Abstract
This study’s integration of molecular dynamics (MD) simulations with non-covalent adaptable networks (NANs) and corroborative wet lab experiments offers a comprehensive approach to understanding the interactions between ritonavir (RTV) and polymers in supersaturated solutions. This multifaceted study not only explored the stabilization mechanisms facilitated by NANs but also examined the influence of polymer selection on the pharmaceutical properties of RTV, a class III compound known for its slow crystallization rate. This research utilized molecular dynamics simulations to model the intermolecular interactions between RTV and two polymers, Polyvinylpyrrolidone (PVP) K30 and Eudragit L100. These simulations were specifically designed to incorporate the effects of NANs, highlighting their dynamic nature and potential to enhance drug stability and solubility. Simultaneously, wet lab experiments were conducted to measure the nucleation induction times and observe the crystallization behavior of RTV under varying conditions of polymer presence. The experimental data demonstrated a significant extension in nucleation induction time, prolonging the duration from 12 to approximately 64 h when PVP K30 and Eudragit L100 were present. This substantial delay in crystallization was attributed to the strong intermolecular interactions between RTV and the polymers, which were effectively stabilized by the non-covalent bonds within the NANs. These findings were consistently confirmed across both computational and experimental settings, illustrating how NANs can effectively inhibit crystallization and enhance the supersaturation state of RTV. This study successfully demonstrates how the physical and chemical properties of polymers influence the crystallization process of poorly water-soluble drugs such as RTV. Leveraging the synergy between computational simulations and empirical laboratory data, this research provides deep insights into the mechanisms at play, ensuring that drug formulations are optimized for both stability and performance.
1. Introduction
Around 70–90% of new drugs have poor water solubility, meaning that they do not work as well when taken orally [1]. Recent studies have shown that about 70% of new drugs have this issue, making it a major hurdle in drug development [2]. To address this, scientists have turned to a technique called drug amorphization. This method works because amorphous drugs, which are not in a crystal form, have higher free energy. This helps them to dissolve better in water and improves their effectiveness when taken orally [3]. However, the downside is that amorphous drugs often revert to crystals over time, especially during storage and when dissolving, due to their natural instability [4]. Non-covalent adaptable networks (NANs) come into play at this point. Also known as vitrimers, NANs use reversible non-covalent bonds that allow the network structure to adjust and recover without falling apart. Recent research by Zhou et al. (2022) shows that adding NANs to drug formulations can help prevent crystallization by keeping the drug in its amorphous state and reducing its instability [5]. Additionally, a study by Yang et al. (2023) found that NANs are better at keeping amorphous drugs soluble compared to traditional methods due to their ability to avoid crystallization during storage [6].
Amorphous solid dispersion (ASD) is a widely used technique to improve the stability and effectiveness of drugs that do not dissolve easily by embedding them in a polymer matrix. The success of ASD depends on how well a drug interacts with a polymer to keep it in an amorphous (non-crystalline) state and prevent crystallization, which otherwise can reduce the drug’s effectiveness [7]. Recently, there have been significant advancements in the integration of NANs into drug–polymer matrices. Moreover, NANs utilize strong but reversible non-covalent interactions, such as hydrogen bonding, electrostatic forces, and hydrophobic interactions. These networks allow the polymer matrix to adapt and maintain stability without the need for covalent changes [8]. By incorporating NANs, researchers can enhance the stabilization of amorphous drugs within ASDs, effectively preventing crystallization and ensuring that these drugs remain highly soluble throughout their storage and use. This approach leverages the dynamic adaptability of non-covalent interactions to improve drug efficacy and stability in pharmaceutical applications [9].
This innovative approach could greatly enhance the performance of ASD formulations by providing added stability and minimizing crystallization risks, leading to more effective treatments for drugs that are difficult to dissolve. Recent studies have suggested that NANs, employing non-covalent adaptable networks, not only enhance the stability of ASD formulations but also extend the shelf life of amorphous drugs by preventing their conversion into crystalline forms [10]. For example, research by Newman and Zografi (2023) demonstrated that NANs can significantly reduce crystallization in drug formulations compared to traditional polymers, resulting in more reliable and effective drug delivery [11]. Furthermore, Liu et al. (2023) highlighted the potential of NANs to improve the solubility and bioavailability of poorly soluble drugs, making them a promising addition to future pharmaceutical formulations [12].
Polymeric materials are frequently employed in the formulation of supersaturated amorphous drugs to prevent crystallization and maintain the high supersaturated solution [13]. However, there is a limited availability of methods for accurately predicting the correct selection of the most suitable polymer for preserving supersaturated drug solutions. The ability of polymers to inhibit nucleation and crystal growth is crucial for preventing crystallization and maintaining supersaturated drug solutions. To achieve the most effective potential prevention of drug crystallization in supersaturated solutions, it is necessary to have a thorough understanding of the nucleation kinetics and mechanisms that occur when polymers inhibit drug crystallization [14].
Molecular dynamics (MD) simulations have become essential for examining molecular structures, particularly in understanding how non-covalent adaptable networks (NANs) reorganize within amorphous formulations [15]. In MD simulations, nanoseconds (ns) are used to capture molecular interactions over very short timescales, allowing researchers to explore early-stage interactions such as the formation and dissociation of non-covalent and hydrogen bonds, as well as hydrophobic interactions between drug molecules and polymers. Although MD simulations cannot directly predict long-term behaviors, they provide valuable insights into initial stabilization mechanisms in amorphous drug formulations [16]. For instance, analyzing interaction energies and hydrogen bonds reveals the strength and type of molecular engagement, which is essential for assessing each polymer’s potential to maintain the drug in a supersaturated state [17,18]. This early understanding serves as a crucial foundation for guiding experimental validation (Figure 1).
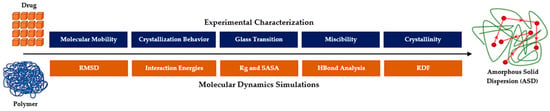
Figure 1.
Integrating experimental and molecular dynamics approaches for characterizing ASD formulations [19,20].
However, to fully capture long-term phenomena like crystallization, these short-term simulations need to be complemented by in-depth experimental characterization [21,22]. Experimental approaches offer a more realistic perspective, helping to validate MD simulation results. By integrating insights from MD simulations with experimental data, researchers gain a comprehensive view of both the initial molecular interactions and the long-term stability of amorphous systems [23]. This combination of simulation and experimental data allows for a more accurate assessment of how NANs contribute to stabilizing amorphous formulations over time. This integrated approach, as highlighted by Pajzderska and Gonzalez (2023), not only deepens our understanding of NANs but also aids in selecting the optimal polymers to maintain drug solubility and stability, resulting in more effective and reliable pharmaceutical formulations [24].
In this study, we applied MD simulations to investigate the interaction between a drug and a polymer in supersaturated drug solutions, which affects crystallization inhibition. RTV was chosen as a model drug due to its low solubility and poor oral absorption, while PVP K30 and Eudragit L100 were selected for their ability to prevent crystallization in such solutions [25]. The statement that “It is difficult to examine these interactions using experimental methods” refers to challenges such as the rapid crystallization process and the dynamic nature of drug–polymer interactions, which are difficult to capture with traditional experimental techniques [26]. Additionally, experimental methods like HPLC and X-ray diffraction cannot fully observe these complex molecular-level interactions, whereas MD simulations provide valuable insights but are limited by assumptions and idealized conditions. These limitations of both experimental and computational approaches should be considered when interpreting the findings.
2. Materials and Methods
2.1. Materials
Ritonavir (RTV), with a molecular weight of 720.95 g/mol, was sourced from ChemShuttle (Hayward, Berkeley Heights, NJ, USA). Polyvinylpyrrolidone (PVP) K30, with an average molecular weight of 40,000–60,000 g/mol, and Eudragit L100, with an average molecular weight of 125,000–135,000 g/mol, were obtained from Merck (Darmstadt, Germany). The choice of PVP K30 and Eudragit L100, along with their respective molecular weights, plays a crucial role in the stabilization of RTV supersaturation. PVP K30, with a lower molecular weight, acts as a stabilizer by forming a network that prevents the crystallization of RTV, thus maintaining its supersaturated state. On the other hand, Eudragit L100, with a higher molecular weight, provides additional stabilization through its ability to form protective films and inhibit the aggregation of RTV molecules. These polymers’ molecular weights influence their interaction with RTV in solution, affecting both the physical stability and bioavailability of RTV in the formulation.
2.2. Analysis of Induction Time in Supersaturated Drug–Polymer Solutions
Induction time studies were conducted with and without the addition of pre-dissolved polymers. Each polymer was dissolved in water at a concentration of 100 μg/mL. Supersaturated solutions were prepared by mixing a stock solution of RTV (1500 μg/mL in methanol) with the polymer solutions, resulting in a final methanol concentration of 2% (v/v) at 25 °C. The solutions were continuously agitated at 25 °C and filtered through a 0.45 μm pore-size filter at various time intervals. After filtration, the samples were mixed with acetonitrile to lower the concentration and subsequently analyzed using high-performance liquid chromatography (HPLC) [27].
2.3. Preparation and Characterization of RTV-Based Polymers
It is essential to accurately describe and optimize the ligand structures before incorporating them into molecular dynamics simulations. In this study, we used RTV (PubChem CID 392622), Eudragit L100 (PubChem CID 65358), and PVP K30 (PubChem CID 481110337) as our ligands, which were obtained from PubChem “http://pubchem.ncbi.nlm.nih.gov (accessed on 28 September 2024)” (Figure 2) [28]. We first created 2D representations of these compounds using ChemDraw Professional 16.0, and then converted them into 3D models with Chem3D 16.0 [29]. We optimized these 3D structures using GaussView 6 and Gaussian16, applying the Density Functional Theory (DFT) method with 6–31 basis sets [30]. The final structures were further refined by adding polar hydrogen atoms and Gasteiger partial charges using AutoDockTools 4.2.6 and MGLTools 1.5.6 [31]. These procedures are crucial for ensuring that the ligand models are both accurate and reliable for simulations. Proper optimization allows for precise predictions of how ligands interact with the target proteins and their binding affinities.
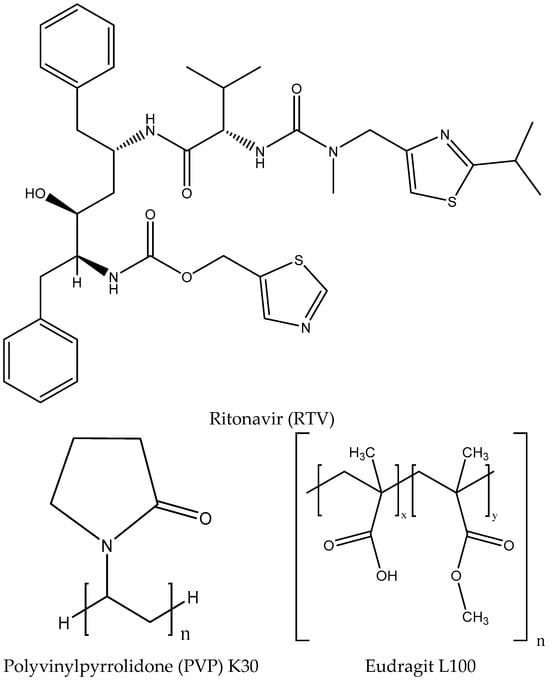
Figure 2.
Two-dimensional visualization of RTV, PVP K30, and Eudragit L100 for the creation of drug–polymer systems.
2.4. Optimizing Induction Time Measurement of RTV
In this study, RTV was dissolved in methanol, while Eudragit L100 and PVP K30 were dissolved in water. The concentrations of the polymers were set at 500 ppm (7 molecules) and 2000 ppm (25 molecules) using the PACKMOL program to generate the initial configurations for optimal packing [32]. The solvent system was then equilibrated for 2 femtoseconds before adding RTV and the polymers. This meticulous preparation ensured a balanced and stable environment for accurate simulations. Distributing methanol and water molecules symmetrically helps create a consistent system for better analysis. Dissolving RTV in methanol and the polymers in water was intended to closely reflect actual conditions and interactions. Using PACKMOL for the initial setup allowed for the precise and efficient configuration of the solvent system. The equilibration step before introducing the solutes was essential for establishing a stable baseline and obtaining reliable simulation results.
2.5. Production Runs and Data Collection Using Molecular Dynamics Simulations
We conducted molecular dynamics simulations on four distinct complexes: RTV with Eudragit L100 at 500 ppm (E1), RTV with Eudragit L100 at 2000 ppm (E2), RTV with PVP K30 at 500 ppm (P1), and RTV with PVP K30 at 2000 ppm (P2). These simulations were carried out using GROMACS 2016.3 software [33,34,35], employing the AMBER99SB-ILDN and AMBER General Force Fields (GAFF) to model the systems [36,37]. The topology and parameterization of RTV, Eudragit L100, and PVP K30 were managed through the AnteChamber Python Parser interface (ACPYPE) [38], and long-range electrostatic interactions were calculated using the Ewald Particle Mesh method [39]. Sodium (Na+) and chloride (Cl−) ions were added to ensure the systems were neutral. The molecular weights of PVP K30 and Eudragit L100 were explicitly calculated to accurately model polymer chains in the simulations. For PVP K30, the average molecular weight (Mw) is 50,000 g/mol, and the molecular weight of its monomer, vinylpyrrolidone, is 111.14 g/mol. The degree of polymerization (DP) was determined using the formula:
Hence, the polymer chain was constructed with 450 monomer units. For Eudragit L100, a random copolymer of methacrylic acid (86.09 g/mol) and methyl methacrylate (100.12 g/mol) in a 1:1 ratio, the molecular weight of the repeating unit was calculated as:
Using the polymer’s average molecular weight of 130,000 g/mol, the degree of polymerization was determined as:
Thus, the polymer chain for Eudragit L100 was modeled with 1396 monomer units, distributed randomly between methacrylic acid and methyl methacrylate. These polymer chains were parameterized using force fields compatible with their chemical properties and integrated into the simulation. The preparation for these simulations involved several key steps. We first performed minimization to resolve any steric clashes and then heated the systems to 25 °C (298 K) to ensure thermal stability. This was followed by the equilibration of both temperature and pressure to create a stable simulation environment. During the production run, a timestep of 2 femtoseconds was used to capture detailed dynamic behavior. Each system was simulated for 100 nanoseconds, and we carefully analyzed energy, pressure, temperature, and Root Mean Square Deviation (RMSD) to assess stability. This thorough approach ensured that the simulations were reliable and provided a solid basis for understanding the interactions within each complex. Additionally, we conducted analyses of RDF, radius of gyration (Rg), Solvent-Accessible Surface Area (SASA), and hydrogen bonding (Hbond) to obtain a deeper understanding of the structural and dynamic properties of the systems, using Grace-5.1.22 and Microsoft Excel 365 for data visualization and analysis.
3. Results
3.1. Investigation of Induction Period in Supersaturated Drug–Polymer Systems
The crystallization tendency of drug molecules in solution can be assessed by analyzing their nucleation induction times [40]. Figure 3 illustrates a comparison of the nucleation induction time measurements for RTV, both with and without a polymer. Remarkably, RTV remained at a high concentration in the buffer for up to 30 min, indicating a slow crystallization rate, thus classifying it as a class III compound according to Taylor’s classification. The addition of polymers significantly extended the induction time of RTV from 12 h (without a polymer) to approximately 64 h when both PVP K30 and Eudragit L100 were present. This considerable increase is likely due to the strong intermolecular interactions between RTV and the polymers, effectively inhibiting RTV crystal nucleation in the supersaturated solution. The induction times reported in Figure 3 were calculated using high-performance liquid chromatography (HPLC) to monitor the concentration of RTV over time. This method allows for the precise tracking of RTV’s concentration and the onset of crystallization. The significant increase in induction time with the addition of polymers highlights their critical role in stabilizing amorphous drug formulations, improving the solubility and stability of poorly water-soluble drugs in supersaturated solutions.
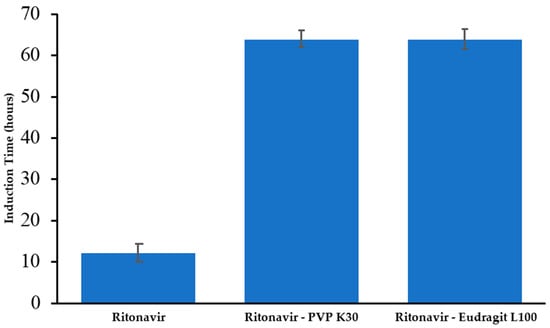
Figure 3.
Comparison of the induction time measurements for RTV in the presence of polymers (PVP K30 and Eudragit L100).
The data highlight how the presence of polymers significantly influences the crystallization process of RTV. Without the polymer additives, RTV begins to crystallize relatively quickly, with an induction time of around 12 h. This suggests that, under these conditions, RTV has a moderate tendency to form crystals. However, the addition of PVP K30 and Eudragit L100 extends the nucleation induction time to approximately 64 h, indicating a substantial delay in the crystallization process. This extension is likely due to the ability of these polymers to interact with RTV, thereby stabilizing the drug molecules in the solution and disrupting the crystal formation process. Such interactions may form complexes or create a more stabilized environment that hinders the nucleation of crystals, allowing RTV to remain in a supersaturated state for a prolonged period [41]. This finding underscores the importance of polymer selection in pharmaceutical formulations, where controlling the crystallization rate can be crucial for optimizing drug stability and performance.
3.2. Characterizing the Dynamic Behavior of RTV Using Molecular Dynamics Simulations
The molecular dynamics of the RTV, Eudragit L100, and PVP K30 complex systems were examined through a detailed trajectory analysis, and the results are summarized in Table 1. We tracked these systems at three time points: the start of the simulation (0 ns), at 50 ns, and at the end (100 ns). The results indicate that both Eudragit L100 and PVP K30 show promise as effective agents for complexing with RTV, thereby maintaining the supersaturation of RTV. At the 50 ns mark, these polymers successfully interacted with RTV, but by 100 ns, this association had broken, probably due to recrystallization. Eudragit L100 is particularly notable for its role in targeted drug delivery; it can break down under specific pH conditions, like those found in the intestines, allowing for a controlled and gradual release of drugs [42]. PVP K30, on the other hand, contributes to drug stability by inhibiting recrystallization and protecting against oxidation, light degradation, and heat, and enhances the physical properties of drugs, such as their crystal stability and solubility, by forming inclusion complexes [43].

Table 1.
Dynamic behavior of the inclusion complex systems in water during molecular dynamics simulations.
These insights highlight the crucial role of polymer selection in drug formulations, as the stability and interaction provided by polymers can greatly affect crystallization and supersaturation, leading to higher drug efficacy. Eudragit L100’s ability to enable controlled release is particularly relevant to modern drug delivery strategies, which seek to achieve precise therapeutic effects. Meanwhile, PVP K30’s protective qualities are essential for extending the drug’s shelf life and maintaining its effectiveness. These simulations provide valuable data on optimizing polymers to boost drug performance, offering a solid foundation for further research and innovation in drug delivery systems [44]. Overall, this study underscores the benefits of adding Eudragit L100 and PVP K30 to pharmaceutical formulations, especially in inhibiting drug recrystallization and maintaining the supersaturation of amorphous drugs, which offers a promising direction for future development in this field.
3.3. Analysis of Conformational Flexibility Using Molecular Dynamics Simulations
We used the Root Mean Square Deviation (RMSD) to assess structural changes during molecular dynamics simulations. RMSD is a critical tool for measuring how closely the simulated structures match the reference structures. It helps track how a molecule’s shape evolves during the simulation and compares these changes to the initial or reference structure. We calculated the RMSD values for the four inclusion complex systems and tracked these changes over time, as shown in Figure 4 and Figure 5. Figure 4 highlights the changes in RTV’s structure during the simulation, revealing that there are no significant differences among the four inclusion complex systems. These results indicate that RTV, Eudragit L100, and PVP K30 maintain a stable structural profile throughout the simulation. The consistent RMSD values across the systems suggest that the inclusion complexes retain their structural integrity [45]. This stability is essential for making reliable predictions about molecular interactions and the effectiveness of drugs. Confirming that the systems stay close to their reference structures validates the accuracy of our simulation results.
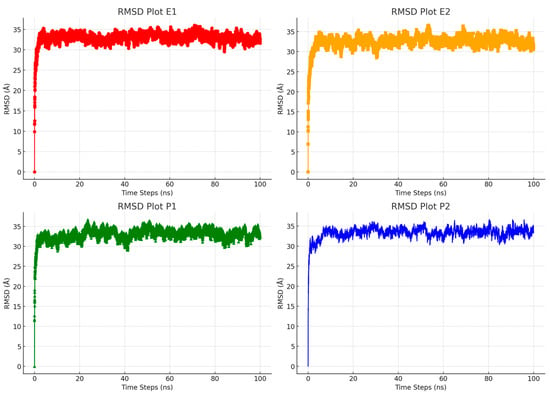
Figure 4.
Evaluation of the Root Mean Square Deviation (RMSD) trends for RTV molecules in water systems during molecular dynamics simulations.
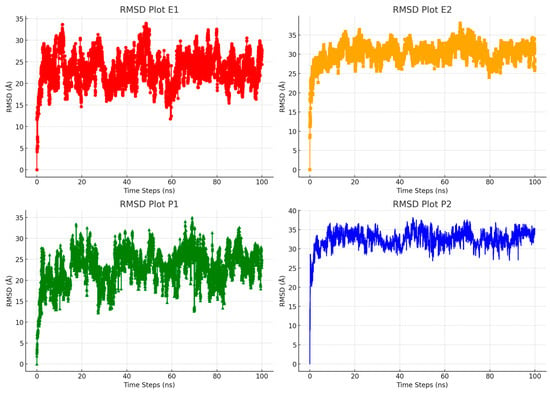
Figure 5.
Comparison of RMSD profiles for Eudragit L100 and PVP K30 molecules in water systems during molecular dynamics simulations.
Figure 5 clearly shows the differences in stability between the RTV complexes. Specifically, the RTV + Eudragit L100 500 ppm (E1) and RTV + PVP K30 500 ppm (P1) complexes are more stable compared to the RTV + Eudragit L100 2000 ppm (E2) and RTV + PVP K30 2000 ppm (P2) complexes. The lower viscosity of the 500 ppm polymer concentration makes these formulations easier to work with and apply. On the other hand, the higher 2000 ppm concentration helps maintain the saturation of the solution, allowing the polymers to stay dissolved in various solvents, including in water. These results emphasize the importance of choosing the right polymer concentration based on the desired application and performance. The greater stability of the 500 ppm complexes suggests they might be more suitable for applications that need long-lasting stability and controlled release.
On the other hand, the ability of 2000 ppm complexes to maintain saturation can be advantageous for formulations that need faster dissolution rates or better compatibility with various solvents. Meanwhile, the lower viscosity of the 500 ppm concentration makes the formulation and handling processes easier, which could lead to more efficient production and smoother integration into different drug delivery systems. Striking the right balance between viscosity and saturation is key to optimizing pharmaceutical products and making sure they meet specific needs. Overall, these results offer valuable insights into how polymer concentration influences the stability and effectiveness of RTV complexes. They highlight the importance of carefully choosing the formulation parameters to achieve the best results in pharmaceutical applications.
3.4. Mechanisms of Polymer-Induced Saturation Stabilization in RTV
A radial distribution function (RDF) analysis was used to see how RTV’s saturation levels change. RDF is a common method for examining how solvent molecules interact in molecular dynamics simulations. By studying RDF, we can understand the attractive forces between solvent molecules and how they work together in a system [46]. Figure 6 shows that RTV’s saturation improves significantly when it forms an inclusion complex with PVP K30. This highlights PVP K30’s role in keeping RTV saturated, which is important for effective drug delivery. The RDF results suggest that PVP K30 helps RTV interact more effectively with solvent molecules, potentially leading to faster dissolution and better bioavailability. Overall, these insights from the RDF analysis can help us design more effective pharmaceutical formulations.
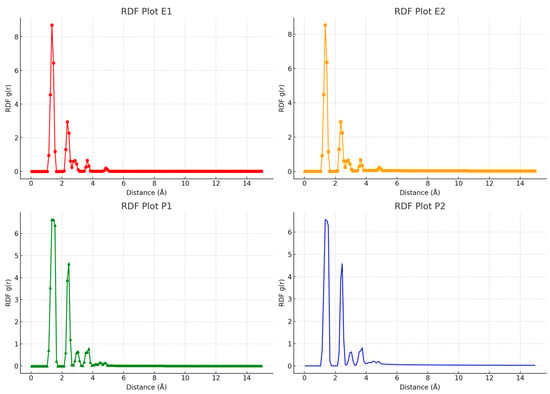
Figure 6.
Comparison of RDF data for Eudragit L100 and PVP K30 in water, as observed using molecular dynamics simulations.
The RDF graph shows that Eudragit L100 helps maintain RTV’s saturation well, but the interaction between RTV and Eudragit L100 is relatively weak at certain distances. This means that while Eudragit L100 is effective at maintaining saturation, its interaction with RTV is not as strong as with some other polymers. On the other hand, PVP K30 does an excellent job at maintaining saturation due to its effective interactions at specific distances, creating a stable, water-friendly environment. PVP K30, being a water-soluble polymer, significantly helps keep RTV saturated by forming interactions that boost its dissolution in water. This highlights how important PVP K30 is in pharmaceutical formulations where maintaining saturation is crucial. These findings stress the need to carefully choose polymers to ensure optimal saturation maintenance and drug effectiveness. The stronger interaction between RTV and PVP K30 suggests that PVP K30 might be better for formulations aimed at increasing drug bioavailability. Overall, these results also help us understand how different polymers can affect the physical properties of drug compounds.
3.5. Effect of Polymer Concentration on the Packaging of RTV Complexes
To assess how well RTV integrates with the polymer matrix, we measured the radius of gyration (Rg) of the molecular inclusion model. Rg helps us understand how particles are dispersed by calculating their average distance from the molecule’s center of mass. This measurement reveals whether the molecule is tightly packed or more dispersed, and provides insights into its flexibility, indicating how easily the molecule can change shape or move [47]. A lower Rg value suggests that the inclusion complex is more compact, stable, and cohesive throughout the simulation, which is beneficial for boosting the effectiveness of the drug in pharmaceutical applications. These results are important for evaluating the structural stability of RTV within a polymer matrix. The data also shed light on how well a polymer stabilizes and encapsulates RTV, which could enhance drug delivery and bioavailability. By tracking the Rg values over time, we can refine the formulation process and better understand how different polymers affect the physical properties of the drug complex. This detailed analysis supports the development of more stable and effective drug formulations, advancing the field of drug delivery systems.
Figure 7 displays the fluctuations in the radius of gyration (Rg) over time in the simulated model. By the 100 ns mark, it becomes clear that the RTV–polymer inclusion complex at a 500 ppm concentration shows greater stability compared to the 2000 ppm concentration. At 500 ppm, each polymer has fewer interactions with other polymers and the methanol–water solvent system, leading to a more compact and stable conformation. Conversely, at 2000 ppm, the increased interactions among the polymers and between the polymers and the solvents result in a more open and flexible conformation. These findings underscore the significant effect of polymer concentration on the structural stability of the inclusion complex. The compact conformation observed at 500 ppm suggests that this concentration is more effective at preserving the structural integrity of the complex, which could enhance its functionality. On the other hand, the increased flexibility at 2000 ppm might indicate potential instability, possibly affecting the formulation’s performance in real-world applications. The data highlight the need to carefully optimize polymer concentration to balance stability and flexibility, which is crucial for improving the effectiveness of drug formulations.
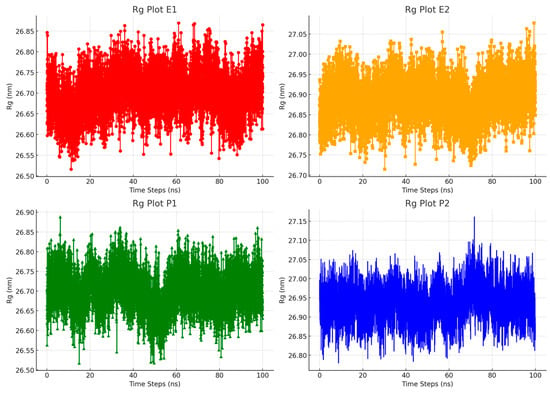
Figure 7.
Comparison of the radius of gyration (Rg) profiles for the inclusion complex systems in water during molecular dynamics simulations.
Additionally, we conducted a Solvent-Accessible Surface Area (SASA) analysis to assess the solubility of molecules in the methanol–water solvent system throughout a 100 ns simulation. An SASA analysis is crucial for understanding how molecules interact with their solvent environment [48]. It reveals which parts of the molecule are accessible to methanol–water and can form hydrogen bonds in contrast to areas that are not interacting. This provides valuable information about these molecules’ hydrophobic and hydrophilic properties under different conditions, such as varying temperatures and pH levels. Figure 8 illustrates that there is a slight difference in solubility levels among the inclusion complexes tested. Overall, these complex systems show effective dissolution in the methanol–water mixture. This is likely due to the increased polarity of both RTV and the polymers, which enhances their stability during the simulation. The similar physical and chemical characteristics of Eudragit L100 and PVP K30 may also contribute to their comparable interactions with the solvents. These results highlight the importance of molecular interactions and solvent characteristics in influencing the solubility and stability of such complex systems.
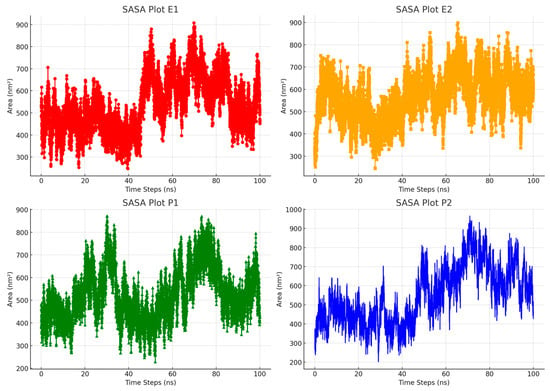
Figure 8.
Comparison of Solvent-Accessible Surface Area (SASA) profiles for the inclusion complex systems in water during molecular dynamics simulations.
3.6. Structural Influences on Binding Affinity in Inclusion Complex Systems
To delve into the binding and release dynamics between RTV and the polymers, we conducted a hydrogen bond (HBond) analysis using molecular dynamics simulations. Hydrogen bonds are crucial for molecular interactions, and their presence and strength can be evaluated using geometric criteria, such as the distances and angles between the atoms involved [49]. A quantitative approach also measures the number of hydrogen bonds formed and their duration throughout the simulation. As shown in Figure 9, PVP K30 at a concentration of 500 ppm demonstrates notable inclusion capability. This is evident from its ability to form stable inclusion complexes between 40 ns and 50 ns, followed by its release from 50 ns onwards until the simulation concludes. These findings underscore the strong binding and release properties of PVP K30, indicating its important role in the stability and functionality of the inclusion complexes. The observed dynamics of complex formation and release suggest that the addition of PVP K30 could be advantageous for controlled drug delivery systems. Moreover, the analysis of hydrogen bond formation and duration offers valuable insights into the robustness of these interactions. Understanding these aspects is key to optimizing polymer formulations for improved performance.
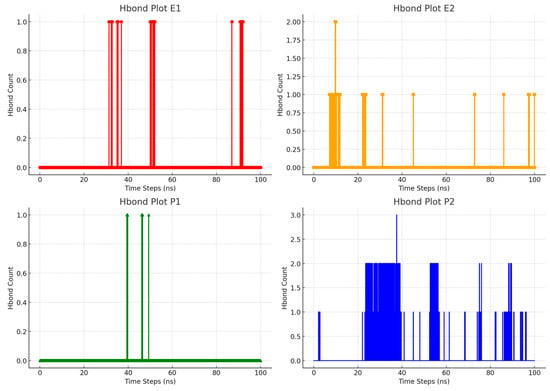
Figure 9.
Graphs of hydrogen bond (HBond) formation in the inclusion complex systems in water during molecular dynamics simulations.
The aim of analyzing the formation of hydrogen bonds during molecular dynamics simulations is to gauge how frequently hydrogen bonds form within a molecular system. This measure, known as hydrogen bond occupancy, reflects the proportion of simulation time during which hydrogen bonds are present. It is a crucial metric for evaluating the strength and durability of molecular interactions. As shown in Table 2, the inclusion complex systems at a 500 ppm concentration exhibit promising binding and release characteristics, with occupancy rates of 1.08% for Eudragit L100 and 0.24% for PVP K30. The data indicate that Eudragit L100 has a higher hydrogen bond occupancy, forming and maintaining hydrogen bonds more consistently than PVP K30. This suggests that Eudragit L100 may provide stronger and more stable interactions, which could be advantageous for applications requiring controlled release of drugs. On the other hand, while PVP K30 has a lower occupancy rate, it still shows significant binding ability. These results offer crucial insights into how different polymers interact within an inclusion complex system, informing the development of pharmaceutical formulations.

Table 2.
Proportion of time that hydrogen bonds are present in the inclusion complex systems in water during molecular dynamics simulations.
The frequency with which hydrogen bonds form in a complex system is affected by several factors, such as the polymer concentration and environmental conditions, including temperature and humidity. Hydrogen bond occupancy reflects how often these bonds are formed over a given period, with higher percentages indicating more frequent bond formation. Raising the polymer concentration from 500 ppm to 2000 ppm increases the amount of polymers available, which can enhance intermolecular interactions and boost hydrogen bond occupancy. However, excessively high concentrations may lead to problems such as polymer aggregation or clumping, potentially compromising the stability and effectiveness of these bonds. Environmental factors such as temperature and humidity also play a significant role in hydrogen bond formation and stability. For example, higher temperatures can increase molecular movement, leading to more transient bond formations but potentially reducing bond stability. On the other hand, carefully controlled environmental conditions can improve the strength and durability of hydrogen bonds, thereby enhancing polymer performance. A thorough understanding of these factors is essential for optimizing polymer formulations to achieve the desired binding properties and stability.
3.7. Dynamic Behavior and Energy Characteristics of Inclusion Complexes
An analysis of potential and kinetic energies plays a fundamental role in molecular dynamics simulations, offering valuable insights into how molecules interact and move within a system. Potential energy is calculated based on interactions such as van der Waals forces and Coulombic attractions, which help us understand the stability and interaction strength of molecular configurations [50]. Kinetic energy, on the other hand, is related to the mass and velocity of atoms, providing information about molecules’ temperature and movement. Examining potential energy allows us to assess the favorability and stability of different molecular arrangements, which is crucial for understanding how interactions are maintained within a complex. Kinetic energy analysis helps reveal molecules’ activity levels and how they affect processes such as reaction rates and diffusion. Monitoring both types of energy throughout a simulation offers a detailed picture of molecular behavior and system dynamics. Fluctuations in these energies can indicate changes in stability and reaction kinetics, which is important for optimizing molecular systems.
The data in Table 3 indicate that there was no significant difference in the performance of the RTV and Eudragit L100 inclusion complex at the two different concentrations. However, a distinct disparity was observed in the RTV and PVP K30 inclusion complex. At a 500 ppm concentration, the complex demonstrated a notably better total energy of −150.00 kJ/mol. This suggests that PVP K30 at 500 ppm likely achieves a more stable molecular configuration and forms more effective intermolecular interactions compared to the higher 2000 ppm concentration. Thus, a 500 ppm concentration of PVP K30 results in a more energetically favorable and stable inclusion complex. These observations highlight that lower PVP K30 concentrations can enhance the stability and performance of the inclusion complex. The superior total energy at 500 ppm indicates improved molecular packing and reduced disruptive interactions. This increased stability will likely improve the reliability and effectiveness of practical applications, such as drug delivery systems. Additionally, the results emphasize the need for careful optimization of polymer concentrations to maximize the efficiency of molecular interactions.

Table 3.
Fraction of hydrogen bond occupancy within the inclusion complex systems in water during molecular dynamics simulations.
3.8. Evaluating the Dynamics of Binding and Release in Inclusion Complexes
In molecular dynamics simulations, a range of techniques can be used to identify interactions, including the analysis of potential energy between the guest and host molecules, evaluation of group correlations and cohesion, and analysis of molecular movements. Understanding interactions within inclusion complex systems provides valuable insights into such systems’ properties and helps predict their behavior more accurately. Identifying these interactions is essential for several reasons: it sheds light on the physical and chemical attributes of the system, allows for the optimization of host molecule designs, and aids in forecasting the system’s overall performance [51]. Moreover, a thorough analysis of these interactions enables precise adjustments to system parameters to achieve specific goals, such as enhancing stability or improving functional characteristics. It also supports the creation of more accurate models for real-world simulations, which can translate into more effective practical applications.
The data in Table 4 show that the Eudragit L100 500 ppm complex interacts with RTV only at specific times, namely, at 30 ns and 40 ns. Conversely, the Eudragit L100 2000 ppm complex maintains consistent interactions throughout the simulation, indicating that RTV remains firmly bound within the system. This stability suggests that the Eudragit L100 2000 ppm complex effectively retains RTV, creating a more stable inclusion complex. For systems involving polymers as guest molecules, which are large and consist of repeating units, the ability to retain these polymers is crucial. Challenges with releasing these polymers from the host can lead to several issues, including decreased separation efficiency and reduced binding capacity. Moreover, if polymers remain bound for too long, it may affect the integrity of the host structure. Understanding these interaction dynamics is vital for optimizing polymer-based inclusion complexes, enhancing their functionality and ensuring that they perform well in practical applications.

Table 4.
Dynamics of the interaction between RTV and Eudragit L100 in water during molecular dynamics simulations.
In contrast to Eudragit L100, PVP K30 at both the 500 ppm and 2000 ppm concentrations demonstrates similar characteristics (Table 5). Its interactions at these concentrations are mainly governed by hydrogen bonds and hydrophobic forces. These types of interactions can make it challenging for the guest molecules to detach from the host, as the strong hydrogen bonds and hydrophobic interactions effectively trap the guest molecules within the complex. Consequently, this can reduce the efficiency of separating the guest molecules from the mixture. To overcome this issue, strategies such as selecting a host with better compatibility with the guest molecules or modifying the environmental conditions could be used to make the release process easier. Additionally, incorporating advanced techniques such as photocatalysis could provide the extra energy needed to break the interactions holding the guest molecules, thus enhancing the release process. These approaches might also improve the overall efficiency of separating the guest molecules, potentially leading to better performance in real-world applications. Gaining a deep understanding of these interaction dynamics is essential for refining polymer-based inclusion systems.

Table 5.
Behavioral analysis of RTV and PVP K30 interactions in a water system using molecular dynamics simulations.
4. Discussion
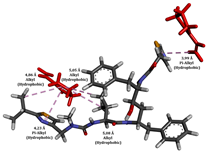
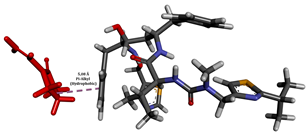
In this study, the addition of polymers such as Eudragit L100 and PVP K30 to supersaturated solution formulations significantly extended the nucleation induction time from 12 to about 64 h, demonstrating their effectiveness in stabilizing ritonavir (RTV). This extension is crucial for preventing the premature crystallization of RTV, thus maintaining its amorphous state and improving its bioavailability. The interaction between the polymers and RTV not only inhibits crystal nucleation but also contributes to a more stable drug environment, which is vital for the effectiveness of amorphous drug formulations. This stabilization mechanism is essential for drugs such as RTV, which have poor water solubility and can greatly benefit from enhanced solubility and delayed crystallization [52]. Integrating non-covalent adaptable networks (NANs) into a polymer-based system can enhance stabilization by adding layers of adaptability and reversibility to the drug–polymer interactions through non-covalent interactions (Figure 10). NANs, which utilize non-covalent interactions such as hydrogen bonding, electrostatic interactions, and hydrophobic interactions, can respond to environmental changes, allowing for more controlled drug release under physiological conditions [5]. This feature enhances the potential to tailor drug release profiles to meet specific therapeutic needs, reducing the likelihood of side effects associated with sudden drug release. Additionally, the adaptability of NANs supports the recovery and reuse of polymers, which is in line with sustainable pharmaceutical practices [8].
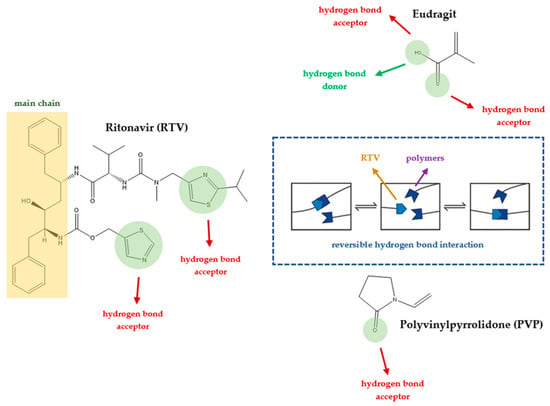
Figure 10.
An illustration of the dynamics of non-covalent adaptable networks (NANs) in drug–polymer systems [4,53].
Furthermore, Figure 5 and Figure 8 offer critical insights into the molecular mechanisms driving this stabilization. Figure 5, which compares the RDF data for Eudragit L100 and PVP K30, clearly shows that these polymers’ effectiveness is concentration-dependent. PVP K30 performs better at lower concentrations, while Eudragit L100 demonstrates superior stabilizing effects at higher concentrations. These findings are corroborated by Figure 8, which tracks the hydrogen bond formation between ritonavir and the polymers over time. The data reveal that higher concentrations of Eudragit L100 result in more frequent and consistent hydrogen bonding, leading to the stronger stabilization of ritonavir. In contrast, PVP K30, while effective at lower concentrations, forms fewer hydrogen bonds, which explains its comparatively reduced performance at higher concentrations. These figures were updated to improve clarity, particularly Figure 5, where the overlapping data points made it difficult to distinguish between trends. The updated visuals clearly show the green curve, representing one of the polymers, which enhances the overall readability and enables a more precise comparison of polymer performance. This clarification reinforces the conclusion that PVP K30 excels at lower concentrations, whereas Eudragit L100 performs better at higher doses, aligning with the findings from the hydrogen bonding analysis shown in Figure 8.
Integrating NANs into molecular dynamics simulations of drug–polymer interactions provides deep insights into the underlying mechanisms that enhance the stability and efficacy of amorphous solid dispersions (ASDs) [9]. This approach allows researchers to not only visualize but also optimize the formation and stability of these complex systems under various conditions. It further facilitates the identification of the optimal combinations of polymers and NANs that maximize drug stability and effectiveness within ASD formulations. Moreover, the adaptability of NANs introduces innovative capabilities into drug formulation technology, enhancing the efficiency and scalability of manufacturing processes [23]. Incorporating NANs into ASD formulations could redefine the synergy between drugs and polymers, particularly for traditional polymers such as Eudragit L100 and PVP K30. Future research could explore the synergistic effects of combining polymers with innovative NANs to develop formulations that exploit the best properties of both components. This strategy could forge the next generation of drug delivery systems that offer improved control over drug release, heightened stability, and superior patient compliance, which is particularly beneficial in ASD contexts where drug solubility and stability are paramount.
5. Conclusions
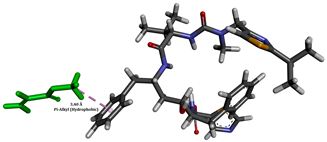
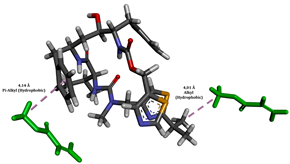
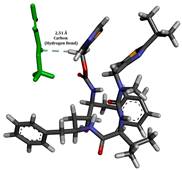
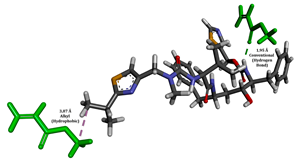
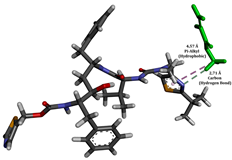
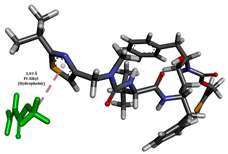
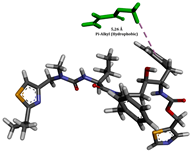
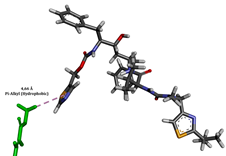
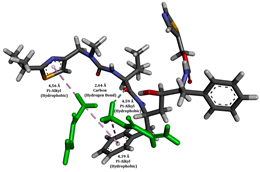
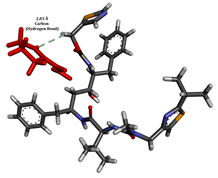
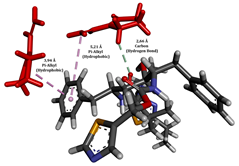
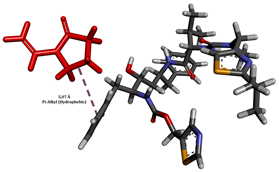
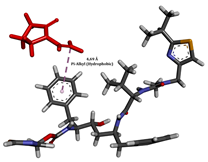
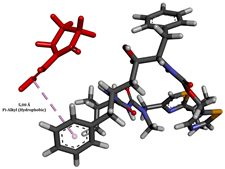
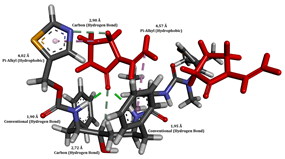
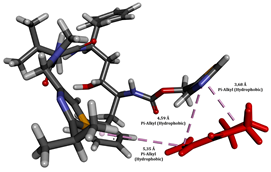
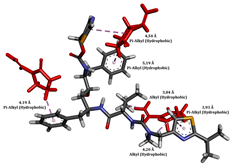
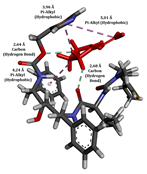
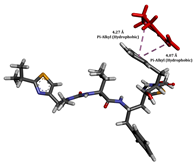
To enhance the understanding of amorphous solid dispersion (ASD) systems, this study integrates molecular dynamics (MD) simulations and wet lab experiments to explore the dispersion of ritonavir (RTV) with Eudragit L100 and Polyvinylpyrrolidone (PVP) K30. This comprehensive approach not only utilizes the advanced capabilities of non-covalent adaptable networks (NANs) within simulations but also confirms the molecular interactions involved based on empirical laboratory findings. The credibility of the MD simulations is validated by analyzing the interactions and structural alterations within the RTV/Eudragit L100 and RTV/PVP K30 systems, with results that are consistent with laboratory observations. These simulations reveal that the translational motion of RTV is effectively restricted by the presence of Eudragit L100 and PVP K30. This restriction occurs due to the formation of pi-alkyl bonds between RTV and the polymers in the solvent, which were observed and directly measured in the wet lab experiments. Simultaneously, hydrogen bonding interactions between RTV and the polymers were also observed throughout the simulations, with similar bonding patterns identified in laboratory-based analyses. The lab experiments provide concrete evidence of how NANs enhance the stability of these interactions. By facilitating adaptable and reversible bonding, NANs prove essential for stabilizing the amorphous drug in a supersaturated solution. Minor differences in the strength and nature of interactions between RTV and each polymer become less critical compared to their overall ability to maintain the supersaturated state. Physical samples from the lab experiments confirm the crucial role of pi-alkyl and hydrogen bonds in maintaining the stability and integrity of amorphous RTV within the formulation. This dual-method investigation—incorporating both computational and experimental techniques—offers profound insights into the effectiveness of NANs in developing amorphous formulations for poorly water-soluble drugs. The findings highlight the importance of MD simulations combined with practical lab tests in assessing intermolecular interactions and selecting the most suitable polymers for ASD formulations. This integrative approach not only validates computational models but also enhances predictive accuracy for the pharmaceutical development of stable and effective drug delivery systems, showcasing the critical synergy between theoretical research and empirical evidence.
Author Contributions
Conceptualization, A.B.; methodology, A.B., T.M.F. and D.L.A.; software, T.M.F.; validation, A.B., S.M. and M.M.; formal analysis, A.B. and T.M.F.; investigation, A.B. and T.M.F.; resources, D.L.A.; data curation, A.B., M.M. and T.M.F.; writing—original draft preparation, A.B. and T.M.F.; writing—review and editing, A.B., D.L.A. and T.M.F.; visualization, S.M. and M.M.; supervision, A.B.; project administration, D.L.A.; funding acquisition, A.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Universitas Padjadjaran (Hibah Riset Data Pustaka dan Daring) to Arif Budiman (No: 1549/UN6.3.1/PT.00/2023, Tanggal 27 Maret 2023).
Data Availability Statement
We just presented this topic in the seminar. The data is not available in any repository.
Acknowledgments
We would like to thank Universitas Padjadjaran for APC, the National Research and Innovation Agency (BRIN, RIIM2), and the Indonesia Endowment Funds for Education (LPDP) for providing ritonavir. The data in this article have been presented in the 6th International Seminar in Pharmaceutical Sciences and Technology (ISPST) Faculty of Pharmacy—Universitas Padjadjaran on 30–31 October 2024.
Conflicts of Interest
The authors declare that they have no conflicts of interest related to the contents of this article.
References
- Budiman, A.; Husni, P.; Shafira; Alfauziah, T.Q. The development of glibenclamide-saccharin cocrystal tablet formulations to increase the dissolution rate of the drug. Int. J. Appl. Pharm. 2019, 11, 359–364. [Google Scholar] [CrossRef]
- Budiman, A.; Aulifa, D.L. Encapsulation of Drug into Mesoporous Silica by Solvent Evaporation: A Comparative Study of Drug Characterization in Mesoporous Silica with Various Molecular Weights. Heliyon 2021, 7, e08627. [Google Scholar] [CrossRef] [PubMed]
- Sabzini, M.; Pourmadadi, M.; Yazdian, F.; Khadiv-Parsi, P.; Rashedi, H. Development of Chitosan/Halloysite/Graphitic-carbon Nitride Nanovehicle for Targeted Delivery of Quercetin to Enhance Its Limitation in Cancer Therapy: An in Vitro Cytotoxicity against MCF-7 Cells. Int. J. Biol. Macromol. 2023, 226, 159–171. [Google Scholar] [CrossRef]
- Laitinen, R.; Löbmann, K.; Strachan, C.J.; Grohganz, H.; Rades, T. Emerging Trends in the Stabilization of Amorphous Drugs. Int. J. Pharm. 2013, 453, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhou, L.; Kang, M.; Zhao, X.; Chang, G.; Chen, M. Tough Non-Covalent Adaptable Networks: Cation-π Cross-Linked Rigid Epoxy. Polymer 2022, 243, 124626. [Google Scholar] [CrossRef]
- Yang, S.; Bai, J.; Sun, X.; Zhang, J. Robust and Healable Poly(Disulfides) Supramolecular Adhesives Enabled by Dynamic Covalent Adaptable Networks and Noncovalent Hydrogen-Bonding Interactions. Chem. Eng. J. 2023, 461, 142066. [Google Scholar] [CrossRef]
- Yu, D.; Li, J.; Wang, H.; Pan, H.; Li, T.; Bu, T.; Zhou, W.; Zhang, X. Role of Polymers in the Physical and Chemical Stability of Amorphous Solid Dispersion: A Case Study of Carbamazepine. Eur. J. Pharm. Sci. 2022, 169, 106086. [Google Scholar] [CrossRef]
- Bookwala, M.; Wildfong, P.L.D. The Implications of Drug-Polymer Interactions on the Physical Stability of Amorphous Solid Dispersions. Pharm. Res. 2023, 40, 2963–2981. [Google Scholar] [CrossRef] [PubMed]
- Bharti, K.; Dubey, G.; Kumar, M.; Jha, A.; Manjit; Upadhyay, M.; Mali, P.S.; Kumar, A.; Bharatam, P.V.; Mishra, B. A Multifaceted Approach for Grading of Polymers for the Development of Stable Amorphous Solid Dispersion of Riluzole. J. Drug Deliv. Sci. Technol. 2023, 90, 105158. [Google Scholar] [CrossRef]
- Bookwala, M.; Buckner, I.S.; Wildfong, P.L.D. Implications of Coexistent Halogen and Hydrogen Bonds in Amorphous Solid Dispersions on Drug Solubility, Miscibility, and Mobility. Mol. Pharm. 2022, 19, 3959–3972. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.; Zografi, G. Considerations in the Development of Physically Stable High Drug Load API- Polymer Amorphous Solid Dispersions in the Glassy State. J. Pharm. Sci. 2023, 112, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, L.; Wu, B.; Chen, F. Improving Solubility of Poorly Water-Soluble Drugs by Protein-Based Strategy: A Review. Int. J. Pharm. 2023, 634, 122704. [Google Scholar] [CrossRef] [PubMed]
- Sarode, A.L.; Wang, P.; Obara, S.; Worthen, D.R. Supersaturation, Nucleation, and Crystal Growth during Single- and Biphasic Dissolution of Amorphous Solid Dispersions: Polymer Effects and Implications for Oral Bioavailability Enhancement of Poorly Water Soluble Drugs. Eur. J. Pharm. Biopharm. 2014, 86, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Bollag, G.; Hirth, P.; Tsai, J.; Zhang, J.; Ibrahim, P.N.; Cho, H.; Spevak, W.; Zhang, C.; Zhang, Y.; Habets, G.; et al. Clinical Efficacy of a RAF Inhibitor Needs Broad Target Blockade in BRAF-Mutant Melanoma. Nature 2010, 467, 596–599. [Google Scholar] [CrossRef] [PubMed]
- Nambiar, A.G.; Singh, M.; Mali, A.R.; Serrano, D.R.; Kumar, R.; Healy, A.M.; Agrawal, A.K.; Kumar, D. Continuous Manufacturing and Molecular Modeling of Pharmaceutical Amorphous Solid Dispersions. AAPS PharmSciTech 2022, 23, 249. [Google Scholar] [CrossRef] [PubMed]
- Muchtaridi, M.; Triwahyuningtyas, D.; Muhammad Fakih, T.; Megantara, S.; Choi, S.B. Mechanistic Insight of α-Mangostin Encapsulation in 2-Hydroxypropyl-β-Cyclodextrin for Solubility Enhancement. J. Biomol. Struct. Dyn. 2023, 42, 3223–3232. [Google Scholar] [CrossRef] [PubMed]
- Bhujbal, S.V.; Mitra, B.; Jain, U.; Gong, Y.; Agrawal, A.; Karki, S.; Taylor, L.S.; Kumar, S.; (Tony) Zhou, Q. Pharmaceutical Amorphous Solid Dispersion: A Review of Manufacturing Strategies. Acta Pharm. Sin. B 2021, 11, 2505–2536. [Google Scholar] [CrossRef]
- Schittny, A.; Huwyler, J.; Puchkov, M. Mechanisms of Increased Bioavailability through Amorphous Solid Dispersions: A Review. Drug Deliv. 2020, 27, 110–127. [Google Scholar] [CrossRef] [PubMed]
- Mansuri, A.; Münzner, P.; Heermant, A.; Patzina, F.; Feuerbach, T.; Winck, J.; Vermeer, A.W.P.; Hoheisel, W.; Böhmer, R.; Gainaru, C.; et al. Molecular Dynamics and Diffusion in Amorphous Solid Dispersions Containing Imidacloprid. Mol. Pharm. 2023, 20, 2067–2079. [Google Scholar] [CrossRef]
- Ma, X.; Williams, R.O. Characterization of Amorphous Solid Dispersions: An Update. J. Drug Deliv. Sci. Technol. 2019, 50, 113–124. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, L.; Ma, D.; Tang, X.; Zhang, Y.; Yin, T.; Gou, J.; Wang, Y.; He, H. Characterizing and Exploring the Differences in Dissolution and Stability Between Crystalline Solid Dispersion and Amorphous Solid Dispersion. AAPS PharmSciTech 2020, 21, 262. [Google Scholar] [CrossRef] [PubMed]
- Moseson, D.E.; Corum, I.D.; Lust, A.; Altman, K.J.; Hiew, T.N.; Eren, A.; Nagy, Z.K.; Taylor, L.S. Amorphous Solid Dispersions Containing Residual Crystallinity: Competition Between Dissolution and Matrix Crystallization. AAPS J. 2021, 23, 69. [Google Scholar] [CrossRef] [PubMed]
- Brunsteiner, M.; Khinast, J.; Paudel, A. Relative Contributions of Solubility and Mobility to the Stability of Amorphous Solid Dispersions of Poorly Soluble Drugs: A Molecular Dynamics Simulation Study. Pharmaceutics 2018, 10, 101. [Google Scholar] [CrossRef]
- Pajzderska, A.; Gonzalez, M.A. Molecular Dynamics Simulations of Selected Amorphous Stilbenoids and Their Amorphous Solid Dispersions with Poly (Vinylpyrrolidone). J. Pharm. Sci. 2023, 112, 2444–2452. [Google Scholar] [CrossRef] [PubMed]
- Budiman, A.; Kalina, K.; Aristawidya, L.; Al Shofwan, A.A.; Rusdin, A.; Aulifa, D.L. Characterizing the Impact of Chitosan on the Nucleation and Crystal Growth of Ritonavir from Supersaturated Solutions. Polymers 2023, 15, 1282. [Google Scholar] [CrossRef] [PubMed]
- Budiman, A.; Citraloka, Z.G.; Muchtaridi, M.; Sriwidodo, S.; Aulifa, D.L.; Rusdin, A. Inhibition of Crystal Nucleation and Growth in Aqueous Drug Solutions: Impact of Different Polymers on the Supersaturation Profiles of Amorphous Drugs—The Case of Alpha-Mangostin. Pharmaceutics 2022, 14, 2386. [Google Scholar] [CrossRef]
- Wilson, V.R.; Mugheirbi, N.A.; Mosquera-Giraldo, L.I.; Deac, A.; Moseson, D.E.; Smith, D.T.; Novo, D.C.; Borca, C.H.; Slipchenko, L.V.; Edgar, K.J.; et al. Interaction of Polymers with Enzalutamide Nanodroplets—Impact on Droplet Properties and Induction Times. Mol. Pharm. 2021, 18, 836–849. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 Update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Brown, T. ChemDraw. Sci. Teach. 2014, 81, 67. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A. Gaussian16, (Revision A.03); Gaussian, Inc.: Wallingford, CT, USA, 2016; Volume 1, p. 572.
- Forli, W.; Halliday, S.; Belew, R.; Olson, A. AutoDock, Version 4.2; The Scripps Research Institute: La Jolla, CA, USA, 2012. Available online: https://autodock.scripps.edu/ (accessed on 28 September 2024).
- Martinez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. PACKMOL: A Package for Building Initial Configurations for Molecular Dynamics Simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef]
- Abraham, M.; Hess, B.; van der Spoel, D.; Lindahl, E. The GROMACS Development Team, GROMACS User Manual; Version 5.0.7; The GROMACS Development Teams at the Royal Institute of Technology and Uppsala University: Uppsala, Sweden, 2015. [Google Scholar]
- Abraham, M.; Hess, B.; van der Spoel, D.; Lindahl, E. Gromacs—Reference Manual Version 2016.3; The Royal Institute of Technology and Uppsala University: Uppsala, Sweden, 2015. [Google Scholar]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Smith, M.D.; Rao, J.S.; Segelken, E.; Cruz, L. Force-Field Induced Bias in the Structure of Aβ21-30: A Comparison of OPLS, AMBER, CHARMM, and GROMOS Force Fields. J. Chem. Inf. Model. 2015, 55, 2587–2595. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, Y.; Cui, Z.; Yin, C. Force Field Benchmark of Amino Acids. 2. Partition Coefficients between Water and Organic Solvents. J. Chem. Inf. Model. 2018, 58, 1669–1681. [Google Scholar] [CrossRef] [PubMed]
- Kagami, L.; Wilter, A.; Diaz, A.; Vranken, W. The ACPYPE Web Server for Small-Molecule MD Topology Generation. Bioinformatics 2023, 39, btad350. [Google Scholar] [CrossRef] [PubMed]
- Isele-Holder, R.E.; Mitchell, W.; Ismail, A.E. Development and Application of a Particle-Particle Particle-Mesh Ewald Method for Dispersion Interactions. J. Chem. Phys. 2012, 137, 174107. [Google Scholar] [CrossRef]
- Wu, J.; Mooter, G. Van den The Influence of Hydrogen Bonding between Different Crystallization Tendency Drugs and PVPVA on the Stability of Amorphous Solid Dispersions. Int. J. Pharm. 2023, 646, 123440. [Google Scholar] [CrossRef] [PubMed]
- Ilie, A.R.; Griffin, B.T.; Brandl, M.; Bauer-Brandl, A.; Jacobsen, A.C.; Vertzoni, M.; Kuentz, M.; Kolakovic, R.; Holm, R. Exploring Impact of Supersaturated Lipid-Based Drug Delivery Systems of Celecoxib on in Vitro Permeation across PermeapadⓇ Membrane and in Vivo Absorption. Eur. J. Pharm. Sci. 2020, 152, 105452. [Google Scholar] [CrossRef]
- Joshi, M. Role of Eudragit in Targeted Drug Delivery. Int. J. Curr. Pharm. Res. 2013, 5, 58–62. [Google Scholar]
- Franco, P.; De Marco, I. The Use of Poly(N-Vinyl Pyrrolidone) in the Delivery of Drugs: A Review. Polymers 2020, 12, 1114. [Google Scholar] [CrossRef]
- Aung, N.N.; Ngawhirunpat, T.; Rojanarata, T.; Patrojanasophon, P.; Opanasopit, P.; Pamornpathomkul, B. Enhancement of Transdermal Delivery of Resveratrol Using Eudragit and Polyvinyl Pyrrolidone-Based Dissolving Microneedle Patches. J. Drug Deliv. Sci. Technol. 2021, 61, 102284. [Google Scholar] [CrossRef]
- Nurisyah; Ramadhan, D.S.F.; Dewi, R.; Asikin, A.; Daswi, D.R.; Adam, A.; Chaerunnimah; Sunarto; Rafika; Artati; et al. Targeting EGFR Allosteric Site with Marine-Natural Products of Clathria Sp.: A Computational Approach. Curr. Res. Struct. Biol. 2024, 7, 100125. [Google Scholar] [CrossRef] [PubMed]
- Aulifa, D.L.; Al Shofwan, A.A.; Megantara, S.; Fakih, T.M.; Budiman, A. Elucidation of Molecular Interactions Between Drug–Polymer in Amorphous Solid Dispersion by a Computational Approach Using Molecular Dynamics Simulations. Adv. Appl. Bioinforma. Chem. 2024, 17, 1–19. [Google Scholar] [CrossRef]
- Liu, P.; Lu, J.; Yu, H.; Ren, N.; Lockwood, F.E.; Wang, Q.J. Lubricant Shear Thinning Behavior Correlated with Variation of Radius of Gyration via Molecular Dynamics Simulations. J. Chem. Phys. 2017, 147, 084904. [Google Scholar] [CrossRef] [PubMed]
- Durham, E.; Dorr, B.; Woetzel, N.; Staritzbichler, R.; Meiler, J. Solvent Accessible Surface Area Approximations for Rapid and Accurate Protein Structure Prediction. J. Mol. Model. 2009, 15, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Zikri, A.T.; Pranowo, H.D.; Haryadi, W. Stability, Hydrogen Bond Occupancy Analysis and Binding Free Energy Calculation from Flavonol Docked in DAPK1 Active Site Using Molecular Dynamic Simulation Approaches. Indones. J. Chem. 2021, 21, 383–390. [Google Scholar] [CrossRef]
- Mishra, A.; Mulpuru, V.; Mishra, N. Exploring the Mechanism of Action of Podophyllotoxin Derivatives through Molecular Docking, Molecular Dynamics Simulation and MM/PBSA Studies. J. Biomol. Struct. Dyn. 2022, 41, 8856–8865. [Google Scholar] [CrossRef]
- Hidayat, A.F.; Fakih, T.M. Self-Assembly of Black Cumin Oil-Based Nanoemulsion on Various Surfactants: A Molecular Dynamics Study. Makara J. Sci. 2021, 25, 8–264. [Google Scholar] [CrossRef]
- Budiman, A.; Aulifa, D.L. A Comparative Study of the Pharmaceutical Properties between Amorphous Drugs Loaded-Mesoporous Silica and Pure Amorphous Drugs Prepared by Solvent Evaporation. Pharmaceuticals 2022, 15, 730. [Google Scholar] [CrossRef]
- Kloxin, C.J.; Bowman, C.N. Covalent Adaptable Networks: Smart, Reconfigurable and Responsive Network Systems. Chem. Soc. Rev. 2013, 42, 7161–7173. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).