Performance of AI-Based Automated Classifications of Whole-Body FDG PET in Clinical Practice: The CLARITI Project
Abstract
1. Introduction
2. Materials and Methods
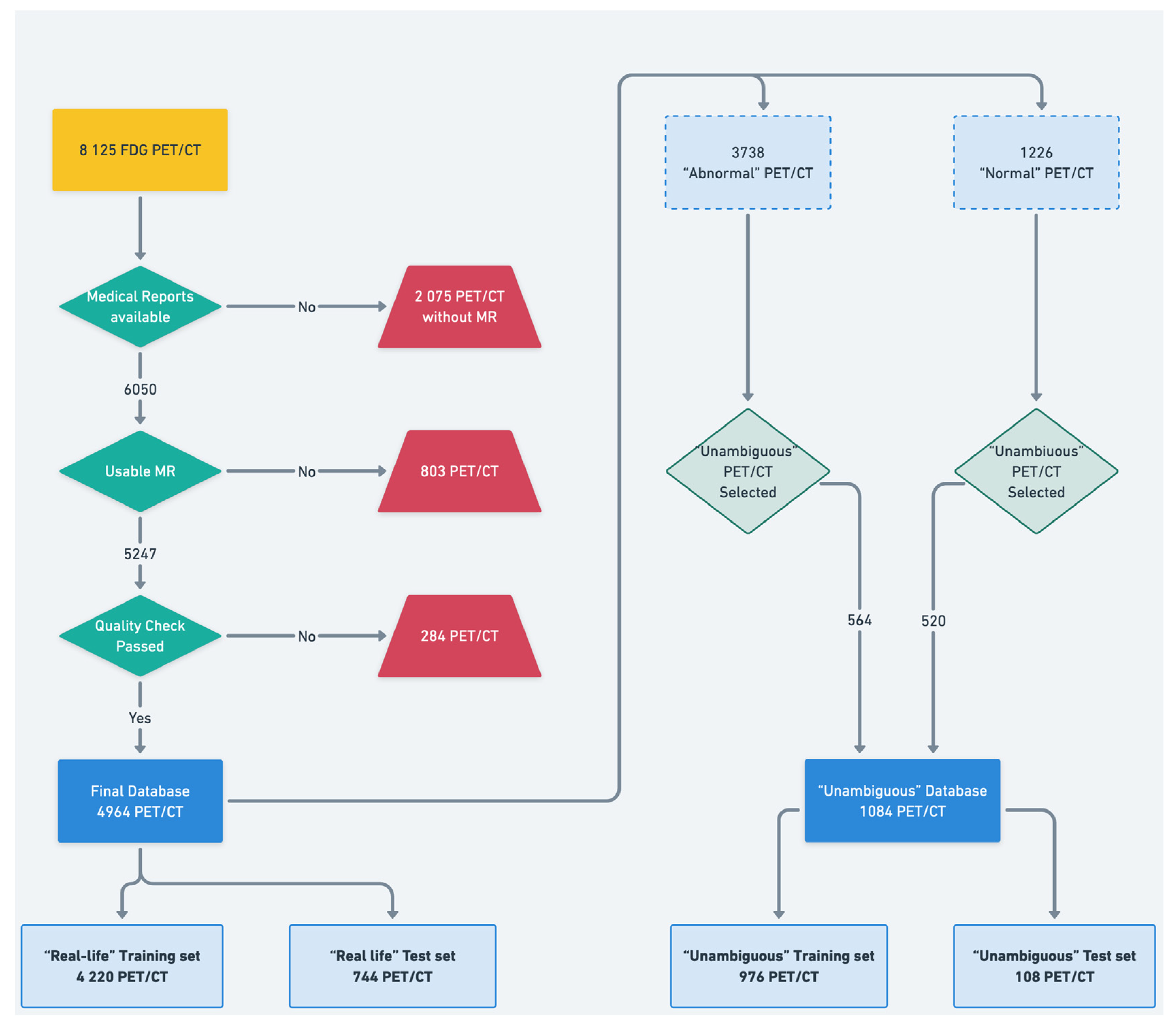
2.1. Dataset
2.2. Data Preprocessing and Labeling
- First, an “in-house” automated text mining procedure was developed to automatically classify the PET scans based on the anonymized written imaging reports.
- Second, the maximum intensity projection of all the PET data was converted into nine 2D projections (from 0° to 180° rotation) to be carefully qualitatively reviewed by an experienced nuclear medicine physician (A.B., who had 5 years of expertise in PET imaging). For this purpose, any well-grounded nonphysiological uptake was considered abnormal.
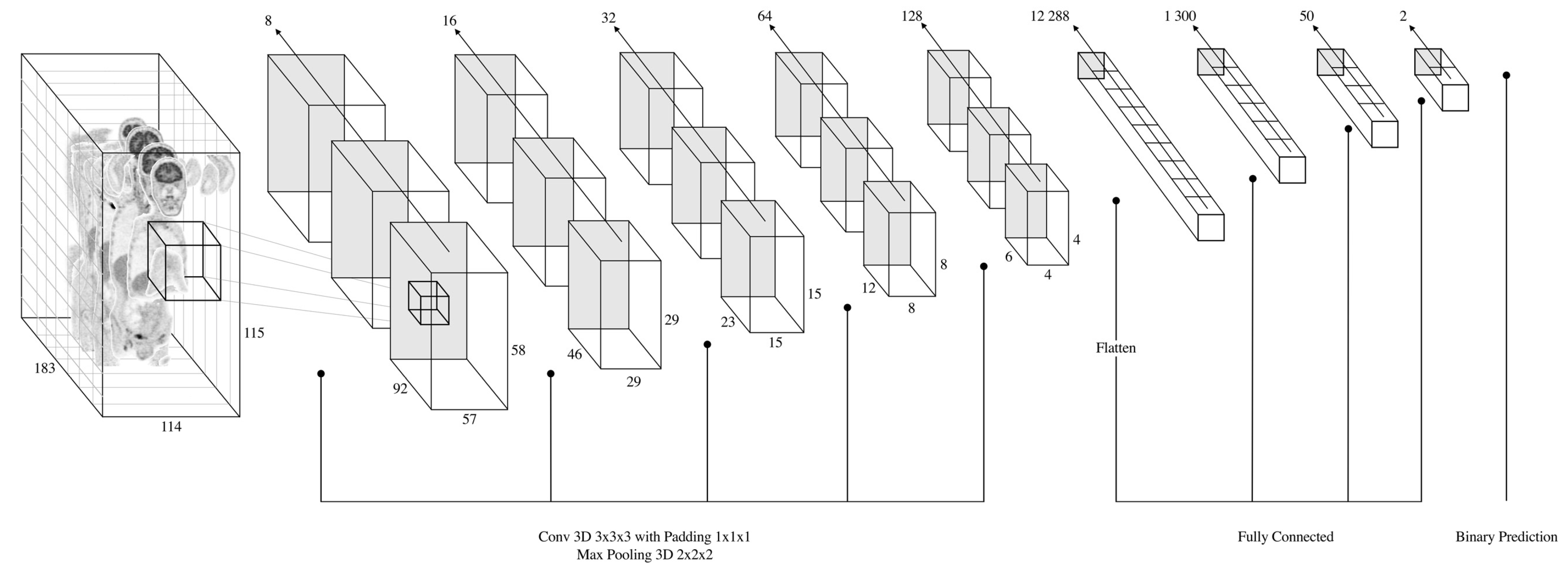
2.3. CNN Architecture, Training and Testing Procedures
2.4. Performance Assessment
3. Results
3.1. ”Unambiguous” Data Sample
3.2. “Real-Life” Data Sample
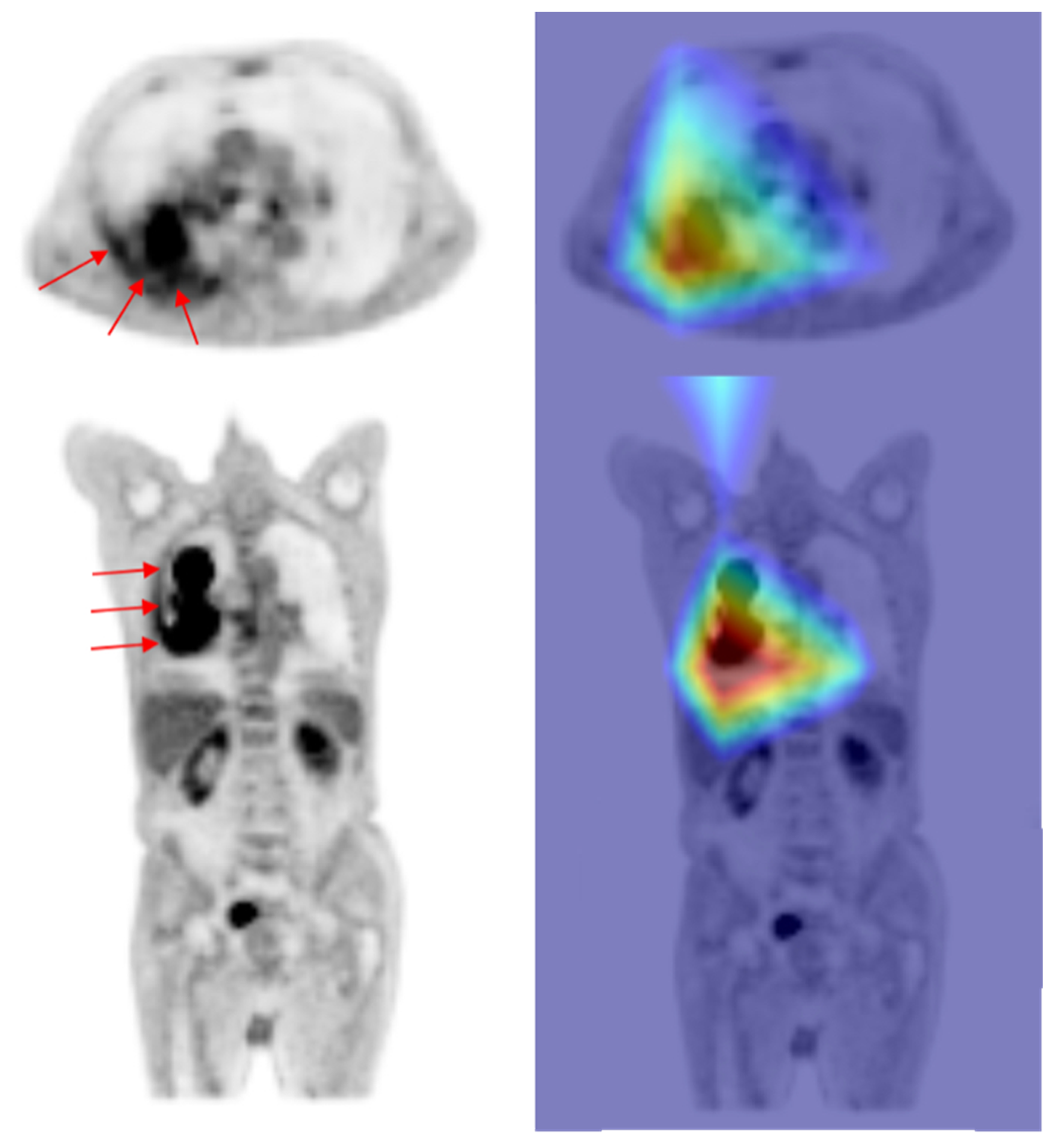
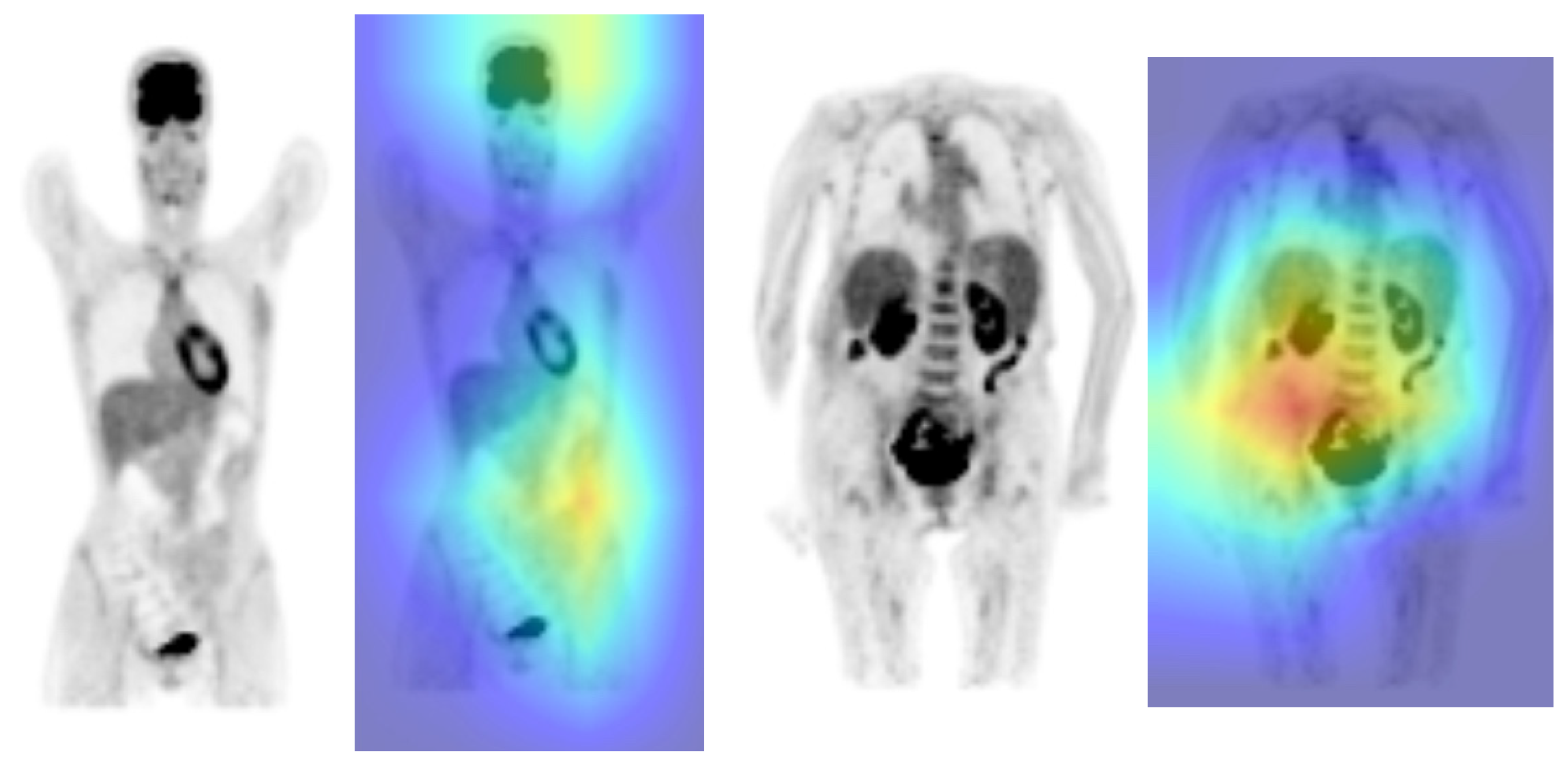
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fletcher, J.W.; Djulbegovic, B.; Soares, H.P.; Siegel, B.A.; Lowe, V.J.; Lyman, G.H.; Coleman, R.E.; Wahl, R.; Paschold, J.C.; Avril, N.; et al. Recommendations on the Use of 18F-FDG PET in Oncology. J. Nucl. Med. 2008, 49, 480–508. [Google Scholar] [CrossRef] [PubMed]
- Jamar, F.; Buscombe, J.; Chiti, A.; Christian, P.E.; Delbeke, D.; Donohoe, K.J.; Israel, O.; Martin-Comin, J.; Signore, A. EANM/SNMMI Guideline for 18F-FDG Use in Inflammation and Infection. J. Nucl. Med. 2013, 54, 647–658. [Google Scholar] [CrossRef]
- Boellaard, R.; Delgado-Bolton, R.; Oyen, W.J.G.; Giammarile, F.; Tatsch, K.; Eschner, W.; Verzijlbergen, F.J.; Barrington, S.F.; Pike, L.C.; Weber, W.A.; et al. FDG PET/CT: EANM Procedure Guidelines for Tumour Imaging: Version 2.0. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 328–354. [Google Scholar] [CrossRef]
- Keraen, J.; Blanc, E.; Besson, F.L.; Leguern, V.; Meyer, C.; Henry, J.; Belkhir, R.; Nocturne, G.; Mariette, X.; Seror, R. Usefulness of 18F-Labeled Fluorodeoxyglucose–Positron Emission Tomography for the Diagnosis of Lymphoma in Primary Sjögren’s Syndrome. Arthritis Rheumatol. 2019, 71, 1147–1157. [Google Scholar] [CrossRef]
- Slart, R.H.J.A.; Writing Group; Reviewer Group; Members of EANM Cardiovascular; Members of EANM Infection & Inflammation; Members of Committees, SNMMI Cardiovascular; Members of Council; PET Interest Group; Members of ASNC & EANM Committee Coordinator. FDG-PET/CT(A) Imaging in Large Vessel Vasculitis and Polymyalgia Rheumatica: Joint Procedural Recommendation of the EANM, SNMMI, and the PET Interest Group (PIG), and Endorsed by the ASNC. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 1250–1269. [Google Scholar] [CrossRef]
- Besson, F.L.; Chaumet-Riffaud, P.; Playe, M.; Noel, N.; Lambotte, O.; Goujard, C.; Prigent, A.; Durand, E. Contribution of 18F-FDG PET in the Diagnostic Assessment of Fever of Unknown Origin (FUO): A Stratification-Based Meta-Analysis. Eur. J. Nucl. Med. Mol. Imaging 2016, 43, 1887–1895. [Google Scholar] [CrossRef] [PubMed]
- Pinochet, P.; Eude, F.; Becker, S.; Shah, V.; Sibille, L.; Toledano, M.N.; Modzelewski, R.; Vera, P.; Decazes, P. Evaluation of an Automatic Classification Algorithm Using Convolutional Neural Networks in Oncological Positron Emission Tomography. Front. Med. 2021, 8, 628179. [Google Scholar] [CrossRef] [PubMed]
- Mlynarski, P.; Delingette, H.; Criminisi, A.; Ayache, N. 3D Convolutional Neural Networks for Tumor Segmentation Using Long-Range 2D Context. Comput. Med. Imaging Graph. 2019, 73, 60–72. [Google Scholar] [CrossRef]
- Lotter, W.; Diab, A.R.; Haslam, B.; Kim, J.G.; Grisot, G.; Wu, E.; Wu, K.; Onieva, J.O.; Boyer, Y.; Boxerman, J.L.; et al. Robust Breast Cancer Detection in Mammography and Digital Breast Tomosynthesis Using an Annotation-Efficient Deep Learning Approach. Nat. Med. 2021, 27, 244–249. [Google Scholar] [CrossRef]
- Lakhani, P.; Sundaram, B. Deep Learning at Chest Radiography: Automated Classification of Pulmonary Tuberculosis by Using Convolutional Neural Networks. Radiology 2017, 284, 574–582. [Google Scholar] [CrossRef]
- Saraf, V.; Chavan, P.; Jadhav, A. Deep Learning Challenges in Medical Imaging. In Advanced Computing Technologies and Applications; Vasudevan, H., Michalas, A., Shekokar, N., Narvekar, M., Eds.; Springer: Singapore, 2020; pp. 293–301. [Google Scholar]
- Daniel, C.; Serre, P.; Orlova, N.; Bréant, S.; Paris, N.; Griffon, N. Initializing a Hospital-Wide Data Quality Program. The AP-HP Experience. Comput. Methods Programs Biomed. 2019, 181, 104804. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Thibeau-Sutre, E.; Diaz-Melo, M.; Samper-González, J.; Routier, A.; Bottani, S.; Dormont, D.; Durrleman, S.; Burgos, N.; Colliot, O. Convolutional Neural Networks for Classification of Alzheimer’s Disease: Overview and Reproducible Evaluation. Med. Image Anal. 2020, 63, 101694. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2017, arXiv:1412.6980. [Google Scholar]
- Refaeilzadeh, P.; Tang, L.; Liu, H. Cross-Validation. In Encyclopedia of Database Systems; Liu, L., Özsu, M.T., Eds.; Springer: Boston, MA, USA, 2009; pp. 532–538. ISBN 978-0-387-35544-3. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef]
- Karimi, D.; Dou, H.; Warfield, S.K.; Gholipour, A. Deep Learning with Noisy Labels: Exploring Techniques and Remedies in Medical Image Analysis. Med. Image Anal. 2020, 65, 101759. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, M.; Biroli, M.; Gelardi, F.; Seregni, E.; Chiti, A.; Sollini, M. Deep Learning in Nuclear Medicine—Focus on CNN-Based Approaches for PET/CT and PET/MR: Where Do We Stand? Clin. Transl. Imaging 2021, 9, 37–55. [Google Scholar] [CrossRef]
- Kawauchi, K.; Furuya, S.; Hirata, K.; Katoh, C.; Manabe, O.; Kobayashi, K.; Watanabe, S.; Shiga, T. A Convolutional Neural Network-Based System to Classify Patients Using FDG PET/CT Examinations. BMC Cancer 2020, 20, 227. [Google Scholar] [CrossRef]
- Sibille, L.; Seifert, R.; Avramovic, N.; Vehren, T.; Spottiswoode, B.; Zuehlsdorff, S.; Schäfers, M. 18F-FDG PET/CT Uptake Classification in Lymphoma and Lung Cancer by Using Deep Convolutional Neural Networks. Radiology 2020, 294, 445–452. [Google Scholar] [CrossRef]
- Roberts, M.; Driggs, D.; Thorpe, M.; Gilbey, J.; Yeung, M.; Ursprung, S.; Aviles-Rivero, A.I.; Etmann, C.; McCague, C.; Beer, L.; et al. Common Pitfalls and Recommendations for Using Machine Learning to Detect and Prognosticate for COVID-19 Using Chest Radiographs and CT Scans. Nat. Mach. Intell. 2021, 3, 199–217. [Google Scholar] [CrossRef]
- Veit-Haibach, P.; Buvat, I.; Herrmann, K. EJNMMI Supplement: Bringing AI and Radiomics to Nuclear Medicine. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2627–2629. [Google Scholar] [CrossRef]
- Eyuboglu, S.; Angus, G.; Patel, B.N.; Pareek, A.; Davidzon, G.; Long, J.; Dunnmon, J.; Lungren, M.P. Multi-Task Weak Supervision Enables Anatomically-Resolved Abnormality Detection in Whole-Body FDG-PET/CT. Nat. Commun. 2021, 12, 1880. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Hu, J. A K-Fold Averaging Cross-Validation Procedure. J. Nonparametric Stat. 2015, 27, 167–179. [Google Scholar] [CrossRef] [PubMed]

| Overall | Training Set | Test Set | ||
|---|---|---|---|---|
| n | 1084 | 976 | 108 | |
| Age in years | 58.5 ± 14.7 | 58.5 ± 14.9 | 58.5 ± 13.5 | |
| Weight in kg | 72.1 ± 16 | 72.2 ± 16 | 71.3 ± 16.3 | |
| Activity in MBq | 251.7 ± 57 | 252 ± 57 | 248.9 ± 57.8 | |
| Sex | F | 481 (44.4%) 603 (55.6%) | 440 (45.1%) 536 (54.9%) | 41 (38%) 67 (62%) |
| M | ||||
| Clinical context | Oncological | 940 (86.7%) | 847 (86.8%) | 93 (86.1%) |
| Non-Oncological | 144 (13.3%) | 129 (13.4%) | 15 (13.9%) | |
| Label | Normal | 520 (48%) | 468 (48%) | 52 (48.1%) |
| Abnormal | 564 (52%) | 508 (52%) | 56 (51.9%) |
| TP | TN | FP | FN | NPV (%) | PPV (%) | Sensibility (%) | Specificity (%) | Accuracy (%) | F1 Score (%) | LR+ | LR− | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fold 1 | 96 | 94 | 0 | 6 | 94.0 | 100 | 94.1 | 100 | 96.9 | 97.0 | >100 | 0.059 |
| fold 2 | 85 | 92 | 1 | 17 | 84.4 | 98.8 | 83.3 | 98.9 | 90.8 | 90.4 | 75.727 | 0.169 |
| fold 3 | 91 | 92 | 1 | 11 | 89.3 | 98.9 | 89.2 | 98.9 | 93.8 | 93.8 | 81.091 | 0.109 |
| fold 4 | 89 | 94 | 0 | 12 | 88.7 | 100 | 88.1 | 100 | 93.8 | 93.7 | >100 | 0.119 |
| fold 5 | 89 | 92 | 2 | 12 | 88.5 | 97.8 | 88.1 | 97.9 | 92.8 | 92.7 | 41.952 | 0.122 |
| Mean | 90 | 92.8 | 0.8 | 11 | 89.0 | 99.1 | 88.6 | 99.1 | 93.6 | 93.5 | 66.257 | 0.116 |
| Variance | 16 | 1.2 | 0.7 | 15 | 0.1 | 0 | 0.1 | 0 | 0 | 0.1 | 450.223 | 0.002 |
| Std deviation | 4.00 | 1.09 | 0.84 | 3.91 | 3.4 | 0.9 | 3.8 | 0.9 | 2.2 | 2.4 | 21.218 | 0.039 |
| Overall | Training Set | Test Set | ||
|---|---|---|---|---|
| n | 4964 | 4220 | 744 | |
| Age in years | 60.7 ± 14.7 | 60.5 ± 14.5 | 62.0 ± 15.5 | |
| Weight in kg | 70.5 ± 16 | 70.4 ± 16 | 71.1 ± 16.4 | |
| Activity in MBq | 246.5 ± 58 | 246 ± 58 | 249.2 ± 58.7 | |
| Sex | F | 2453 (49.4%) | 2111 (50.0%) | 342 (46%) |
| M | 2511 (50.6%) | 2109 (50.0%) | 402 (54%) | |
| Clinical context | Oncological | 4370 (88.0%) | 3739 (88.6%) | 631 (84.8%) |
| Non-oncological | 594 (12.0%) | 481 (11.4%) | 113 (15.2%) | |
| Label | Normal | 1226 (24.7%) | 1042 (24.7%) | 184 (24.7%) |
| Abnormal | 3738 (75.3%) | 3178 (75.3%) | 560 (75.3%) | |
| TP | TN | FP | FN | NPV (%) | PPV (%) | Sensibility (%) | Specificity (%) | Accuracy (%) | F1 Score (%) | LR+ | LR− | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fold 1 | 340 | 181 | 27 | 296 | 37.9 | 92.6 | 53.5 | 87.0 | 61.7 | 67.8 | 4.1183 | 0.5348 |
| fold 2 | 367 | 161 | 47 | 269 | 37.4 | 88.6 | 57.7 | 77.4 | 62.6 | 69.9 | 2.5537 | 0.5464 |
| fold 3 | 422 | 147 | 61 | 214 | 40.7 | 87.4 | 66.4 | 70.7 | 67.4 | 75.4 | 2.2625 | 0.4761 |
| fold 4 | 391 | 161 | 48 | 244 | 39.8 | 89.1 | 61.6 | 77.0 | 65.4 | 72.8 | 2.6811 | 0.4988 |
| fold 5 | 406 | 139 | 70 | 229 | 37.8 | 85.3 | 63.9 | 66.5 | 64.6 | 73.1 | 1.909 | 0.5422 |
| Mean | 385.2 | 157.8 | 50 | 250.4 | 38.7 | 88.6 | 60.6 | 75.7 | 64.3 | 71.8 | 2.705 | 0.52 |
| Variance | 1048.7 | 257.2 | 265 | 1062.3 | 0.0 | 0.1 | 0.3 | 0.6 | 0.1 | 0.1 | 0.713 | 0.001 |
| Std deviation | 32.38 | 16.04 | 16.29 | 32.59 | 1.4 | 2.7 | 5.1 | 7.8 | 2.3 | 3.0 | 0.844 | 0.031 |
| Total | TP | TN | FP | FN | NPV | PPV | Sensibility | Specificity | Accuracy (CI95%) | F1 Score | LR+ | LR− | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All test sets | 744 | 403 | 143 | 41 | 403 | 0.26 | 0.91 | 0.50 | 0.78 | 0.73 ± 0.032 | 0.64 | 2.24 | 0.64 |
| Unambiguous PET studies | 177 | 99 | 67 | 11 | 99 | 0.40 | 0.90 | 0.50 | 0.86 | 0.94 ± 0.036 | 0.64 | 3.54 | 0.58 |
| Ambiguous PET studies | 567 | 304 | 76 | 30 | 304 | 0.20 | 0.91 | 0.50 | 0.72 | 0.67 ± 0.039 | 0.65 | 1.77 | 0.70 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berenbaum, A.; Delingette, H.; Maire, A.; Poret, C.; Hassen-Khodja, C.; Bréant, S.; Daniel, C.; Martel, P.; Grimaldi, L.; Frank, M.; et al. Performance of AI-Based Automated Classifications of Whole-Body FDG PET in Clinical Practice: The CLARITI Project. Appl. Sci. 2023, 13, 5281. https://doi.org/10.3390/app13095281
Berenbaum A, Delingette H, Maire A, Poret C, Hassen-Khodja C, Bréant S, Daniel C, Martel P, Grimaldi L, Frank M, et al. Performance of AI-Based Automated Classifications of Whole-Body FDG PET in Clinical Practice: The CLARITI Project. Applied Sciences. 2023; 13(9):5281. https://doi.org/10.3390/app13095281
Chicago/Turabian StyleBerenbaum, Arnaud, Hervé Delingette, Aurélien Maire, Cécile Poret, Claire Hassen-Khodja, Stéphane Bréant, Christel Daniel, Patricia Martel, Lamiae Grimaldi, Marie Frank, and et al. 2023. "Performance of AI-Based Automated Classifications of Whole-Body FDG PET in Clinical Practice: The CLARITI Project" Applied Sciences 13, no. 9: 5281. https://doi.org/10.3390/app13095281
APA StyleBerenbaum, A., Delingette, H., Maire, A., Poret, C., Hassen-Khodja, C., Bréant, S., Daniel, C., Martel, P., Grimaldi, L., Frank, M., Durand, E., & Besson, F. L. (2023). Performance of AI-Based Automated Classifications of Whole-Body FDG PET in Clinical Practice: The CLARITI Project. Applied Sciences, 13(9), 5281. https://doi.org/10.3390/app13095281






