Finite Element Modeling of Cells Adhering to a Substrate: An Overview
Abstract
1. Introduction
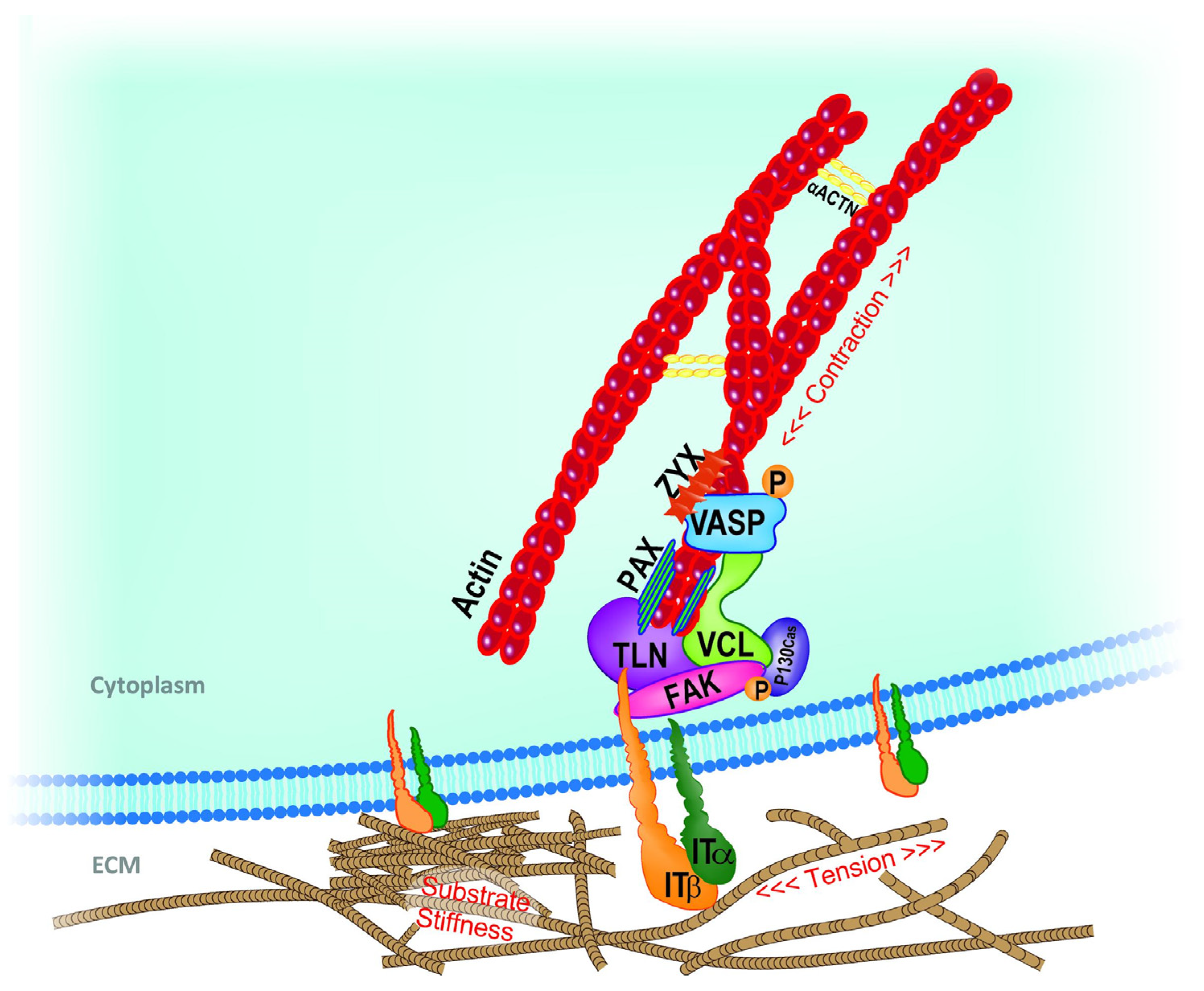
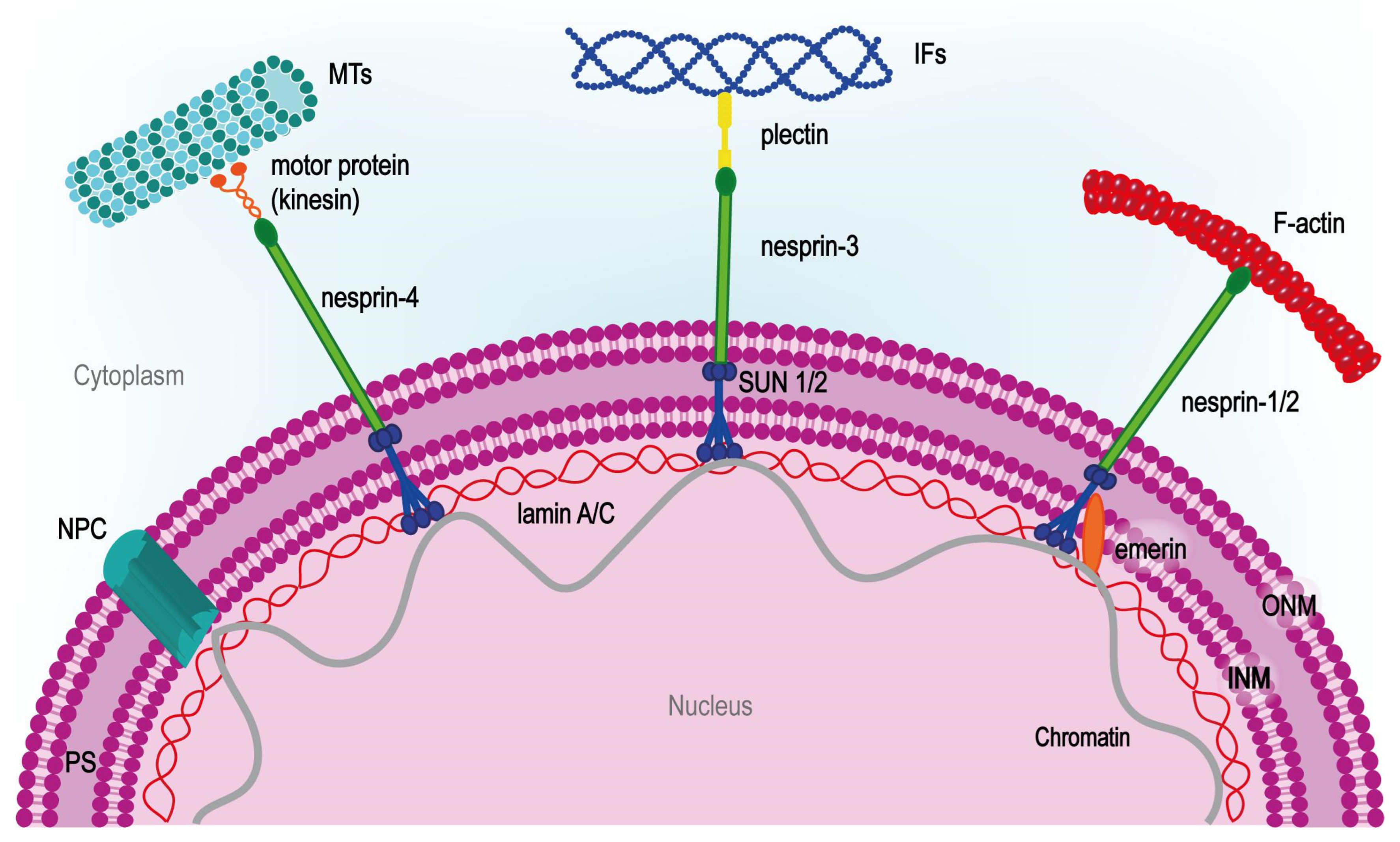
1.1. Theoretical Background (Principles) of Cell Adhesion and Mechanotransduction Mechanism
1.2. Finite Element Modeling (FEM)
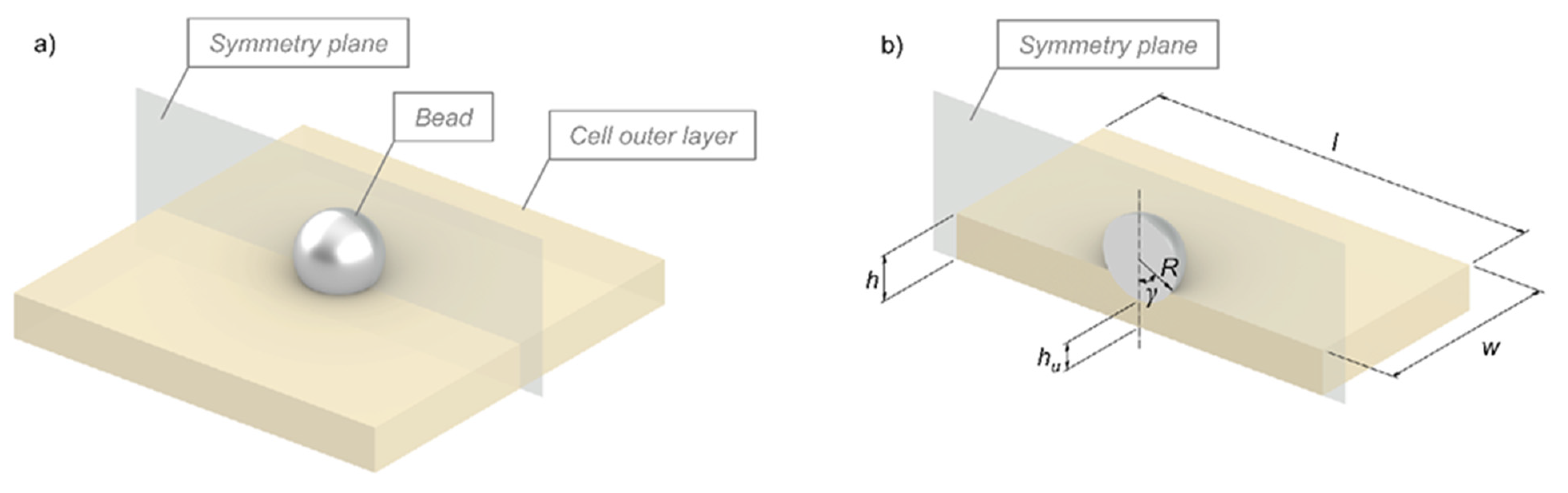
2. Physical Modeling of Cells in Adhesion Phenomena
3. Discussion
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Guilak, F.; Cohen, D.M.; Estes, B.T.; Gimble, J.M.; Liedtke, W.; Chen, C.S. Control of Stem Cell Fate by Physical Interactions with the Extracellular Matrix. Cell Stem Cell 2009, 5, 17–26. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Schwarz, U.S. A Kinetic Model for RNA-Interference of Focal Adhesions. BMC Syst. Biol. 2013, 7, 2. [Google Scholar] [CrossRef]
- Milan, J.-L.; Lavenus, S.; Pilet, P.; Louarn, G.; Wendling, S.; Heymann, D.; Layrolle, P.; Chabrand, P. Computational Model Combined with in Vitro Experiments to Analyse Mechanotransduction during Mesenchymal Stem Cell Adhesion. Eur. Cells Mater. 2012, 25, 97–113. [Google Scholar] [CrossRef]
- Paglia, E.B.; Baldin, E.K.K.; Freitas, G.P.; Santiago, T.S.A.; Neto, J.B.M.R.; Silva, J.V.L.; Carvalho, H.F.; Beppu, M.M. Circulating Tumor Cells Adhesion: Application in Biosensors. Biosensors 2023, 13, 882. [Google Scholar] [CrossRef]
- van Stalborch, A.-M.D.; Clark, A.G.; Sonnenberg, A.; Margadant, C. Imaging and Quantitative Analysis of Integrin-Dependent Cell-Matrix Adhesions. STAR Protoc. 2023, 4, 102473. [Google Scholar] [CrossRef]
- Montes, A.R.; Gutierrez, G.; BuganzaTepole, A.; Mofrad, M.R.K. Multiscale Computational Framework to Investigate Integrin Mechanosensing and Cell Adhesion. J. Appl. Phys. 2023, 134, 114702. [Google Scholar] [CrossRef]
- Giverso, C.; Jankowiak, G.; Preziosi, L.; Schmeiser, C. The Influence of Nucleus Mechanics in Modelling Adhesion-Independent Cell Migration in Structured and Confined Environments. Bull. Math. Biol. 2023, 85, 88. [Google Scholar] [CrossRef] [PubMed]
- Rasouli, M.; Soleimani, M.; Hosseinzadeh, S.; Ranjbari, J. Bacterial Cellulose as Potential Dressing and Scaffold Material: Toward Improving the Antibacterial and Cell Adhesion Properties. J. Polym. Environ. 2023, 31, 4621–4640. [Google Scholar] [CrossRef]
- Huber, M.; Casares-Arias, J.; Fässler, R.; Müller, D.J.; Strohmeyer, N. In Mitosis Integrins Reduce Adhesion to Extracellular Matrix and Strengthen Adhesion to Adjacent Cells. Nat. Commun. 2023, 14, 2143. [Google Scholar] [CrossRef]
- Nellinger, S.; Kluger, P.J. How Mechanical and Physicochemical Material Characteristics Influence Adipose-Derived Stem Cell Fate. Int. J. Mol. Sci. 2023, 24, 3551. [Google Scholar] [CrossRef]
- Koushki, N.; Ghagre, A.; Srivastava, L.K.; Molter, C.; Ehrlicher, A.J. Nuclear Compression Regulates YAP Spatiotemporal Fluctuations in Living Cells. Proc. Natl. Acad. Sci. USA 2023, 120, e2301285120. [Google Scholar] [CrossRef] [PubMed]
- Vassaux, M.; Milan, J.L. Stem Cell Mechanical Behaviour Modelling: Substrate’s Curvature Influence during Adhesion. Biomech. Model Mechanobiol. 2017, 16, 1295–1308. [Google Scholar] [CrossRef] [PubMed]
- Khalili, A.A.; Ahmad, M.R. A Review of Cell Adhesion Studies for Biomedical and Biological Applications. Int. J. Mol. Sci. 2015, 16, 18149–18184. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Wu, C.; Yang, W.; Liang, W.; Yu, H.; Liu, L. Recent Advance in Surface Modification for Regulating Cell Adhesion and Behaviors. Nanotechnol. Rev. 2020, 9, 971–989. [Google Scholar] [CrossRef]
- Harjunpää, H.; Asens, M.L.; Guenther, C.; Fagerholm, S.C. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front. Immunol. 2019, 10, 1078. [Google Scholar] [CrossRef] [PubMed]
- Shinde, A.; Illath, K.; Gupta, P.; Shinde, P.; Lim, K.T.; Nagai, M.; Santra, T.S. A Review of Single-cell Adhesion Force Kinetics and Applications. Cells 2021, 10, 577. [Google Scholar] [CrossRef]
- Lin, X.; Zhang, K.; Wei, D.; Tian, Y.; Gao, Y.; Chen, Z.; Qian, A. The Impact of Spaceflight and Simulated Microgravity on Cell Adhesion. Int. J. Mol. Sci. 2020, 21, 3031. [Google Scholar] [CrossRef]
- Ohashi, K.; Fujiwara, S.; Mizuno, K. Roles of the Cytoskeleton, Cell Adhesion and Rho Signalling in Mechanosensing and Mechanotransduction. J. Biochem. 2017, 161, 245–254. [Google Scholar] [CrossRef]
- Maurer, M.; Lammerding, J. The Driving Force: Nuclear Mechanotransduction in Cellular Function, Fate, and Disease. Annu. Rev. Biomed. Eng. 2019, 21, 443–468. [Google Scholar] [CrossRef]
- Davidson, P.M.; Cadot, B. Actin on and around the Nucleus. Trends Cell Biol. 2021, 31, 211–223. [Google Scholar] [CrossRef]
- Wang, N.; Tytell, J.; Ingber, D. Mechanotransduction at a Distance: Mechanically Coupling the Extracellular Matrix with the Nucleus. Nat. Rev. Mol. Cell Biol. 2009, 10, 75–82. [Google Scholar] [CrossRef]
- Tan, X.; Yan, Y.; Song, B.; Zhu, S.; Mei, Q.; Wu, K. Focal Adhesion Kinase: From Biological Functions to Therapeutic Strategies. Exp. Hematol. Oncol. 2023, 12, 83. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Guo, X.; Liang, Z.; Guo, R.; Liang, S.; Liu, H. Hax1 Regulate Focal Adhesion Dynamics through IQGAP1. Cell Commun. Signal. 2023, 21, 182. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liao, J.; Vlashi, R.; Chen, G. Focal Adhesion Kinase (FAK): Its Structure, Characteristics, and Signaling in Skeletal System. Cell. Signal. 2023, 111, 110852. [Google Scholar] [CrossRef]
- Fujiwara, T.K.; Tsunoyama, T.A.; Takeuchi, S.; Kalay, Z.; Nagai, Y.; Kalkbrenner, T.; Nemoto, Y.L.; Chen, L.H.; Shibata, A.C.E.; Iwasawa, K.; et al. Ultrafast Single-Molecule Imaging Reveals Focal Adhesion Nano-Architecture and Molecular Dynamics. J. Cell Biol. 2023, 222, e202110162. [Google Scholar] [CrossRef]
- Walter, K.; Bourquin, J.; Amiri, A.; Scheer, N.; Dehnert, M.; Eichhorn, A.L.; Dietz, C. Probing Local Lateral Forces of Focal Adhesions and Cell-Cell Junctions of Living Cells by Torsional Force Spectroscopy. Soft Matter. 2023, 19, 4772–4779. [Google Scholar] [CrossRef] [PubMed]
- Dumbauld, D.W.; Shin, H.; Gallant, N.D.; Michael, K.E.; Radhakrishna, H.; García, A.J. Contractility Modulates Cell Adhesion Strengthening through Focal Adhesion Kinase and Assembly of Vinculin-Containing Focal Adhesions. J. Cell. Physiol. 2010, 223, 746–756. [Google Scholar] [CrossRef]
- Fraley, S.I.; Feng, Y.; Krishnamurthy, R.; Kim, D.-H.; Celedon, A.; Longmore, G.D.; Wirtz, D. A Distinctive Role for Focal Adhesion Proteins in Three-Dimensional Cell Motility. Nat. Cell Biol. 2010, 12, 598–604. [Google Scholar] [CrossRef]
- Wozniak, M.A.; Modzelewska, K.; Kwong, L.; Keely, P.J. Focal Adhesion Regulation of Cell Behavior. Biochim. Biophys. Acta Mol. Cell Res. 2004, 1692, 103–119. [Google Scholar] [CrossRef]
- Seetharaman, S.; Etienne-Manneville, S. Microtubules at Focal Adhesions—A Double-Edged Sword. J. Cell Sci. 2019, 132, jcs232843. [Google Scholar] [CrossRef]
- Bauer, M.S.; Baumann, F.; Daday, C.; Redondo, P.; Durner, E.; Jobst, M.A.; Milles, L.F.; Mercadante, D.; Pippig, D.A.; Gaub, H.E.; et al. Structural and Mechanistic Insights into Mechanoactivation of Focal Adhesion Kinase. Proc. Natl. Acad. Sci. USA 2019, 116, 6766–6774. [Google Scholar] [CrossRef]
- Mavrakis, M.; Juanes, M.A. The Compass to Follow: Focal Adhesion Turnover. Curr. Opin. Cell Biol. 2023, 80, 102152. [Google Scholar] [CrossRef]
- Chen, S.; He, T.; Zhong, Y.; Chen, M.; Yao, Q.; Chen, D.; Shao, Z.; Xiao, G. Roles of Focal Adhesion Proteins in Skeleton and Diseases. Acta Pharm. Sin. B 2023, 13, 998–1013. [Google Scholar] [CrossRef]
- Martino, F.; Perestrelo, A.R.; Vinarský, V.; Pagliari, S.; Forte, G. Cellular Mechanotransduction: From Tension to Function. Front. Physiol. 2018, 9, 824. [Google Scholar] [CrossRef] [PubMed]
- Ingber, D.E. Tensegrity I. Cell Structure and Hierarchical Systems Biology. J. Cell Sci. 2003, 116, 1157–1173. [Google Scholar] [CrossRef]
- Vaiani, L.; Uva, A.E.; Boccaccio, A. Structural and Topological Design of Conformal Bilayered Scaffolds for Bone Tissue Engineering. Thin-Walled Struct. 2023, 192, 111209. [Google Scholar] [CrossRef]
- Rodríguez-Montaño, Ó.L.; Cortés-Rodríguez, C.J.; Naddeo, F.; Uva, A.E.; Fiorentino, M.; Naddeo, A.; Cappetti, N.; Gattullo, M.; Monno, G.; Boccaccio, A. Irregular Load Adapted Scaffold Optimization: A Computational Framework Based on Mechanobiological Criteria. ACS Biomater. Sci. Eng. 2019, 5, 5392–5411. [Google Scholar] [CrossRef] [PubMed]
- Perier-Metz, C.; Duda, G.N.; Checa, S. A Mechanobiological Computer Optimization Framework to Design Scaffolds to Enhance Bone Regeneration. Front. Bioeng. Biotechnol. 2022, 10, 980727. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Montaño, Ó.L.; Cortés-Rodríguez, C.J.; Uva, A.E.; Fiorentino, M.; Gattullo, M.; Monno, G.; Boccaccio, A. Comparison of the Mechanobiological Performance of Bone Tissue Scaffolds Based on Different Unit Cell Geometries. J. Mech. Behav. Biomed. Mater. 2018, 83, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Byrne, D.P.; Lacroix, D.; Planell, J.A.; Kelly, D.J.; Prendergast, P.J. Simulation of Tissue Differentiation in a Scaffold as a Function of Porosity, Young’s Modulus and Dissolution Rate: Application of Mechanobiological Models in Tissue Engineering. Biomaterials 2007, 28, 5544–5554. [Google Scholar] [CrossRef] [PubMed]
- Pobloth, A.M.; Checa, S.; Razi, H.; Petersen, A.; Weaver, J.C.; Chmidt-Bleek, K.; Windolf, M.; Tatai, A.A.; Roth, C.P.; Schaser, K.D.; et al. Mechanobiologically Optimized 3D Titanium-Mesh Scaffolds Enhance Bone Regeneration in Critical Segmental Defects in Sheep. Sci. Transl. Med. 2018, 10, eaam8828. [Google Scholar] [CrossRef] [PubMed]
- Velasco Peña, M.A. Mechanobiological Model of Bone Tissue Regeneration on Scaffolds; Universidad Nacional de Colombia: Bogotá, Colombia, 2016. [Google Scholar]
- Sun, W.; Starly, B.; Nam, J.; Darling, A. Bio-CAD Modeling and Its Applications in Computer-Aided Tissue Engineering. CAD Comput. Aided Des. 2005, 37, 1097–1114. [Google Scholar] [CrossRef]
- Bahraminasab, M. Challenges on Optimization of 3D-Printed Bone Scaffolds. Biomed. Eng. Online 2020, 19, 69. [Google Scholar] [CrossRef]
- Zhang, S.; Vijayavenkataraman, S.; Lu, W.F.; Fuh, J.Y.H. A Review on the Use of Computational Methods to Characterize, Design, and Optimize Tissue Engineering Scaffolds, with a Potential in 3D Printing Fabrication. J. Biomed. Mater. Res. B Appl. Biomater. 2019, 107, 1329–1351. [Google Scholar] [CrossRef]
- Lv, Y.; Wang, B.; Liu, G.; Tang, Y.; Lu, E.; Xie, K.; Lan, C.; Liu, J.; Qin, Z.; Wang, L. Metal Material, Properties and Design Methods of Porous Biomedical Scaffolds for Additive Manufacturing: A Review. Front. Bioeng. Biotechnol. 2021, 9, 641130. [Google Scholar] [CrossRef]
- Castro, A.P.G.; Lacroix, D. Micromechanical Study of the Load Transfer in a Polycaprolactone–Collagen Hybrid Scaffold When Subjected to Unconfined and Confined Compression. Biomech. Model. Mechanobiol. 2018, 17, 531–541. [Google Scholar] [CrossRef]
- Sandino, C.; Checa, S.; Prendergast, P.J.; Lacroix, D. Simulation of Angiogenesis and Cell Differentiation in a CaP Scaffold Subjected to Compressive Strains Using a Lattice Modeling Approach. Biomaterials 2010, 31, 2446–2452. [Google Scholar] [CrossRef]
- Metz, C.; Duda, G.N.; Checa, S. Towards Multi-Dynamic Mechano-Biological Optimization of 3D-Printed Scaffolds to Foster Bone Regeneration. Acta Biomater. 2019, 101, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Checa, S.; Prendergast, P.J. A Mechanobiological Model for Tissue Differentiation That Includes Angiogenesis: A Lattice-Based Modeling Approach. Ann. Biomed. Eng. 2009, 37, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Sandino, C.; Planell, J.A.; Lacroix, D. A Finite Element Study of Mechanical Stimuli in Scaffolds for Bone Tissue Engineering. J. Biomech. 2008, 41, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Bose, S.; Roy, M.; Bandyopadhyay, A. Recent Advances in Bone Tissue Engineering Scaffolds. Trends Biotechnol. 2012, 30, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Chanda, S.; Chakraborty, D. Application of Finite Element Analysis to Tissue Differentiation and Bone Remodelling Approaches and Their Use in Design Optimization of Orthopaedic Implants: A Review. Int. J. Numer. Method Biomed. Eng. 2022, 38, e3637. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z. Computational Modelling of Biomechanics and Biotribology in the Musculoskeletal System: Biomaterials and Tissues; Elsevier: Amsterdam, The Netherlands, 2014; ISBN 9780857096739. [Google Scholar]
- Vignesh, S.; Vijayakumar, P.; Anbarasan, B. Review on Finite Element Method in Biomedical Engineering. J. Adv. Res. Dyn. Control. Syst. 2019, 11, 1028–1032. [Google Scholar]
- Jean, R.P.; Chen, C.S.; Spector, A.A. Finite-Element Analysis of the Adhesion-Cytoskeleton-Nucleus Mechanotransduction Pathway during Endothelial Cell Rounding: Axisymmetric Model. J. Biomech. Eng. 2005, 127, 594–600. [Google Scholar] [CrossRef] [PubMed]
- Huiskes, R.; Chao, E.Y.S. A Survey of Finite Element Analysis in Orthopedic Biomechanics: The First Decade. J. Biomech. 1983, 16, 385–409. [Google Scholar] [CrossRef]
- Barreto, S.; Perrault, C.M.; Lacroix, D. Structural Finite Element Analysis to Explain Cell Mechanics Variability. J. Mech. Behav. Biomed. Mater. 2014, 38, 219–231. [Google Scholar] [CrossRef]
- Vaiani, L.; Migliorini, E.; Cavalcanti-Adam, E.A.; Uva, A.E.; Fiorentino, M.; Gattullo, M.; Manghisi, V.M.; Boccaccio, A. Coarse-Grained Elastic Network Modelling: A Fast and Stable Numerical Tool to Characterize Mesenchymal Stem Cells Subjected to AFM Nanoindentation Measurements. Mater. Sci. Eng. C 2021, 121, 111860. [Google Scholar] [CrossRef]
- Boccaccio, A.; Frassanito, M.C.; Lamberti, L.; Brunelli, R.; Maulucci, G.; Monaci, M.; Papi, M.; Pappalettere, C.; Parasassi, T.; Sylla, L.; et al. Nanoscale Characterization of the Biomechanical Hardening of Bovine Zona Pellucida. J. R. Soc. Interface 2012, 9, 2871–2882. [Google Scholar] [CrossRef]
- Boccaccio, A.; Lamberti, L.; Papi, M.; De Spirito, M.; Pappalettere, C. Effect of AFM Probe Geometry on Visco-Hyperelastic Characterization of Soft Materials. Nanotechnology 2015, 26, 325701. [Google Scholar] [CrossRef]
- Ficarella, E.; Minooei, M.; Santoro, L.; Toma, E.; Trentadue, B.; De Spirito, M.; Papi, M.; Pruncu, C.I.; Lamberti, L. Visco-HyperelasticCharacterization of the Equine Immature Zona Pellucida. Materials 2021, 14, 1223. [Google Scholar] [CrossRef]
- Lin, D.C.; Shreiber, D.I.; Dimitriadis, E.K.; Horkay, F. Spherical Indentation of Soft Matter beyond the Hertzian Regime: Numerical and Experimental Validation of Hyperelastic Models. Biomech. Model Mechanobiol. 2009, 8, 345–358. [Google Scholar] [CrossRef]
- Hsia, C.C.W.; Chuong, C.J.C.; Johnson, R.L., Jr. Effect of Shape Distortion of Red Blood Cell on Pulmonary Capillary Diffusive Uptake of Carbon Monoxide: A Finite Element Analysis. FASEB J. 1996, 10, A362. [Google Scholar]
- Boccaccio, A.; Uva, A.E.; Papi, M.; Fiorentino, M.; De Spirito, M.; Monno, G. Nanoindentation Characterisation of Human Colorectal Cancer Cells Considering Cell Geometry, Surface Roughness and Hyperelastic Constitutive Behaviour. Nanotechnology 2017, 28, 45703. [Google Scholar] [CrossRef]
- Migliorini, E.; Cavalcanti-Adam, E.A.; Uva, A.E.; Fiorentino, M.; Gattullo, M.; Manghisi, V.M.; Vaiani, L.; Boccaccio, A. Nanoindentation of Mesenchymal Stem Cells Using Atomic Force Microscopy: Effect of Adhesive Cell-Substrate Structures. Nanotechnology 2021, 32, 215706. [Google Scholar] [CrossRef]
- Vargas-Pinto, R.; Gong, H.; Vahabikashi, A.; Johnson, M. The Effect of the Endothelial Cell Cortex on Atomic Force Microscopy Measurements. Biophys. J. 2013, 105, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Kamm, R.D.; McVittie, A.K.; Bathe, M. On the Role of Continuum Models in Mechanobiology. In Proceedings of the ASME International Mechanical Engineering Congress and Exposition, Orlando, FL, USA, 5–10 November 2000; Proceedings (IMECE). Volume 2000-Z, pp. 1–11. [Google Scholar]
- Thoumine, O.; Cardoso, O.; Meister, J.-J. Changes in the Mechanical Properties of Fibroblasts during Spreading: A Micromanipulation Study. Eur. Biophys. J. 1999, 28, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Frisch, T.; Thoumine, O. Predicting the Kinetics of Cell Spreading. J. Biomech. 2002, 35, 1137–1141. [Google Scholar] [CrossRef] [PubMed]
- Thoumine, O.; Ott, A. Time Scale Dependent Viscoelastic and Contractile Regimes in Fibroblasts Probed by Microplate Manipulation. J. Cell Sci. 1997, 110, 2109–2116. [Google Scholar] [CrossRef] [PubMed]
- Kamgoué, A.; Ohayon, J.; Tracqui, P. Estimation of Cell Young’s Modulus of Adherent Cells Probed by Optical and Magnetic Tweezers: Influence of Cell Thickness and Bead Immersion. J. Biomech. Eng. 2007, 129, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Hénon, S.; Lenormand, G.; Richert, A.; Gallet, F. A New Determination of the Shear Modulus of the Human Erythrocyte Membrane Using Optical Tweezers. Biophys. J. 1999, 76, 1145–1151. [Google Scholar] [CrossRef]
- Lenormand, G.; Hénon, S.; Richert, A.; Siméon, J.; Gallet, F. Elasticity of the Human Red Blood Cell Skeleton. Biorheology 2002, 40, 247–251. [Google Scholar]
- Bausch, A.R.; Möller, W.; Sackmann, E. Measurement of Local Viscoelasticity and Forces in Living Cells by Magnetic Tweezers. Biophys. J. 1999, 76, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Feneberg, W.; Aepfelbacher, M.; Sackmann, E. Microviscoelasticity of the Apical Cell Surface of Human Umbilical Vein Endothelial Cells (HUVEC) within Confluent Monolayers. Biophys. J. 2004, 87, 1338–1350. [Google Scholar] [CrossRef] [PubMed]
- Phan-Thien, N. Rigid Spherical Inclusion: The Multipole Expansion. J. Elast. 1993, 32, 243–252. [Google Scholar] [CrossRef]
- Mijailovich, S.M.; Kojic, M.; Zivkovic, M.; Fabry, B.; Fredberg, J.J. A Finite Element Model of Cell Deformation during Magnetic Bead Twisting. J. Appl. Physiol. 2002, 93, 1429–1436. [Google Scholar] [CrossRef] [PubMed]
- Ohayon, J.; Tracqui, P. Computation of Adherent Cell Elasticity for Critical Cell-Bead Geometry in Magnetic Twisting Experiments. Ann. Biomed. Eng. 2005, 33, 131–141. [Google Scholar] [CrossRef]
- Laurent, V.M.; Hénon, S.; Planus, E.; Fodil, R.; Balland, M.; Isabey, D.; Gallet, F. Assessment of Mechanical Properties of Adherent Living Cells by Bead Micromanipulation: Comparison of Magnetic Twisting Cytometry vs Optical Tweezers. J. Biomech. Eng. 2002, 124, 408–421. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, C.F.; Gasperini, L.; Marques, A.P.; Reis, R.L. The Stiffness of Living Tissues and Its Implications for Tissue Engineering. Nat. Rev. Mater. 2020, 5, 351–370. [Google Scholar] [CrossRef]
- Cui, Q.; Liu, T.; Li, X.; Zhao, L.; Wu, Q.; Wang, X.; Song, K.; Ge, D. Validation of the Mechano-Bactericidal Mechanism of Nanostructured Surfaces with Finite Element Simulation. Colloids Surf. B Biointerfaces 2021, 206, 111929. [Google Scholar] [CrossRef]
- Ananthakrishnan, R.; Guck, J.; Wottawah, F.; Schinkinger, S.; Lincoln, B.; Romeyke, M.; Moon, T.; Käs, J. Quantifying the Contribution of Actin Networks to the Elastic Strength of Fibroblasts. J. Theor. Biol. 2006, 242, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Tracqui, P.; Ohayon, J. Transmission of Mechanical Stresses within the Cytoskeleton of Adherent Cells: A Theoretical Analysis Based on a Multi-Component Cell Model. Acta Biotheor. 2004, 52, 323–341. [Google Scholar] [CrossRef] [PubMed]
- McGarry, J.G.; Prendergast, P.J.; Ashton, B.; Klein-Nulend, J. A Three-Dimensional Finite Element Model of an Adherent Eukaryotic Cell. Eur. Cell Mater. 2004, 7, 27–34. [Google Scholar] [CrossRef] [PubMed]


| Cell Type | References |
|---|---|
| Stem Cells | [4,13,40,41,49,51,54,60,67] |
| Endothelial Cells | [49,51,57,68,73,79] |
| Fibroblasts | [59,70,71,79,84,86] |
| Epithelial Cells | [69,73,79,80,85] |
| Blood Cells | [8,65,75] |
| Cancer Cells | [66,84] |
| Oocytes | [61,62,63] |
| Connective Tissue Cells (Chondrocytes, Osteoblasts) | [48,64] |
| Bacteria | [83] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santoro, L.; Vaiani, L.; Boccaccio, A.; Lamberti, L.; Lo Muzio, L.; Ballini, A.; Cantore, S. Finite Element Modeling of Cells Adhering to a Substrate: An Overview. Appl. Sci. 2024, 14, 2596. https://doi.org/10.3390/app14062596
Santoro L, Vaiani L, Boccaccio A, Lamberti L, Lo Muzio L, Ballini A, Cantore S. Finite Element Modeling of Cells Adhering to a Substrate: An Overview. Applied Sciences. 2024; 14(6):2596. https://doi.org/10.3390/app14062596
Chicago/Turabian StyleSantoro, Lorenzo, Lorenzo Vaiani, Antonio Boccaccio, Luciano Lamberti, Lorenzo Lo Muzio, Andrea Ballini, and Stefania Cantore. 2024. "Finite Element Modeling of Cells Adhering to a Substrate: An Overview" Applied Sciences 14, no. 6: 2596. https://doi.org/10.3390/app14062596
APA StyleSantoro, L., Vaiani, L., Boccaccio, A., Lamberti, L., Lo Muzio, L., Ballini, A., & Cantore, S. (2024). Finite Element Modeling of Cells Adhering to a Substrate: An Overview. Applied Sciences, 14(6), 2596. https://doi.org/10.3390/app14062596








