.22 | 59.34 ± 1.44 |
| MEL | 59.31 ± 2.11 | 5.25 ± 0.44 | 94.75 ± 0.44 | 62.07 ± 1.57 | 90.34 ± 0.32 | 60.39 ± 1.26 |
| BCC | 75.16 ± 1.18 | 2.93 ± 0.29 | 97.07 ± 0.29 | 78.54 ± 1.76 | 94.35 ± 0.29 | 76.73 ± 1.16 |
| BKL | 71.68 ± 1.71 | 3.94 ± 0.31 | 96.06 ± 0.31 | 72.61 ± 1.32 | 92.99 ± 0.28 | 72.01 ± 1.06 |
| VASC | 94.77 ± 0.99 | 0.97 ± 0.10 | 99.03 ± 0.10 | 93.37 ± 0.60 | 98.50 ± 0.12 | 94.03 ± 0.50 |
| DF | 97.54 ± 0.72 | 1.17 ± 0.13 | 98.83 ± 0.13 | 92.29 ± 0.79 | 98.66 ± 0.17 | 94.83 ± 0.65 |
| SCC | 97.01 ± 0.83 | 1.59 ± 0.17 | 98.41 ± 0.17 | 89.71 ± 1.00 | 98.23 ± 0.22 | 93.20 ± 0.82 |
| AK | 98.62 ± 0.58 | 0.98 ± 0.16 | 99.02 ± 0.16 | 93.64 ± 0.95 | 98.97 ± 0.18 | 96.05 ± 0.65 |
| Overall Average | 81.24 ± 2.19 | 2.67 ± 0.22 | 97.33 ± 0.22 | 80.77 ± 1.64 | 95.33 ± 0.45 | 80.82 ± 1.90 |
| Model | Dataset | Recall | FP Rate | Specificity | Precision | Accuracy | F1 Score |
|---|---|---|---|---|---|---|---|
| 2D superpixels + RCNN [28] | HAM-10000 | 0.8450 | - | - | 0.8349 | 0.8550 | 0.8530 |
| ResNeXt101 [29] | ISIC-2019 | 0.8810 | - | - | 0.8740 | 0.8850 | 0.8830 |
| MobileNetV2 [30] | ISIC-2019 | 0.8633 | - | - | 0.7890 | 0.8530 | - |
| VGG19 [31] | ISIC-2019, Derm-IS | 0.8666 | - | - | 0.9070 | 0.8857 | 0.8765 |
| ConvNet [32] | ISIC-2019, Derm-IS | 0.8747 | - | - | 0.8614 | 0.8690 | - |
| Inception-v2 [33] | ISIC-2019 | 0.9015 | - | - | 0.8737 | 0.8904 | 0.8876 |
| This work | ISIC-2019 | 0.8124 | 0.0267 | 0.9733 | 0.8077 | 0.9533 | 0.8082 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Ávila, L.F.; Álvarez-Borrego, J. Hybrid Transform-Based Feature Extraction for Skin Lesion Classification Using RGB and Grayscale Analysis. Appl. Sci. 2025, 15, 5860. https://doi.org/10.3390/app15115860
López-Ávila LF, Álvarez-Borrego J. Hybrid Transform-Based Feature Extraction for Skin Lesion Classification Using RGB and Grayscale Analysis. Applied Sciences. 2025; 15(11):5860. https://doi.org/10.3390/app15115860
Chicago/Turabian StyleLópez-Ávila, Luis Felipe, and Josué Álvarez-Borrego. 2025. "Hybrid Transform-Based Feature Extraction for Skin Lesion Classification Using RGB and Grayscale Analysis" Applied Sciences 15, no. 11: 5860. https://doi.org/10.3390/app15115860
APA StyleLópez-Ávila, L. F., & Álvarez-Borrego, J. (2025). Hybrid Transform-Based Feature Extraction for Skin Lesion Classification Using RGB and Grayscale Analysis. Applied Sciences, 15(11), 5860. https://doi.org/10.3390/app15115860






