Abstract
Survival analysis is widely used in fields such as medical research and reliability engineering to analyze data where not all subjects experience the event of interest by the end of the study. It requires dedicated methods capable of handling censored cases. This paper extends the collection of techniques applicable to censored data by introducing a novel algorithm for interpretable recommendations based on a set of survival action rules. Each action rule contains recommendations for changing the values of attributes describing examples. As a result of applying the action rules, an example is moved from a group characterized by a survival curve to another group with a significantly different survival rate. In practice, an example can be covered by several induced rules. To decide which attribute values should be changed, we propose a recommendation algorithm that analyzes all actions suggested by the rules covering the example. The efficiency of the algorithm has been evaluated on several benchmark datasets. We also present a qualitative analysis of the generated recommendations through a case study. The results indicate that the proposed method produces high-quality recommendations and leads to a significant change in the estimated survival time.
1. Introduction
Action rule induction is a machine learning technique that enables the creation of action models. Based on the input feature vector, the action model generates recommendations for changes in attribute values. The objective is to change the assignment of a given example, representing a particular group (referred to as the source group) so that it matches examples from another group (referred to as the target group). Typically, this approach does not take into account the specific actions required to change the values of a particular attribute. In particular, action rules previously have only been defined for classification data where each example belongs to one of several decision classes.
An important feature of rule-based action models is their interpretability. However, the large number of rules in the model can naturally lead to comprehension problems.
Many algorithms have been proposed for action rule induction. In particular, Sikora et al. [1] introduce a sequential covering action rule induction algorithm for classification problems and a method for verifying the quality of the generated rules.
In [2], we presented an algorithm for the induction of action rules for censored data, which is the first proposal for the induction of survival action rules. The algorithm can be used, for example, in medical data and reliability analysis. The article [2] focuses solely on knowledge discovery through the induction of a set of survival action rules without addressing prediction. Prediction here involves making specific recommendations for changing attribute values for a given example (e.g., a test example). The application of these changes should result in the considered example being moved to an area of the attribute space where there are examples with a significantly different survival rate than the considered example before the changes.
If each example were covered by only one rule, the procedure would be straightforward: simply make the changes recommended by the rule applicable to that particular example. In practice, however, examples are covered by many rules, sometimes containing conflicting recommendations. This scenario represents one of the main scientific problems addressed in this paper. Consequently, there is a need to develop a method for resolving conflicts between rules, similar to how conflicts are resolved in classification systems.
This article presents a proposal for an algorithm for generating recommendations based on a set of survival action rules. The main contribution of this article is the proposal of such an algorithm. Based on the induced survival rules, the algorithm builds a special meta-table containing information about all elementary conditions (elementary actions) that constitute the induced rules. In this meta-table, conflict resolution between rules covering the example is solved by inducing a special meta-action survival rule. The meta-action survival rule contains the actions that maximally differentiate the survival rates, characterizing the example before and after the application of these actions.
The efficiency of the algorithm was evaluated on several benchmark datasets. In addition, we present a qualitative analysis of the generated recommendations through a case study.
The recommendation algorithm introduced in this study can be considered to be a specialized counterfactual explanation method tailored for survival data analysis. So far, only one counterfactual explanation method dedicated to survival data has been proposed [3]. Furthermore, this method operates with datasets that exclusively contain numerical attributes. It generates recommendations for a specific instance without generating a global action model.
The recommendation algorithm presented in the article has several unique features:
- it is based on a global action model represented by survival action rules,
- recommendations for a specific instance are generated based on the fusion of information contained in the action model,
- the method can be applied to datasets containing numerical as well as categorical attributes,
- the algorithm uses a computationally efficient hill climbing strategy to search recommendations.
The article also presents a proposal for verifying the quality of the recommendation algorithm. The proposed verification method does not require observing an example after the recommendations have been applied. An independent arbiter is used to verify whether the application of the recommendations actually changes the value of the survival rate estimated for the example. The arbiter is trained on a training dataset and has very good predictive abilities for estimating the survival rate (in the article, we use gradient boosting [4]).
For an unseen (e.g., test) example x, we denote by the example whose attribute values were changed according to the recommendation algorithm. If there is no statistical difference between the survival rates for estimated by the recommendation algorithm and the arbiter model (using the log-rank test), we assume that the attribute value changes suggested by the recommendation algorithm were successful.
The existing literature on counterfactual explanation and action rule induction has not emphasis the evaluation of the recommendations produced by these techniques. Incorporating an external arbiter enables the validation of whether the modifications proposed by recommendation algorithms effectively impact the estimated survival time or rate.
The article is organized as follows. The Section 2 contains a brief literature review of survival analysis and action rule induction. Section 3 introduces survival action rule induction and then explains the principles of defining a meta-table and the recommendation induction algorithm. Section 4 presents both quantitative and qualitative experimental results and discusses the implications of the findings obtained from these results. Section 5 concludes the article and includes suggestions for future work.
2. Related Work
The approach presented in this article is based on survival analysis, action rules, and, to some extent, a novel technique for interpreting machine learning models—counterfactual explanations.
Survival analysis is used to study processes in which the time that elapses before a particular event occurs is of interest. It is based on statistical and machine learning methods. Statistical methods use a univariate approach to assess the impact of a single factor on a process [5,6]. Multivariate statistical methods, on the other hand, take into account more than one factor and allow the strength of the impact of each factor to be determined [7]. Statistical methods can be further classified into non-parametric approaches, such as Kaplan–Meier survival curves [8] or the log-rank test [9]; semi-parametric approaches, such as the Cox regression model [10]; and parametric approaches, including proportional hazard models [11].
Machine learning methods used in survival analysis include [12]: decision trees [13,14,15], survival rules [16,17], support vector machines [18,19], neural networks [20,21], and Bayesian networks [22,23].
Ensemble methods have also been proposed for survival data analysis, such as random forests [24,25] and gradient boosting [25,26]. Recently, several deep neural network architectures have been proposed for analyzing survival data [27,28,29,30].
To date, only Badura et al. [2] have addressed the challenge of action rule mining for survival analysis. In contrast, there are numerous methods for applying action rules to classification problems. The concept of action rules for classification was first introduced by Raś and Wieczorkowska in [31]. They proposed a rough set-based approach that generated rules using reducts and then created action rules based on them. The rough set-based method was also employed by Tsay and Raś in the DEAR system [32,33]. Generally, two approaches are commonly used for action rule learning. The first approach involves inducing classification rules first and then deriving action rules based on these rules. This category includes the previously mentioned rough set-based methods. In the latter approach, action rules are generated directly from data. The direct induction of action rules is based on a priori-like algorithms [34,35,36], heuristic strategies [1,37,38], or evolutionary strategies [39,40].
In most cases, the aforementioned approaches to action rule learning cannot efficiently handle large datasets. To address this problem, some authors have proposed distributed, partition-based methods. Bagavathi et al. [41] focus on vertical (i.e., by columns) data splitting using information granules. Benedict and Raś [42] utilize vertical partitioning based on attribute correlation. Tarnowska et al. [43] use both vertical and horizontal (i.e., by rows) data partitions.
It is worth noting that decision trees [44,45,46] and decision tree ensembles [47,48] have also been used for action and recommendation mining.
Works such as [49,50] focus on measuring the expected effects of interventions (actions) suggested by decision rules.
A methodology similar to our approach is the counterfactual explanation. This is an example-based explainability technique used to explain a particular outcome of a machine learning model. The task of counterfactual explanations is to identify the necessary changes that would change the model’s decision. Counterfactual explanations provide a post hoc interpretation of a particular decision made by machine learning models.
Wachter et al. [51] defined the counterfactual explanation problem as an optimization problem with two criteria. The loss function measures the following: the distance between the model prediction for the counterfactual and the desired outcome , as well as the distance between the instance to be explained and its counterfactual outcome. Dandl et al. [52] consider additional criteria, such as the number of features to be changed. A comprehensive review of the literature on counterfactual explanation methods can be found in [53,54]. Most counterfactual explanation methods are defined for classification and regression problems, but some articles also present counterfactual explanation methods for survival models [3,55,56].
The main differences between counterfactual explanations and action rules lie in the level of explanation and searching strategies.
Counterfactual explanations are generated separately for each example, making it a local explanation method. These methods mainly use global optimization methods (e.g., genetic algorithms). Rule-based methods generally use heuristic strategies (e.g., hill climbing) and allow the generation of an interpretable action model for the entire dataset.
3. Methods
In survival models, when the data lacks complete information on the timing of an event, we are dealing with so-called censored data [57]. There are three main types of censoring [58]: right censoring occurs when the true event time exceeds the observation period; left censoring happens when the event takes place before the observation begins without a precise time; and interval censoring arises when the event is known to have occurred within a specific period, but the exact moment remains unknown.
Right-censored data are most commonly used in survival analysis. There are several reasons for right censoring: the observation period of the phenomenon only covers a certain period of time, the object of observation is removed from the study, or the observation of the object is interrupted for external reasons.
The main applications of survival analysis include estimating and interpreting the survival function, comparing survival curves across different groups of observations, and evaluating how conditional attributes affect survival time.
The survival function, together with the hazard function, is of particular interest in the analysis of survival data. To estimate a survival function from observed survival times without assuming an underlying probability distribution, the Kaplan–Meier estimator is recommended [6]. For each time interval, the probability of survival is calculated as the number of surviving subjects divided by the number of subjects at risk.
3.1. Basic Notions
Consider a set of examples. Each example is described with a set of attributes , an observation time T, and a survival status .
A straightforward and understandable form of knowledge representation rules. In general, rules take the form of an implication: IF THEN , where is the premise of the rule and is the conclusion. In rule-based reasoning, the premise determines the conditions that must be satisfied for the conclusion to be true. The rule premise takes the form of a conjunction of elementary conditions , where is an element of the domain of the attribute , and ⊙ is a relation (= for symbolic attributes or for ordinal and numerical attributes). An example is considered to be covered by a rule if it satisfies the conditions specified in the rule’s premise. The conclusion of a rule varies based on its type, including classification, association, regression, survival, or action rules.
In the case of survival rules, the conclusion is the estimator of a survival function (e.g., the Kaplan–Meier estimator). A survival rule, therefore, has the following form:
The survival function resulting from the conclusion of an action rule describes the survival time of all examples covered by the rule. A survival action rule is a combination of two action rules:
An action rule’s premise is composed of a conjunction of elementary actions. Each elementary action, denoted as →, signifies a modification in the attribute ’s value. This includes the elementary action’s premise, , outlining the initial value range of the attribute, and the elementary action’s conclusion, , detailing the desired target value range.
The survival action rule can be decomposed into two survival rules (called source (3) and target (4) rules):
Applying the actions contained in the premise of the survival action rule (2) results in a change in the estimated survival time for the examples covered by the source and target rules.
For an elementary action , three cases can be distinguished: , , and the last case where the range of the target part is not specified (i.e., ). The first case represents a real change in the value of the attribute , while the second represents the preservation of the attribute value. It can be said that, in the second case, the attribute value should not be changed, and in the third case, the attribute can take any value.
Example 1.
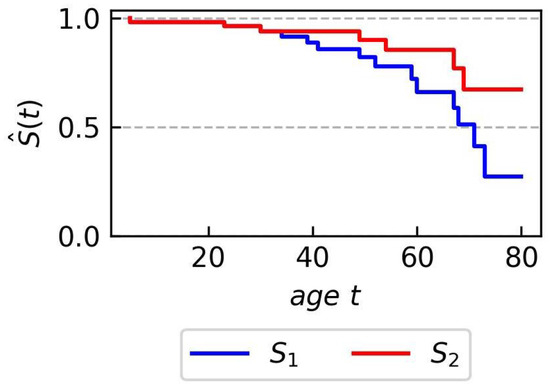
To illustrate the concept of survival action rules, let us consider the body mass index (BMI), which measures the proportionality of body weight in relation to height and age. It is well known that people classified as overweight or obese have a higher risk of premature death than those with a normal BMI.
A normal BMI is between 18.5 and 24.9. A BMI between 25.0 and 29.9 is considered overweight, while a BMI of 30.0 or higher is considered obese.
Now, let us define two groups of people characterized by BMI values as follows: and
These groups are characterized by two different survival curves, as shown in Figure 1. The survival action rule (5) states that a reduction in BMI to the range [19,25] will increase the estimated survival time, as shown by the curve .

Figure 1.
Synthetic data—Kaplan-Meier plots for two groups and used in the illustrative example of survival action rules concept.
3.2. Survival Action Rule Induction
In this section, we provide a brief overview of our algorithm proposed in [2] for generating an action rule set to be applied to the analysis of censored data (see Algorithm 1). The rule set consists of survival action rules. The algorithm is essential for understanding the subsequent content.
| Algorithm 1 An action rule set induction algorithm for censored data. |
| Require: —a dataset described by attributes A, observation time T, and survival status P—a set of input parameters —a set of stable attributes from the set A Ensure: R—a set of survival action rules
|
Algorithm 1 outlines the main loop that implements rule set induction. In each loop, a new rule is formed through a two-step process: first, the rule is specialized (referenced in line 5 of Algorithm 1), and then it is pruned (mentioned in line 6 of the same algorithm). The loop run ends with the elimination of the examples that the newly formed rule covers from the pool of examples yet to be covered (line 9). If the process of creating a rule yields one with no conditions in its premise, such a rule is excluded from the final set of rules (line 7), signaling that it is impossible to derive a new actionable rule. The process of rule induction terminates when such an empty rule, which applies to all examples in the dataset D, is formed.
When comparing the source and target survival curves, it is clear that their relative positions are not constant. The log-rank measure [6] does not take into account the relative position of these curves. It is a symmetric measure that only gives the maximum distance between the curves. In contrast, the proposed algorithm can generate three types of rules: improving (i.e., seeking actions that increase survival), worsening (i.e., seeking rules that decrease survival), and arbitrary rules (i.e., allowing the simultaneous induction of both types of rules). The type of rules generated is determined by the run mode—an algorithm’s parameter. During the induction of a new action rule, the area under the survival curve and the survival probability (using the Kaplan–Meier estimator) are measured. For the improving rule, both the area and the probability of the target rule should be higher, whereas for the worsening rule, both parameters should be lower.
The most important phase in the induction of a survival action rule is the specialization phase, illustrated in Algorithm 2.
The process of rule specialization involves adding successive actions into the rule’s premise, which is initially empty (lines 10–13 of Algorithm 2). These actions derive from two elementary conditions identified during the specialization. The first is the best elementary condition to include in the rule’s source part (line 5), and the second is the best counter-condition or the best elementary condition for the rule’s target part (line 9). Specialization finishes when no further best elementary condition can be found for the rule’s source.
Identifying the best elementary condition (line 5) involves finding a condition that, once added to the action rule’s source premise, yields the highest quality survival rule. This is determined by maximizing the log-rank statistic’s value by comparing the example subset covered by the action rule against those not covered. If multiple conditions equally maximize the log-rank value, the condition expanding the uncovered example set the most is selected.
| Algorithm 2 Specialization of the action rule in the survival analysis. |
| Require: D—a dataset —a collection of examples that are not yet covered —the minimum number of examples that the new rule must cover — the maximum rule coverage —the maximum percentage of examples that can be covered by both the left and right rules —a set of stable attributes from the set A Ensure: r—the action rule
|
Line 6 indicates that the best-identified elementary condition is added to the set, which contains conditions already evaluated. This addition ensures that the condition is not repeatedly selected in the loop if a counter-condition is not identified, thus preventing its redundant inclusion in the rule’s premise.
The search for the best counter-condition (line 9) follows the principle that, upon finding the best condition for attribute , a counter-condition is sought. This counter-condition, for the same attribute, aims to maximize the log-rank test value between subsets defined by the initial source premise with and the target premise with .
Once rule specialization is complete, the rule’s generalization phase begins. During this phase, actions are systematically removed from the premise to assess whether the log-rank measure’s value for the revised rule improves or remains unchanged. Pruning finishes when no further actions can be removed or when any modification would diminish the rule’s quality. This phase may also end if the rule covers all examples from the set.
Action removal is governed by two principles. An action is retained if its removal would increase the proportion of examples covered by a single rule beyond the parameter value, preventing overly general rules. The second principle involves the maximum shared example proportion between the source and target rules, which, if exceeding the parameter, prevents the action’s removal, ensuring significant differentiation between the rule’s source and target example groups.
For missing values, a strategy of ignoring such values is employed. This approach focuses on creating rules based only on available, observed values, excluding observations with missing attribute values from rule coverage. This method is comparably effective to more sophisticated methods for handling missing value [59] and avoids the complexities of imputation techniques.
The obtained rule-based action model not only describes the dependencies occurring in the analyzed dataset but can also be used for prediction. To estimate the expected survival time, we can apply the changes specified by a rule in the set whose source part covers the example under consideration to a given example.
The problem in prediction arises when a given example for which we want to take an action is covered by several rules. This leads to a conflict about which actions to apply. Our proposed solution involves creating a meta-table based on the rules obtained. Then, within this meta-table, an action rule that gives the final recommendation is induced. The rule indicates which attribute values need to be changed and to what extent.
An action rule contains recommendations for changing attribute values describing examples, with the aim of moving an example from a group defined by a particular survival curve to another group that has a statistically significant survival rate after the action rules have been applied. In practice, a single example may be covered by numerous introduced rules. To make the final decision about which attribute values should be changed, we propose a recommendation algorithm that analyzes all the actions suggested by all the rules covering the example. Subsequently, an action rule is identified that specifies the final recommendation and indicates which attribute should be changed, including the specific attribute values and their range.
Illustrative Example
Below, the stages of the presented rule induction approach are outlined to elucidate the functioning of this method. This explanation is confined to the specialization (growth) phase, as indicated in line 5 of Algorithm 1.
Consider a synthetic dataset with two symbolic attributes . Attribute a can take three values: . Attribute b can also take three values: . Survival curves corresponding to these attribute values are plotted in Figure 2, showing the survival rates of different groups.
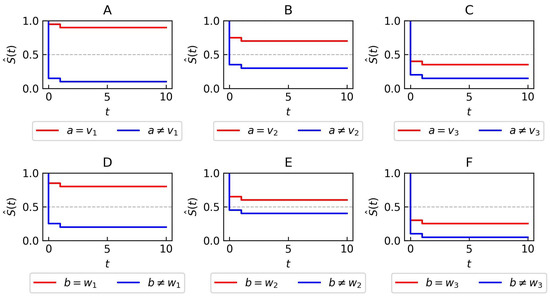
Figure 2.
Synthetic data—Kaplan–Meier plots for two attributes a and b used in the illustrative example of rule generation. Each subfigure presents two distinct curves. The red curve corresponds to data points that have either attribute a (A–C) or b (D–F) set to a particular value. The blue curve represents all other data points that don’t fit the criteria specified for the red curve (A–F).
The specialization of the survival action rule is given in line 5 of Algorithm 1. For the synthetic data, this task is performed by the following steps, performed iteratively in a loop:
- The initial step involves searching for the best elementary condition to add to the left side of the rule (line 5 in Algorithm 2). In the first iteration, the condition achieves the highest score. This condition is considered the best because it has the largest log-rank test value when comparing the survival curve where with the curve where (panel A in Figure 2).
- The identified elementary condition is then added to the set of previously tested conditions (line 6 in Algorithm 2).
- The best counter-condition is searched for (line 9 in Algorithm 2). In the first iteration, condition achieves the highest score. This condition is considered the best because it has the largest log-rank test value when comparing the survival curve where with the curve where (panels A and C in Figure 2).
- An action , where denotes the Kaplan–Meier estimate for examples that fulfill the condition , is created from the condition and the counter-condition (line 10 in Algorithm 2). This action is then added to the premise of the rule (line 11 in Algorithm 2).
The second loop iteration is performed as follows:
- The initial step involves searching for the best elementary condition to add to the left side of the rule (line 5 in Algorithm 2). In the second iteration, the condition achieves the highest score. This condition is considered the best because it has the largest log-rank test value when comparing the survival curve where with the curve where (panel B in Figure 3).
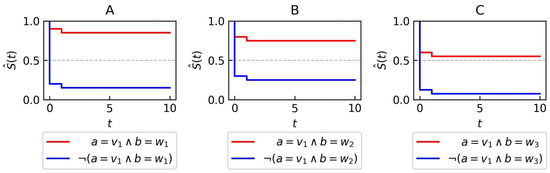 Figure 3. Syntheticdata—Kaplan–Meier plots for two attributes a and b used in the illustrative example of rule generation. Each subfigure presents two distinct curves. The red curve corresponds to data points that have attribute a set to the value and b set to value (A), (B), or (C). The blue curve represents all other data points that don’t fit the criteria specified for the red curve (A–C).
Figure 3. Syntheticdata—Kaplan–Meier plots for two attributes a and b used in the illustrative example of rule generation. Each subfigure presents two distinct curves. The red curve corresponds to data points that have attribute a set to the value and b set to value (A), (B), or (C). The blue curve represents all other data points that don’t fit the criteria specified for the red curve (A–C). - The identified elementary condition is then added to the set of conditions previously considered (line 6 in Algorithm 2).
- The best counter-condition is searched for (line 9 in Algorithm 2). In the second iteration, the condition achieves the highest score. This condition is considered the best because it has the largest log-rank test value when comparing the survival curve where with the curve where (panel B in Figure 4).
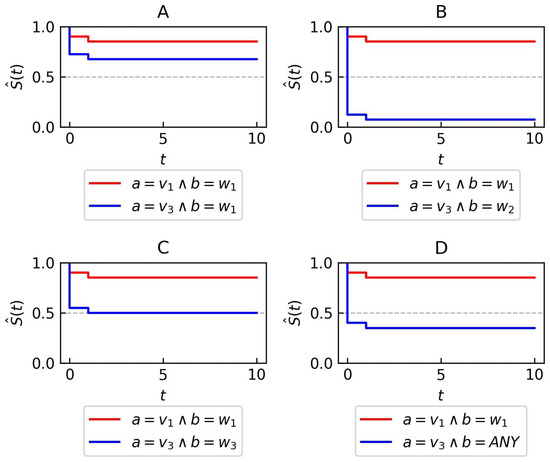 Figure 4. Syntheticdata—Kaplan–Meier plots for two attributes a and b used in the illustrative example of rule generation. Each subfigure presents two distinct curves. The red curve corresponds to data points that have attribute a set to the value and b set to value (A–D). The blue curve corresponds to data points that have attribute a set to the value and b set to value (A), (B), (C) or (D) value.
Figure 4. Syntheticdata—Kaplan–Meier plots for two attributes a and b used in the illustrative example of rule generation. Each subfigure presents two distinct curves. The red curve corresponds to data points that have attribute a set to the value and b set to value (A–D). The blue curve corresponds to data points that have attribute a set to the value and b set to value (A), (B), (C) or (D) value. - An action is created from the condition and counter-condition (line 10 in Algorithm 2). This action is then added to the premise of the rule (line 11 in Algorithm 2).
The result of the specialization phase is a rule of the form
3.3. Recommendation Induction
The newly discovered knowledge is expressed by an induced set of survival action rules (). These rules show that by changing certain attribute values, as described in the rule’s premise, there is a consequential shift in the survival curve. This shift is observable as the survival curve transitions from its original state, denoted by , to a new state, denoted by .
However, an example x, especially a new (unseen) example, may be covered by multiple survival action rules from . In such a case, the challenge is to determine which specific actions are necessary to significantly change the survival rate for x.
To address this issue, a unique type of rule is derived from a special dataset. This dataset, known as the “meta-example set” or “meta-table”, is formulated based on the survival action rules already induced in . In scenarios where multiple recommendations are required, it is possible to extract several different rules from this dataset to meet these specific requirements.
3.3.1. Meta-Table
Suppose there is a set of examples. Each example is characterized by a set of attributes , an observation time T, and a survival status .
Suppose a set of survival action rules is induced from a .
The meta-table is a set of meta-examples, where each meta-example is described by a set of meta-attributes , where .
If does not occur in any of the elementary actions in , then the attribute corresponding to a is not a member of the set .
If an attribute is of the symbolic type and occurs in at least one elementary action in , then .
If an attribute is of numeric type and occurs in at least one elementary action in , then the values for the corresponding meta-attribute are defined as follows. Let denote a set of all the source rules (3) and —a set of all the target rules. Suppose that the sets and contain all elementary conditions from and , respectively, and are built on the attribute a. The set contains all elementary conditions that are based on a. Each of these conditions is of the form or , where v is a particular value of a. To determine the value set for , all such values v occurring in are sorted. The values of the meta-attribute are then defined as sequential identifiers representing each interval created by this sorting process.
Observation time T and survival status are not defined in the meta-table because they are not needed for further consideration.
Example 2.

Let us assume that two attributes a (symbolic) and b (numeric) are given.
Consider a set of tree survival action rules:
From these rules, we can construct the following sets of elementary conditions:
In , there are only two values of b, so the set of values of is . From , we obtain the following partition of the set of values of a:
where and are the minimum and maximum values of a. The partition is a sequence of intervals and defines the set of values of , which consists of five values. Each value corresponds to one of the intervals in the partition (12). For example, the interval is assigned to the value 3 within because it is the third element in a sequence of intervals.
A meta-table is defined as the Cartesian product of the value sets of and , as shown in Table 1.

Table 1.
An exemplary meta-table. The table columns display examples from the metatable, along with the values these examples have for the attributes and .
Let us consider an example and . This example is covered by the meta-example .
In general, if a meta-table has n attributes, each attribute ( has values, then the meta-table has examples.
3.3.2. Final Recommendation
The final recommendation is generated by applying a customized version of the survival rule induction algorithm (Algorithm 1) to the meta-table. This ensures that the method can be applied to any previously unseen example due to the structure of the meta-table. The algorithm works on a single test example x associated with a survival curve . There are two methods for constructing a KM curve to characterize test examples. The first method uses the meta-table, and the second method uses an arbiter model. The principle of the method is illustrated in the Figure 5.
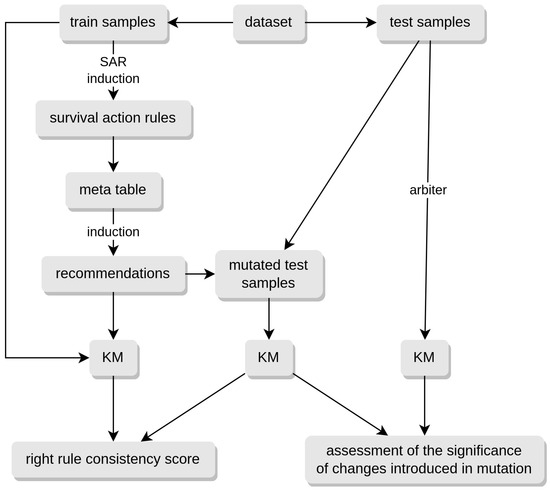
Figure 5.
Diagram illustrating principle of the method.
In the first case, the survival curve is constructed from training examples using a meta-table. Each meta-example is covered by several examples from the initial (training) set of examples , while each example from this set is covered by only a single meta-example. To define , a meta-example is identified that covers the example x. Finally, the Kaplan–Meier estimator () is defined based on all training examples from D that are covered by .
In some cases, the number of training examples covering may be small. This, in turn, affects the confidence in the estimated curve . In such a case, the arbiter model method is used to associate x with . The arbiter model is a survival data model trained on the entire set of training examples D. In the article, an implementation of gradient boosting with a regression tree-based learner [25] is used as the arbiter model. The trained arbiter model generates the survival curve associated with the example x.
Regardless of the method used, the survival curve is treated as the conclusion of a hypothetical rule (3) covering the test example x.
The recommendation process involves suggesting specific changes to the attribute values of example x. The aim of these changes is to align the test example with a meta-example (or multiple meta-examples) that will move the test example from its current Kaplan–Meier curve, denoted as , to a different target Kaplan–Meier curve, . The algorithm aims to minimize the number of attribute changes required for this reassignment. The range of values into which a test example is carried is determined by more than one meta-example as long as the ranges of attribute values in these meta-examples are adjacent and together produce a curve of better quality than any of them individually. The process described above is equivalent to the induction of an (4) rule as described in Algorithms 1 and 2. In the induction process, the algorithm searches for a rule built on a set of meta-examples whose conclusion differs the most from the survival curve . To determine the degree of difference, the two curves are compared using the Kolmogorov–Smirnov test with two samples [60].
Example 3.
Let us assume that the induction of recommendations is conducted in the meta-table presented in the previous example and that a test example x = is given. The example x covers the meta-example . The survival curve associated with x is determined based on all examples from the training set that covers the meta-example . Let us further assume that the recommendation algorithm suggests only one action . This recommendation implies that there is no need to change the value of the attribute (b). The value represents the interval . The recommendation is, therefore, to change the value of the attribute a from its current value of 10 to a new value within the interval , since in the meta-table corresponds to in the training set.
4. Experiments and Results
The experiments were conducted on 19 datasets in a 10-fold stratified cross-validation mode. The detailed characteristics of the datasets are presented in Table 2. The minimum number of examples that the new rule must cover was set to 30%, and the maximum percentage of examples that can be covered by both the source and target rules was set to 10%. For simplicity and due to the large number of datasets, all attributes were treated as flexible. An extensive case study was carried out for two scenarios. The first involves the application of our method to a specific dataset. In this study, it was assumed that the values of all attributes can be changed (i.e., no stable attributes—whose values cannot be changed—were defined in the datasets). The second case study involves a comparative analysis between the recommendations of our method and those of a counterfactual explanation approach using a specific test example.

Table 2.
Description of datasets employed in experimental research. Columns are as follows: the dataset name and source, the number of examples (), the number of conditional attributes (), the percentage of missing values (Miss), and the percentage of censored observations in the set (Cens).
The quality of the induced rules and recommendations was assessed comprehensively. Table 3 presents the quantitative characteristics of the induced rule sets. The column () shows the percentage of statistically significant induced rules. A rule is considered statistically significant if the difference between its source and target Kaplan–Meier curves is statistically significant, indicating that the rule suggests changes that could contribute to real changes in the survival of the examples covered by its premise. The log-rank test was used for this comparison.

Table 3.
Rules statistics. The columns represent the average values of the following parameters: the number of rules (); the number of actions in a rule (); the number of ANY actions in a rule (); the percentage of rules with a p-value less than 0.05 (); the percentage of rules with a p-value less than 0.01 (); the percentage of examples covered by the source rule (); the percentage of examples covered by the target rule (); the number of BETTER rules (); the number of WORSE rules ().
The analysis of Table 3 shows that the number of action rules generated is not large, averaging about 8 rules. For most datasets, the number of BETTER rules generated is greater than the number of WORSE rules, meaning that more rules define conditions that lead to an increase in the average survival time. On average, the rules contain 3 actions, with 2 to 3 of these actions being different from ANY actions, suggesting specific changes in attribute values. There is a notable variation in the coverage of source and target rules across the datasets. For example, actg320 shows a significant difference in coverage between source and target (22.48% vs. 39.29%), indicating a substantial extension of the applicability of the rules from source to target.
For the two datasets, “std” and “lung”, the average number of “WORSE” type rules exceeded the number of “BETTER” type rules. WORSE type rules give information about what actions to pay attention to in order not to worsen survival time. Depending on the needs, the algorithm can be parameterized to obtain only BETTER type rules, only WORSE type rules, or rules of any type, as in the considered scenario.
It is noteworthy that the vast majority of rules (more than on average) are statistically significant at both the 0.05 and 0.01 significance levels. The exception here is the zinc set, where only half of the induced rules are significant at the 0.01 level and at the 0.05 level.
Table 4 shows the predictive capacity of the recommendation algorithm. To measure this feature, an arbiter model is used to test whether performing the actions suggested by the recommendation algorithm for a given example causes a significant change in the estimated survival time. The arbiter model chosen is the XGBoost algorithm, which is suitable for the analysis of censored data. In the literature, the XGBoost algorithm is reported to have high predictive performance in solving classification and regression problems and in survival analysis [73]. The model in our study is based on gradient-boosted Cox proportional hazard loss, with regression trees as base learners, and is available in the scikit-survival Python package. The validation procedure is as follows:

Table 4.
Predictive capabilities of the recommendation algorithm. Columns Cs represent the average values of the consistency score at different significance levels.
- The XGBoost model is trained on the training data.
- For each test example x, the survival curve is determined, which represents the conclusion of the recommendation.
- The example is passed to the trained XGBoost model with attribute values modified according to the recommendation algorithm.
- The XGBoost model assigns the survival curve to the example .
- The difference between the curves and is examined; if the curves are not significantly different, then the rule action model is assumed to be working correctly.
The validation process presented above is called the consistency score (column in Table 4). The column shows the average percentage of examples for which the p-value is less than 0.05, testing the null hypothesis that the curve for the target part of the recommendation is identical to the curve derived from the arbiter model for examples with mutated attribute values.
The log-rank test was found to be inappropriate for the comparison because it requires raw survival data. Therefore, an alternative approach was taken using the two-sample Kolmogorov–Smirnov test (two-sample K–S test) [74]. This method was chosen specifically for cases where at least one of the survival curves being compared is derived from an arbiter model using a single test example, resulting in the output of callable step functions. These step functions, although not compatible with the analysis criteria of the log-rank test, are effectively analyzed using the two-sample K–S test.
The two-sample K–S test evaluates the maximum distance between the cumulative distribution functions (CDFs) of two samples. The test statistic is defined as:
where and are empirical distribution functions of the first and second samples, with n and m observations, respectively, and represents the supremum of the set of distances over all possible values of x. In the context of survival analysis, this approach allows a non-parametric comparison of the empirical survival distributions.
The quantitative analysis of the determined rules (Table 3) has shown that most of them are statistically significant. The recommendation algorithm is based on the set of induced rules. The datasets used do not allow for verifying whether the value changes suggested by the recommendation algorithm lead to an increase/decrease in survival time for test examples. This verification would be possible if it were possible to “track the fate” of a given test example over time. Therefore, an independent arbiter model was used to verify whether an alternative model for estimating survival time would be consistent with the estimation made by the recommendation algorithm.
The results of this verification are presented in Table 4. As can be seen, there is a high correspondence between the recommendation algorithm and the XGBoost algorithm running on mutated test examples. It is important to note that the recommendation algorithm and the XGBoost algorithm do not necessarily base their decisions on the same set of attributes. For example, in an extreme case, the recommendation algorithm might suggest changing the values of attributes a and b, while the XGBoost-based model uses a different set of features. In such a case, there will be a discrepancy between the decisions of the recommendation algorithm and the XGBoost algorithm. For the XGBoost algorithm, an example before changing the attribute values—where only attributes a and b are modified—is "identical" to the example after the change. This situation occurred in some examples from the FD001, FD003, and maintenance datasets. These discrepancies do not necessarily imply that one of these algorithms is wrong; they simply make decisions in different ways.
Table 5 presents the results, showing shows the average number of actions suggested by the recommendation algorithm and the percentage of conditional attributes for which actions were defined.

Table 5.
Attribute statistics for the generated recommendations. The columns show the following information: the average number of actions per recommendation () and the average percentage of attributes that were mutated ().
To conclude this part of the analysis, Table 5 presents information on the average number of elementary actions required—the values in Table 5 represent the result illustrating the operation of the recommendation algorithm that merges information from all the induced rules. As observed, the recommendation algorithm forces the execution of an average of two elementary actions for each example. This means that the algorithm suggests changes in the values of two attributes. The column shows the percentage of all attributes that are involved in the elementary actions suggested by the recommendation algorithm. On average, of the attributes are involved. Specifically, for the std set, half of the attributes are involved, and for the zinc set, just under of the attributes are involved. These statistics show that the induced action rules and the recommendation algorithm based on them also lead to a significant reduction in the number of features whose values need to be changed.
4.1. Case Studies
The case study examined two scenarios. The first scenario focused on the application of our method to the maintenance dataset. In the second scenario, a comparative analysis was performed, contrasting the recommendations from our approach with those derived from a counterfactual explanation method, using a selected test example from the same dataset.
4.1.1. Maintenance Dataset
The maintenance dataset is available as part of the popular Python package, PySurvival [68]. Its attributes are well defined, so the values are easy to understand. Most of the conditional attributes in this dataset are of the flexible type, so they can be subjected to actions. The maintenance dataset contains five conditional attributes and two decision attributes:
- pressureInd
- moistureInd
- temperatureInd
- provider
- team
- time
- status
The pressure index (pressureInd) is used to quantify the flow of liquid through pipes, as a sudden drop in pressure may indicate a leak. The moisture index (moistureInd) is a measure of the relative humidity in the air. It is important to monitor this as excessive humidity can lead to mold and equipment damage. The temperature index (temperatureInd) of the machine is calculated using voltage devices called thermocouples, which convert a change in voltage into a measure of temperature. It is recorded to prevent damage to electrical circuits, fire, or even explosion. The team (team) and machine indicator (provider) specify which team was using the machine and the name of the machine manufacturer. These two attributes have been omitted to simplify the analysis. Survival time (time) indicates the number of weeks the machine has been active. Survival status (status) specifies whether the machine was broken or remained intact during the corresponding weeks of activity.
This experiment was designed to see if the generated rules would show actions that would generate recommendations to change the working conditions of the machine to increase the length of the activity period.
It was assumed that the values of all attributes could be changed (i.e., no stable attributes were defined in the dataset). The experiments were run in a 10-fold stratified cross-validation mode. For the case study, the first fold was selected.
Table 6 provides a summary of the results from the experiment involving the induction of survival action rules. Chosen metrics representing the quality of the rule sets across different versions of the experiment are displayed. Rule induction was conducted in ANY mode, where both types of rules—improving and worsening—can be formed. The experiment produced a relatively large number of rules—23. Each rule had at least one action, with the majority having three actions. The absence of differences between the average number of conditions () and the average number of actions () suggests that no supporting actions appeared in the rules. For every rule, the p-value resulting from the log-rank test comparing the Kaplan–Meier curves of the left and right sides was below . Only one rule had a p-value greater than .

Table 6.
Characteristics of the induced survival action rules.
To present the sample results of the experiment, the selected rules obtained are presented in Table 7. They give information on what to focus on when maintaining the machine so that its active period does not decrease. The table shows only the premise part of the rules.

Table 7.
Selected survival action rules for the maintenance dataset.

Table 8.
Characteristics of the selected action rules presented in Table 7. The columns indicate the following: rule identifier (id), the count of conditions (), the count of actions (), and the count of any actions (), the coverage percentage of examples by either the left or right rule (/), p-value of the log-rank test comparing the Kaplan–Meier curves of the left and right rules (p), and the median survival times for examples under the left or right rule (/), the difference in median survival times between the left and right rules ().
- number of conditions,
- number of actions,
- the percentage of all examples that the left part of the rule covers,
- the percentage of all examples that the right part of the rule covers,
- p-value of the log-rank test between the Kaplan–Meier curves of the left and right sides of the rule,
- median survival time for examples covered by the left rule (time for probability = ),
- median survival time for examples covered by the right rule,
- difference between the median survival times of the left side of the rule and the right side of the rule.
An important measure is the p-value of the log-rank test between the Kaplan–Meier curves of the left and right sides of the rule. The lower the value of this measure, the further apart the two curves are.
A meta-table was created based on all the generated rules, and recommendations were generated for all test examples. Selected recommendations are shown in Table 9.

Table 9.
Selected recommendations for examples from the maintenance dataset.
4.1.2. Comparative Study
The second case study involves a comparative analysis between the recommendations of our method and those of a counterfactual explanation approach [3]. This comparison encompasses a thorough evaluation using the maintenance dataset in cross-validation mode. Additionally, an in-depth comparison is provided for a particular test example x from the same dataset, as shown in Table 10.

Table 10.
Characteristics of the test example x.
Dataset
Both counterfactual and recommendation methods were applied to the entire maintenance dataset. Each experiment was conducted using a 10-fold stratified cross-validation approach. With a total of 1000 samples in the dataset, the training set consisted of 900 samples, and the test set consisted of 100 samples for each fold. Subsequent analyses were performed on a combined total of 1000 test samples from all folds.
The recommendation algorithm was applied using the same parameters as in the previous experiments, specifically setting the minimum number of examples covered by a single rule to 30. The counterfactual explanation method was applied for both values of and 1, with the Random Survival Forest model selected for analysis. This resulted in three sets of results: one from the recommendation algorithm, one from the counterfactual explanations with , and one from the counterfactual explanations with .
For each test sample, a survival curve was constructed using the Random Survival Forest method. Similarly, for each recommendation, a mutated sample was obtained, and a survival curve was created using the same method. These curves were then compared using the two-sample K–S test, which provided a pair of results: the p-value and the test statistic. Small p-values indicate that the survival curve of the test sample is different from the survival curve of the test sample mutated according to the recommendation, which means that the change applied by the method is significant.
In this experimental setup, our goal was to obtain a p-value below the significance threshold. A test yielding such a result indicates a significant difference between the survival curve of the original test sample and the curve of the modified test sample, based on the recommendation. This indicates that the recommended change is substantial.
The results are presented in Figure 6. The histograms show a predominance of lower p-values for all methods. Detailed results in the form of a share of recommendations that achieved a p-value lower than are presented in Table 11. Better results were achieved for the counterfactual method, but the computation time was significantly longer, as shown in Table 12.
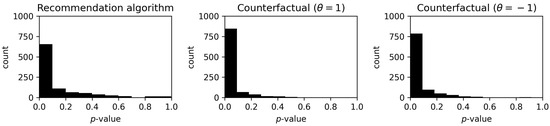
Figure 6.
Distribution of p-values for the recommendation algorithm and counterfactual methods. The left panel shows the p-value frequency for the recommendation algorithm, the middle panel displays the distribution for the counterfactual method with the parameter set to 1, and the right panel presents the distribution for counterfactual method with parameter set to . The x-axis represents the p-value range from to , and the y-axis shows the count of recommendations.

Table 11.
Comparison of methods based on the significance of p-value. The column represents the percentage of recommendations with a p-value less than 0.05.

Table 12.
Comparison of methods based on execution time.
Detailed Example
The example has been mutated based on the recommendation from Table 13, resulting in its modified version—Table 14.

Table 13.
Recommendation for the example x.

Table 14.
Example x after mutation—recommendation algorithm.
For the example x, a counterexample was generated using the method described in [3,24]. The running parameters were maintained at default values, except for the parameter , which determines whether the survival curve for the counterfactual should be improving or worsening relative to that of the initial example. The parameter was set to 1, corresponding to the improving scenario. A counterfactual generated for the example x is shown in Table 15.

Table 15.
Example x after mutation—a counterfactual explanation.
In the example being examined, the recommendation algorithm suggests changing the value of only one attribute—moistureInd. The counterfactual explanations algorithm forces the values of all three conditional attributes to change. In both cases, implementing the suggested changes results in an increase in the estimated survival time, but the increase is greater for the recommendation algorithm as shown in Figure 7.
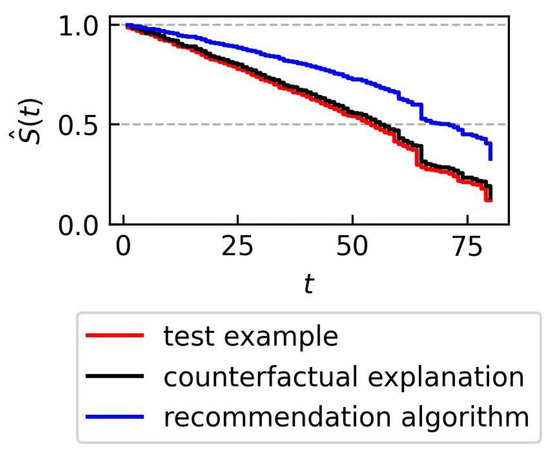
Figure 7.
Kaplan–Meier plots for three samples: the original test example x, the example x after recommendation-based mutation, and a counterfactual for x.
5. Conclusions
The article presents a recommendation algorithm based on survival action rules. The recommendation algorithm is based on previously induced survival action rules. The motivation for the development of this algorithm arises from situations where decisions about changing the values of a specific example are complicated by the fact that the example is covered by numerous action rules. The recommendation algorithm, which relies on the induction of action rules in a specially prepared meta-table, addresses this issue. The meta-table represents a kind of discretization for continuous attributes, as defined by the previously determined action rules.
The experiments conducted show that the algorithm is a useful tool for generating recommendations. The presented algorithm is the first of its kind designed for censored data. Survival action rules and the recommendation algorithm based on them offer a slightly different solution than the counterfactual explanations (CF) methodology. To our knowledge, there are at least two implementations of CF dedicated to censored data. However, CF is a method that works locally to identify the minimal changes required for the estimated survival time of a test example to differ from the currently estimated time. The action rule induction algorithm generates a model of value changes for the entire training dataset, and then the recommendation algorithm generates change recommendations for new (or training) examples. The recommendation algorithm also looks for changes that cause the most significant change in the estimated survival time.
The algorithm is available in the GitHub repository (https://github.com/adaa-polsl/action-mining-based-on-sar, accessed on 26 March 2024) along with detailed results from the conducted experiments.
Our further work will focus on the development of an algorithm for the induction of exception rules [75] tailored to the task of action planning. Exception rules allow the identification of specific actions within induced action rules (source rules) whose application does not positively affect the reassignment of an example to the intended target class. The identification of such rules is expected to result in an action model that provides more precise recommendations. We also aim to integrate software that enables the induction of action rules with the RuleKit package [76] we are developing, further enhancing its capabilities.
Author Contributions
Conceptualization, M.S.; methodology, M.S. and M.H.; software, M.H.; validation, M.H. and M.S.; formal analysis, M.H.; investigation, M.H., B.S., M.S. and Ł.W.; resources, M.H. and B.S.; data curation, M.H. and B.S.; writing—original draft preparation, M.S., M.H. and B.S.; writing—review and editing, M.S., B.S. and Ł.W.; visualization, M.H.; supervision, M.S.; project administration, B.S. and Ł.W.; funding acquisition, Ł.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by the Polish National Centre for Research and Development, Poland, under the Operational Program Intelligent Development (grant POIR.01.01.01-00-0304/19). The publication was supported by the rector’s grant for scientific quality improvement. Silesian University of Technology, grant number: 02/120/RGJ23/0028 and 02/120/RGJ23/0027.
Data Availability Statement
The original data presented in the study are openly available. For dataset access and relevant literature, see Table 2.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| BMI | Body Mass Index |
| CF | counterfactual explanations |
| KM | Kaplan–Meier |
| RUL | survival action rules |
References
- Sikora, M.; Matyszok, P.; Wróbel, Ł. Scari: Separate and conquer algorithm for action rules and recommendations induction. Inf. Sci. 2022, 607, 849–868. [Google Scholar] [CrossRef]
- Badura, J.; Hermansa, M.; Kozielski, M.; Sikora, M.; Wróbel, Ł. Separate-and-conquer survival action rule learning. Knowl.-Based Syst. 2023, 280, 110981. [Google Scholar] [CrossRef]
- Kovalev, M.; Utkin, L.; Coolen, F.; Konstantinov, A. Counterfactual Explanation of Machine Learning Survival Models. Informatica 2021, 32, 817–847. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Clark, T.; Bradburn, M.; Love, S.; Altman, D. Survival Analysis Part I: Basic concepts and first analyses. Br. J. Cancer 2003, 89, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Bewick, V.; Cheek, L.; Ball, J. Statistics review: Survival analysis. Crit. Care 2004, 8, 1–6. [Google Scholar]
- Bradburn, M.; Clark, T.; Love, S.; Altman, D. Survival analysis Part II: Multivariate data analysis–an introduction to concepts and methods. Br. J. Cancer 2003, 89, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, E.; Meier, P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Harrington, D.; Fleming, T. A class of rank test procedures for censored survival data. Biometrika 1982, 69, 553–566. [Google Scholar] [CrossRef]
- Cox, D. Regression models and life-tables. J. R. Stat. Soc. Ser. B (Methodol.) 1972, 34, 187–220. [Google Scholar] [CrossRef]
- Schober, P.; Vetter, T. Survival analysis and interpretation of time-to-event data: The tortoise and the hare. Anesth. Analg. 2018, 127, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, Y.; Reddy, C.K. Machine learning for survival analysis: A survey. ACM Comput. Surv. (CSUR) 2019, 51, 1–36. [Google Scholar] [CrossRef]
- Segal, M. Regression trees for censored data. Biometrics 1988, 44, 35–47. [Google Scholar] [CrossRef]
- LeBlanc, M.; Crowley, J. Survival trees by goodness of split. J. Am. Stat. Assoc. 1993, 88, 457–467. [Google Scholar] [CrossRef]
- Bou-Hamad, I.; Larocque, D.; Ben-Ameur, H. A review of survival trees. Stat. Surv. 2011, 5, 44–71. [Google Scholar] [CrossRef]
- Wróbel, Ł.; Gudyś, A.; Sikora, M. Learning rule sets from survival data. BMC Bioinform. 2017, 18, 285. [Google Scholar] [CrossRef] [PubMed]
- Sikora, M.; Wróbel, Ł.; Gudyś, A. GuideR: A guided separate-and-conquer rule learning in classification, regression, and survival settings. Knowl.-Based Syst. 2019, 173, 1–14. [Google Scholar] [CrossRef]
- Van Belle, V.; Pelckmans, K.; Van Huffel, S.; Suykens, J. Improved performance on high-dimensional survival data by application of Survival-SVM. Bioinformatics 2011, 27, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Pölsterl, S.; Navab, N.; Katouzian, A. Fast training of support vector machines for survival analysis. In Machine Learning and Knowledge Discovery in Databases; Machine Learning and Knowledge Discovery in Databases. ECML PKDD 2015. Lecture Notes in Computer Science 9285; Springer International Publishing: Cham, Switzerland, 2015; pp. 243–259. [Google Scholar]
- Faraggi, D.; Simon, R. A neural network model for survival data. Stat. Med. 1995, 14, 73–82. [Google Scholar] [CrossRef]
- Ripley, R.; Harris, A.; Tarassenko, L. Non-linear survival analysis using neural networks. Stat. Med. 2004, 23, 825–842. [Google Scholar] [CrossRef]
- Štajduhar, I.; Dalbelo-Bašić, B.; Bogunović, N. Impact of censoring on learning bayesian networks in survival modelling. Artif. Intell. Med. 2009, 47, 199–217. [Google Scholar] [CrossRef]
- Štajduhar, I.; Dalbelo-Bašić, B. Learning bayesian networks from survival data using weighting censored instances. J. Biomed. Inform. 2010, 43, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Ishwaran, H.; Kogalur, U.B.; Blackstone, E.H.; Lauer, M.S. Random survival forests. Ann. Appl. Stat. 2008, 2, 841–860. [Google Scholar] [CrossRef]
- Hothorn, T.; Bühlmann, P.; Dudoit, S.; Molinaro, A.; Van Der Laan, M.J. Survival ensembles. Biostatistics 2005, 7, 355–373. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jia, Z.; Mercola, D.; Xie, X. A gradient boosting algorithm for survival analysis via direct optimization of concordance index. Comput. Math. Methods Med. 2013, 2013, 873595. [Google Scholar] [CrossRef] [PubMed]
- Shashikumar, S.P.; Josef, C.S.; Sharma, A.; Nemati, S. DeepAISE—An interpretable and recurrent neural survival model for early prediction of sepsis. Artif. Intell. Med. 2021, 113, 102036. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Fridgeirsson, E.; Wingen, G.V.; Welling, M. Transformer-Based Deep Survival Analysis. In Proceedings of the AAAI Spring Symposium on Survival Prediction—Algorithms, Challenges, and Applications 2021, Palo Alto, CA, USA, 22–24 March 2021; Volume 146, pp. 132–148. [Google Scholar]
- Fotso, S. Deep neural networks for survival analysis based on a multi-task framework. arXiv 2018, arXiv:1801.05512. [Google Scholar]
- Zhao, L.; Feng, D. Deep neural networks for survival analysis using pseudo values. IEEE J. Biomed. Health Inform. 2020, 24, 3308–3314. [Google Scholar] [CrossRef]
- Ras, Z.W.; Wieczorkowska, A. Action-rules: How to increase profit of a company. In Proceedings of the European Conference on Principles of Data Mining and Knowledge Discovery, Lyon, France, 13–16 September 2000; pp. 587–592. [Google Scholar]
- Tsay, L.S.; Raś, Z. Action rules discovery: System DEAR2, method and experiments. J. Exp. Theor. Artif. Intell. 2005, 17, 119–128. [Google Scholar] [CrossRef]
- Raś, Z.; Tsay, L.S. Mining E-action rules, System DEAR. In Intelligent Information Processing and Web Mining; Data Mining: Foundations and Practice. Studies in Computational Intelligence 118; Springer: Berlin/Heidelberg, Germany, 2008; pp. 289–298. [Google Scholar]
- Agrawal, R.; Srikant, R. Fast algorithms for mining association rules. In Proceedings of the 20th International Conference on Very Large Data Bases—VLDB 1994, Santiago de Chile, Chile, 12–15 September1994; pp. 487–499. [Google Scholar]
- He, Z.; Xu, X.; Deng, S.; Ma, R. Mining action rules from scratch. Expert Syst. Appl. 2005, 29, 691–699. [Google Scholar] [CrossRef]
- Im, S.; Raś, Z. Action rule extraction from a decision table: ARED. In Foundations of Intelligent Systems, Proceedings of the ISMIS 2008, Cosenza, Italy, 3–5 October 2008; Lecture Notes in Computer Science 4994; Springer: Berlin/Heidelberg, Germany, 2008; pp. 160–168. [Google Scholar]
- Matyszok, P.; Wróbel, Ł.; Sikora, M. Bidirectional action rule learning. In Computer and Information Sciences, Proceedings of the ISCIS 2018, Poznan, Poland, 20–21 September 2018; Communications in Computer and Information Science 935; Springer: Cham, Switzerland, 2018; pp. 220–228. [Google Scholar]
- Raś, Z.; Dardzińska, A. Action rules discovery without pre-existing classification rules. In Rough Sets and Current Trends in Computing, RSCTC 2008, Akron, OH, USA, 23–25 October 2008; Lecture Notes in Computer Science 5306; Springer: Berlin/Heidelberg, Germany, 2008; pp. 181–190. [Google Scholar]
- Daly, G.; Benton, R.; Johnsten, T. A multi-objective evolutionary action rule mining method. In Proceedings of the 2018 IEEE Congress on Evolutionary Computation (CEC), Rio de Janeiro, Brazil, 8–13 July 2018. [Google Scholar]
- Hashemi, S.; Shamsinejad, P. Ga2rm: A ga-based action rule mining method. Int. J. Comput. Intell. Appl. 2021, 20, 2150012. [Google Scholar] [CrossRef]
- Bagavathi, A.; Tripathi, A.; Tzacheva, A.A.; Ras, Z.W. Actionable pattern mining-a scalable data distribution method based on information granules. In Proceedings of the 17th IEEE International Conference on Machine Learning and Applications (ICMLA), Orlando, FL, USA, 17–20 December 2018; pp. 32–39. [Google Scholar]
- Benedict, A.C.; Ras, Z.W. Distributed Action-Rule Discovery Based on Attribute Correlation and Vertical Data Partitioning. Appl. Sci. 2024, 14, 1270. [Google Scholar] [CrossRef]
- Tarnowska, K.A.; Bagavathi, A.; Ras, Z.W. High-Performance Actionable Knowledge Miner for Boosting Business Revenue. Appl. Sci. 2022, 12, 12393. [Google Scholar] [CrossRef]
- Yang, Q.; Yin, J.; Ling, C.; Pan, R. Extracting actionable knowledge from decision trees. IEEE Trans. Knowl. Data Eng. 2007, 19, 43–56. [Google Scholar] [CrossRef]
- Subramani, S.; Wang, H.; Balasubramaniam, S.; Zhou, R.; Ma, J.; Zhang, Y.; Whittaker, F.; Zhao, Y.; Rangarajan, S. Mining actionable knowledge using reordering based diversified actionable decision trees. In Proceedings of the International Conference on Web Information Systems Engineering, Shanghai, China, 8–10 November 2016; pp. 553–560. [Google Scholar]
- Su, P.; Yang, J.; Li, Z.; Liu, Y. Mining actionable behavioral rules based on decision tree classifier. In Proceedings of the 13th International Conference on Semantics, Knowledge and Grids (SKG), Beijing, China, 14–15 August 2017; pp. 139–143. [Google Scholar]
- Cui, Z.; Chen, W.; He, Y.; Chen, Y. Optimal action extraction for random forests and boosted trees. In Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Sydney, Australia, 10–13 August 2015; pp. 179–188. [Google Scholar]
- Tolomei, G.; Silvestri, F. Generating actionable interpretations from ensembles of decision trees. IEEE Trans. Knowl. Data Eng. 2019, 33, 1540–1553. [Google Scholar] [CrossRef]
- Greco, S.; Matarazzo, B.; Pappalardo, N.; Słowinski, R. Measuring expected effects of interventions based on decision rules. J. Exp. Theor. Artif. Intell. 2005, 17, 103–118. [Google Scholar] [CrossRef]
- Słowiński, R.; Greco, S. Measuring attractiveness of rules from the viewpoint of knowledge representation, prediction and efficiency of intervention. In Proceedings of the International Atlantic Web Intelligence Conference, Lodz, Poland, 6–9 June 2005; pp. 11–22. [Google Scholar]
- Wachter, S.; Mittelstadt, B.; Russell, C. Counterfactual Explanations without Opening the Black Box: Automated Decisions and the GDPR (6 October 2017). Harv. J. Law Technol. 2018, 31, 841–887. [Google Scholar]
- Dandl, S.; Molnar, C.; Binder, M.; Bischl, B. Multi-objective counterfactual explanations. In Proceedings of the International Conference on Parallel Problem Solving from Nature, Leiden, The Netherlands, 5–9 September 2020; pp. 448–469. [Google Scholar]
- Guidotti, R. Counterfactual explanations and how to find them: Literature review and benchmarking. Data Min. Knowl. Discov. 2022. [Google Scholar] [CrossRef]
- Verma, S.; Boonsanong, V.; Hoang, M.; Hines, K.; Dickerson, J.; Shah, C. Counterfactual Explanations and Algorithmic Recourses for Machine Learning: A Review. arXiv 2022, arXiv:2010.10596v3. [Google Scholar]
- Kovalev, M.S.; Utkin, L.V.; Kasimov, E.M. SurvLIME: A method for explaining machine learning survival models. Knowl.-Based Syst. 2020, 203, 106164. [Google Scholar] [CrossRef]
- Chapfuwa, P.; Assaad, S.; Zeng, S.; Pencina, M.J.; Carin, L.; Henao, R. Enabling Counterfactual Survival Analysis with Balanced Representations. In Proceedings of the ACM Conference on Health, Inference, and Learning, New York, NY, USA, 30 March 2021. [Google Scholar]
- Leung, K.M.; Elashoff, R.; Afifi, A. Censoring issues in survival analysis. Annu. Rev. Public Health 1997, 18, 83–104. [Google Scholar] [CrossRef]
- Stevenson, M. An Introduction to Survival Analysis; EpiCentre, IVABS, Massey University: Palmerston North, New Zealand, 2007. [Google Scholar]
- Wohlrab, L.; Fürnkranz, J. A review and comparison of strategies for han1050 dling missing values in separate-and-conquer rule learning. J. Intell. Inf. Syst. 2011, 36, 73–98. [Google Scholar] [CrossRef]
- Hodges, J., Jr. The significance probability of the Smirnov two-sample test. Arkiv Mat. 1958, 3, 469–486. [Google Scholar] [CrossRef]
- Saxena, A.; Goebel, K.F.; Simon, D.L.; Eklund, N.H.W. Damage propagation modeling for aircraft engine run-to-failure simulation. In Proceedings of the International Conference on Prognostics and Health Management, Denver, CO, USA, 6–9 October 2008; pp. 1–9. [Google Scholar]
- Schumacher, M.; Bastert, G.; Bojar, H.; Hübner, K.; Olschewski, M.; Sauerbrei, W.; Schmoor, C.; Beyerle, C.; Neumann, R.; Rauschecker, H.; et al. Randomized 2 x 2 trial evaluating hormonal treatment and the duration of chemotherapy in node-positive breast cancer patients. German Breast Cancer Study Group. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 1994, 12, 2086. [Google Scholar] [CrossRef] [PubMed]
- Andersen, P.K.; Borgan, O.; Gill, R.D.; Keiding, N. Statistical Models Based on Counting Processes; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Hosmer, D.W.; Lemeshow, S.; May, S. Applied Survival Analysis: Regression Modeling of Time to Event Data; Wiley-Interscience: Hoboken, NJ, USA, 2008. [Google Scholar]
- Kałwak, K.; Porwolik, J.; Mielcarek, M.; Gorczyńska, E.; Owoc-Lempach, J.; Ussowicz, M.; Dyla, A.; Musiał, J.; Paździor, D.; Turkiewicz, D.; et al. Higher CD34(+) and CD3(+) cell doses in the graft promote long-term survival, and have no impact on the incidence of severe acute or chronic graft-versus-host disease after in vivo T cell-depleted unrelated donor hematopoietic stem cell transplantation in children. Biol. Blood Marrow Transpl. 2010, 16, 1388–1401. [Google Scholar] [CrossRef]
- Pintilie, M. Competing Risks: A Practical Perspective; John Wiley & Sons: Hoboken, NJ, USA, 2006; Volume 58. [Google Scholar]
- Lange, N.; Ryan, L.; Billard, L.; Brillinger, D.; Conquest, L.; Greenhouse, J. Case Studies in Biometry; Wiley Series in Probability and Mathematical Statistics: Applied Probability And Statistics; Wiley: Hoboken, NJ, USA, 1994. [Google Scholar]
- Fotso, S. PySurvival: Open Source Package for Survival Analysis Modeling. 2019. Available online: https://square.github.io/pysurvival (accessed on 26 March 2024).
- Kyle, R.A. ”Benign” monoclonal gammopathy—After 20 to 35 years of follow-up. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 1993; Volume 68, pp. 26–36. [Google Scholar]
- Fleming, T.R.; Harrington, D.P. Counting Processes and Survival Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2011; Volume 169. [Google Scholar]
- Klein, J.P.; Moeschberger, M.L. Survival Analysis: Techniques for Censored and Truncated Data; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Abnet, C.C.; Lai, B.; Qiao, Y.L.; Vogt, S.; Luo, X.M.; Taylor, P.R.; Dong, Z.W.; Mark, S.D.; Dawsey, S.M. Zinc concentration in esophageal biopsy specimens measured by x-ray fluorescence and esophageal cancer risk. J. Natl. Cancer Inst. 2005, 97, 301–306. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Pratt, J.W.; Gibbons, J.D. Kolmogorov-Smirnov Two-Sample Tests. In Concepts of Nonparametric Theory; Springer: New York, NY, USA, 1981; pp. 318–344. [Google Scholar] [CrossRef]
- Suzuki, E.; Żytkow, J.M. Unified algorithm for undirected discovery of exception rules. Int. J. Intell. Syst. 2005, 20, 673–691. [Google Scholar] [CrossRef]
- Gudyś, A.; Sikora, M.; Wróbel, Ł. RuleKit: A comprehensive suite for rule-based learning. Knowl.-Based Syst. 2020, 194, 105480. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).