CNN-Based Pill Image Recognition for Retrieval Systems
Abstract
1. Introduction
- We investigate the challenging problem of image retrieval, specifically targeting pill images.
- We develop an efficient image retrieval system based on deep learning and the k-Nearest Neighbor (k-NN) classifier.
- We employ a real-life dataset of pill images to evaluate the proposed system against accuracy and runtime, as well as compare the results with relevant image retrieval systems from the literature.
- Our proposed model increased the accuracy of identifying pills form images by 10% while maintaining the same runtime as comparable methods.
2. Related Work
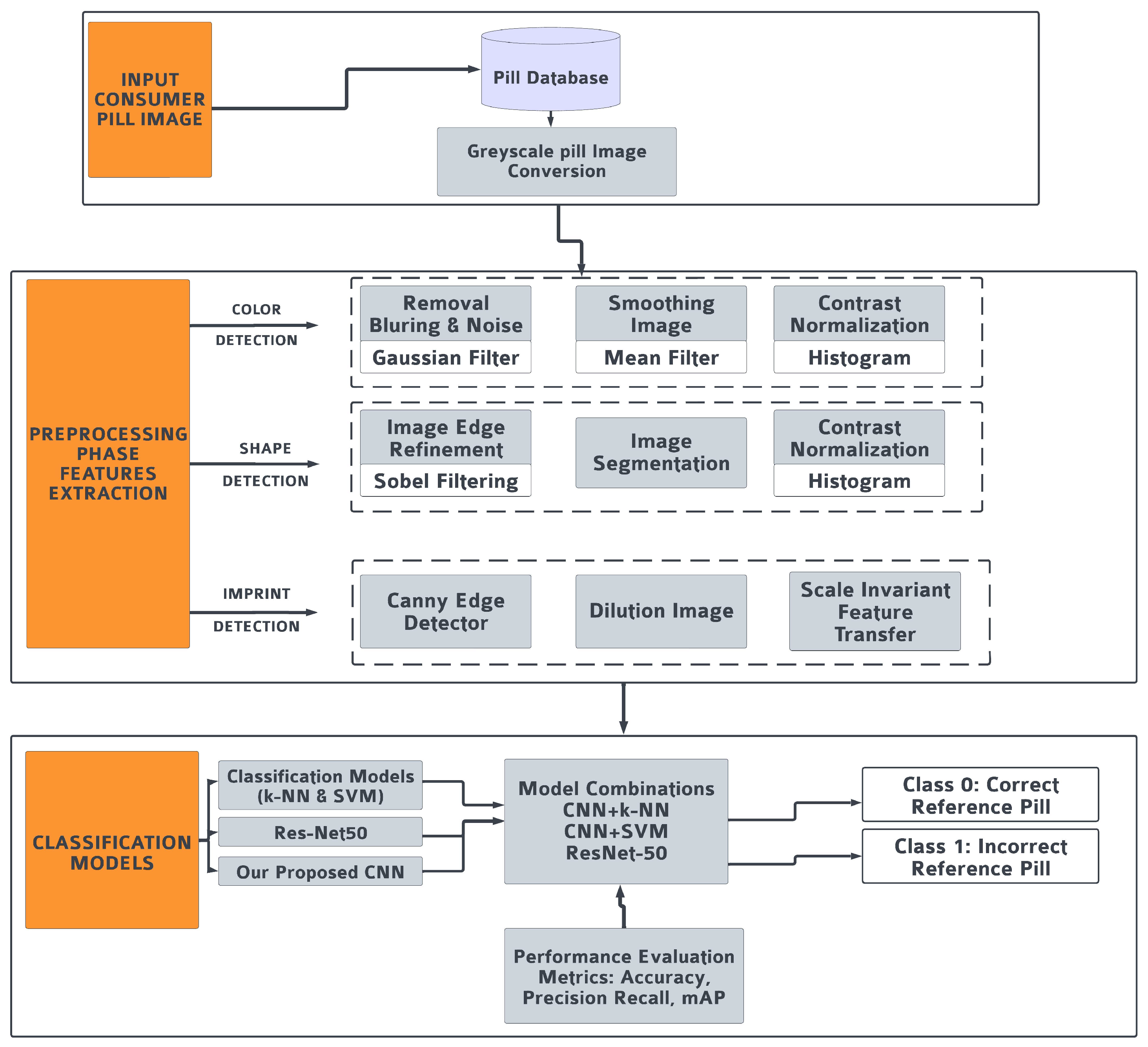
3. Methodology
3.1. Preprocessing and Features Extraction
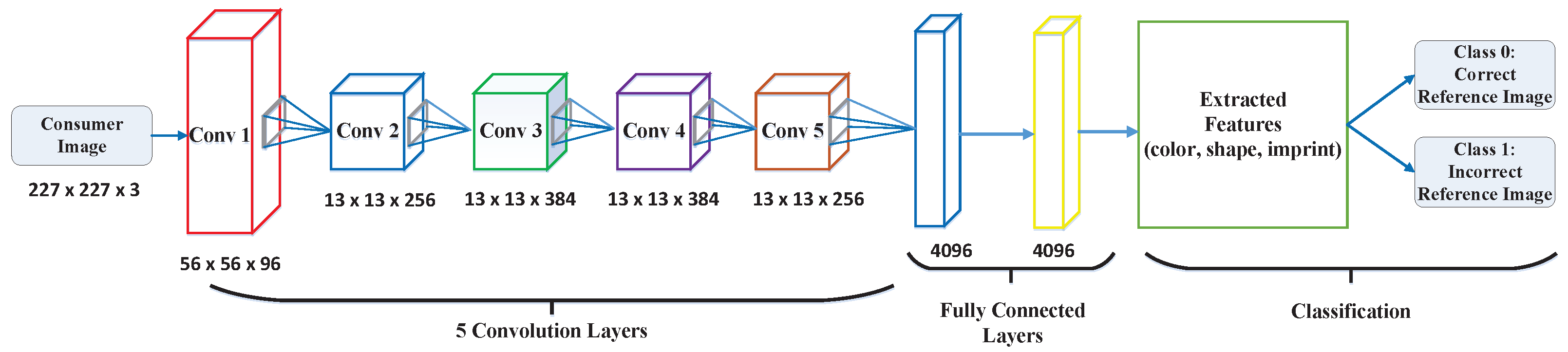
3.2. Proposed CNN Architecture
- Pill images go through one convolutional layer (Conv1) with 56 × 56 × 96, which means that the input to the layer is a pill image with a height and width of 56 pixels and that has 96 color channels.
- The resulting tensors (images) go through four additional convolution layers with a smaller height and width (13 pixels) than the previous layer, and the number of input channels is increased to 256 color channels.
- The resulting feature maps are converted to a fully connected (FC) layer of 4096 neurons, which is connected to a second fully connected layer of 4096 neurons.
- Then, the extracted features (color, shape, and imprint) are fed into the classification layers; we then employ a k-NN classifier to handle the prediction more accurately and with less runtime.
3.3. Classification
- k-Nearest Neighbors (k-NN) [13,14] is a non-parametric classifier that assumes similar objects (i.e., data points) are usually “closer” to one another in comparison to dissimilar objects. k-NN measures similarity between data points using distance metrics. One of the most common distance metrics is the Euclidean distance and is measured by the following function:where X and Y are two data points in the n-dimensional space, and and are Euclidean vectors from the point of origin. When our proposed model receives a query image, the model converts the image to feature vectors, which the classifier will use to predict the pill type in the query image. We set k to 5 for all our experimental analysis in Section 4.
- The Support Vector Machine (SVM) [48] is a classifier that, when given a set of input objects, creates an imaginary wall that separates dissimilar objects. This imaginary wall is called a hyperplane, because it can separate data points represented in spaces beyond three-dimensional. Given a set of input data points, there are several potential hyperplanes that the SVM can create. The SVM creates the best separation between the data points, i.e., it only keeps the hyperplane that minimizes the classification error.
- Residual Network [31], or otherwise known as ResNet, is a neural network-based model that can be used as a final identifier in a convolutional layout. ResNet can accommodate more than 50 layers and be used to classify and extract features in an image. This technique makes use of skip connections to reduce the training error and help add the output of earlier layers to later layers without losing the image quality.
4. Results
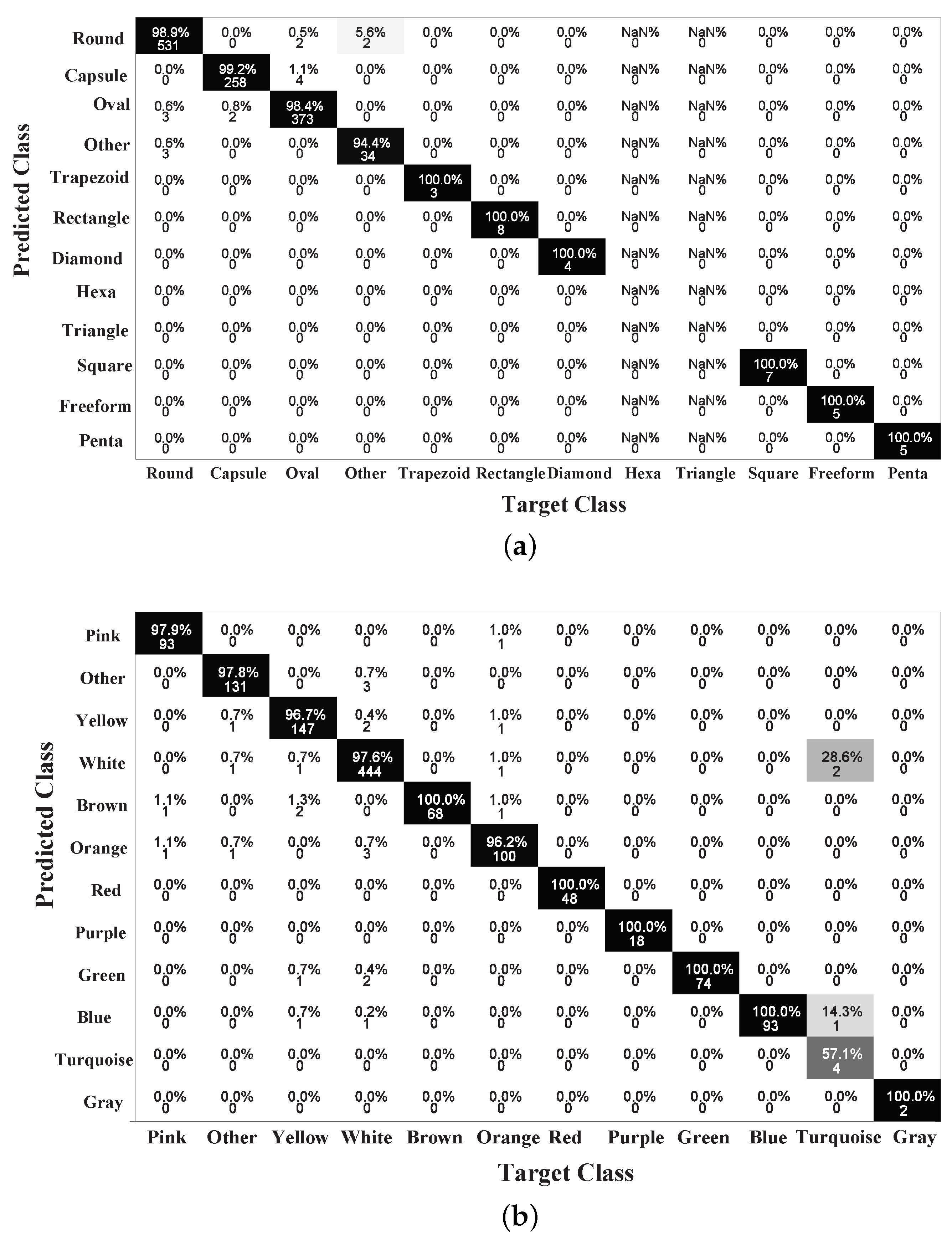
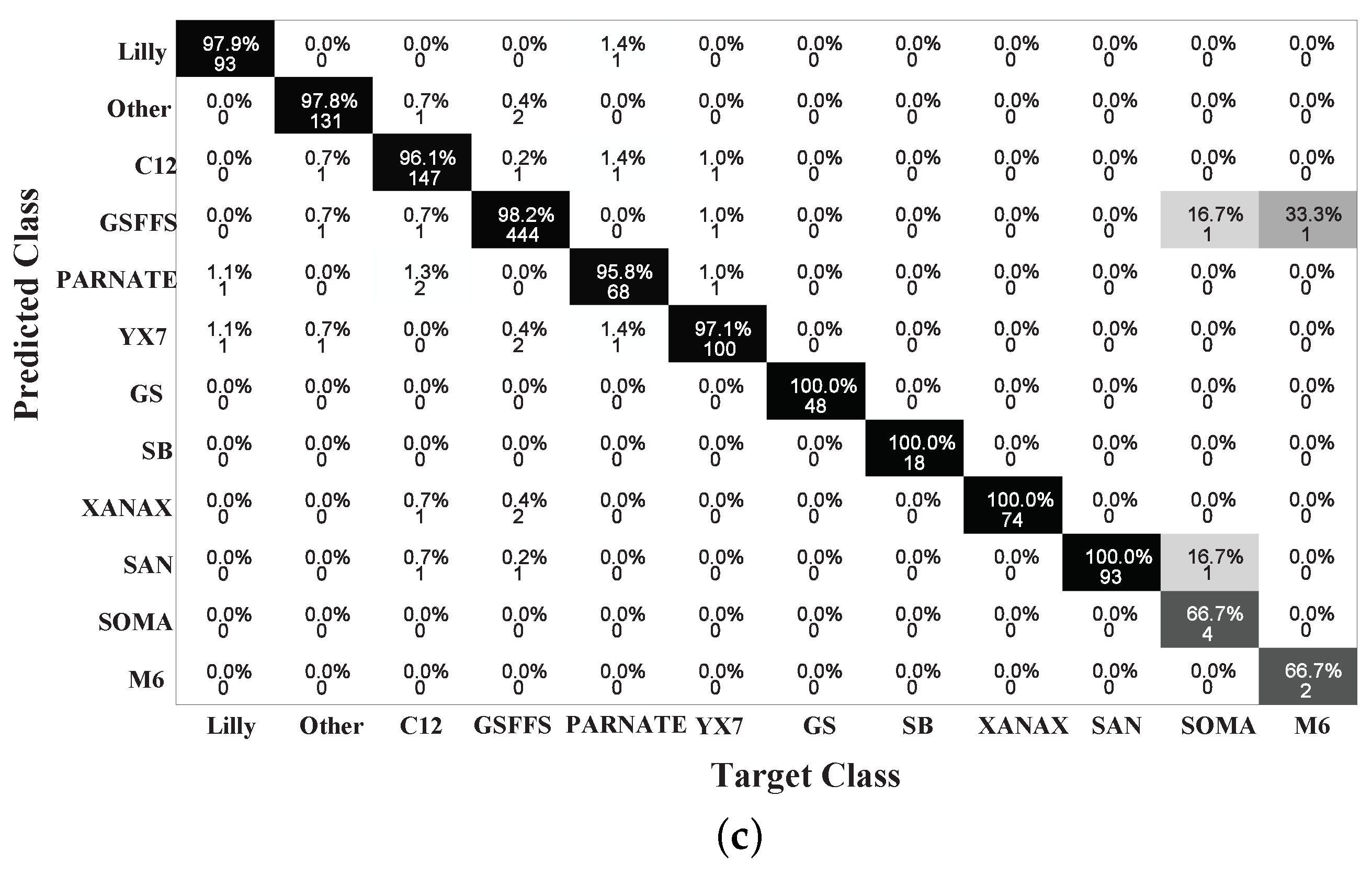
4.1. Dataset
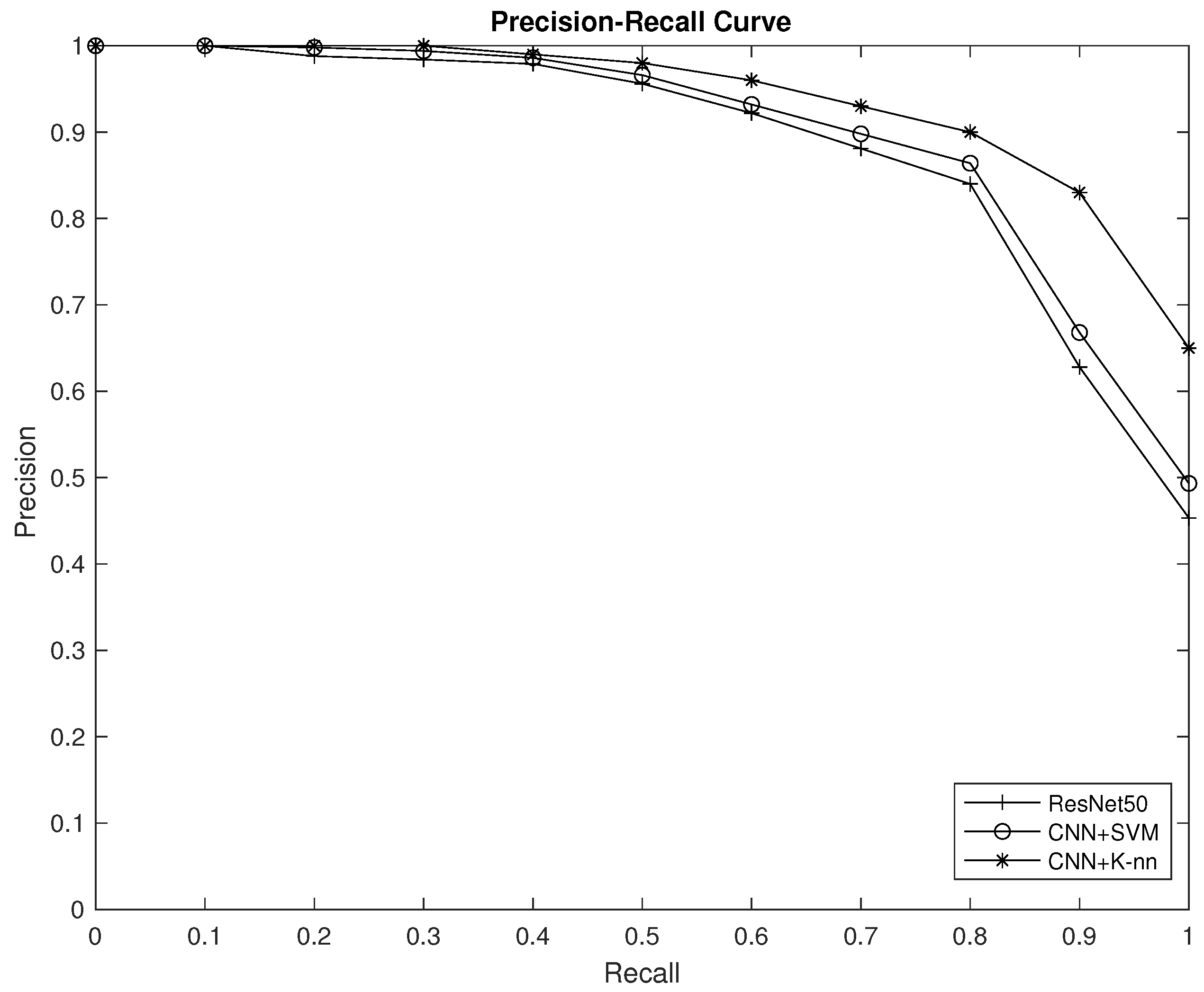
4.2. Performance Analysis
- For each object class, calculate the Average Precision (AP) as:where (n) is the total number of relevant items in the dataset for the given object class, and the Precision at each relevant k-object is the Precision calculated at the position of the relevant item in the ranked list of predicted items.
- Calculate the mAP as the mean of the AP scores for all object classes:where (N) is the total number of object classes in the dataset.
4.3. Efficiency
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CBIR | Content-Based Information Retrieval |
| WHO | World Health Organization |
| CNN | Convolutional Neural Network |
| ResNet | Residual Network |
| SVM | Support Vector Machine |
| NLM | National Library of Medicine |
| NIH | National Institutes of Health |
| k-NN | k-Nearest Neighbor |
References
- Crestani, F.; Mizzaro, S.; Scagnetto, I. Mobile Information Retrieval, 1st ed.; Springer International Publishing: Cham, Switzerland, 2017. [Google Scholar]
- Celik, C.; Bilge, H.S. Content based image retrieval with sparse representations and local feature descriptors: A comparative study. Pattern Recognit. 2017, 68, 1–13. [Google Scholar] [CrossRef]
- Madduri, A. Content based Image Retrieval System using Local Feature Extraction Techniques. Int. J. Comput. Appl. 2021, 183, 16–20. [Google Scholar] [CrossRef]
- Dubey, S.R. A Decade Survey of Content Based Image Retrieval Using Deep Learning. IEEE Trans. Circuits Syst. Video Technol. 2022, 32, 2687–2704. [Google Scholar] [CrossRef]
- Makary, M.A.; Daniel, M. Medical error—the third leading cause of death in the US. BMJ 2016, 353, i2139. [Google Scholar] [CrossRef] [PubMed]
- World Health Assembly. Patient Safety: Global Action on Patient Safety: Report by the Director-General; World Health Assembly: Geneva, Switzerland, 2019; p. 8. [Google Scholar]
- WHO. Medication Without Harm: Real-Life Stories; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- WHO. 10 Facts on Patient Safety. 2019. Available online: https://www.who.int/news-room/photo-story/photo-story-detail/10-facts-on-patient-safety (accessed on 31 July 2022).
- Larios Delgado, N.; Usuyama, N.; Hall, A.K.; Hazen, R.J.; Ma, M.; Sahu, S.; Lundin, J. Fast and accurate medication identification. NPJ Digit. Med. 2019, 2, 10. [Google Scholar] [CrossRef]
- Yu, J.; Chen, Z.; Kamata, S.i. Pill Recognition Using Imprint Information by Two-Step Sampling Distance Sets. In Proceedings of the 2014 22nd International Conference on Pattern Recognition, Stockholm, Sweden, 24–28 August 2014; pp. 3156–3161. [Google Scholar] [CrossRef]
- RxList. Pill Identifier (Pill Finder Wizard). 2022. Available online: https://www.rxlist.com/pill-identification-tool/article.htm (accessed on 31 July 2022).
- Healthline. Medication Safety: Pill Identification, Storage, and More. 2021. Available online: https://www.healthline.com/health/pill-identification (accessed on 31 July 2022).
- Guo, G.; Wang, H.; Bell, D.; Bi, Y.; Greer, K. KNN Model-based Approach in Classification. In On The Move to Meaningful Internet Systems 2003: CoopIS, DOA, and ODBASE; Springer: Berlin/Heidelberg, Germany, 2003; pp. 986–996. Volume 2888. [Google Scholar] [CrossRef]
- Altman, N.S. An Introduction to Kernel and Nearest-Neighbor Nonparametric Regression. Am. Stat. 1992, 46, 175–185. [Google Scholar]
- Maron, M.E.; Kuhns, J.L. On Relevance, Probabilistic Indexing and Information Retrieval. J. ACM 1960, 7, 216–244. [Google Scholar] [CrossRef]
- Adel’son-Vel’skii, G.M.; Landis, E.M. An algorithm for organization of information. Dokl. Akad. Nauk. 1962, 146, 263–266. [Google Scholar]
- Chang, S.K.; Liu, S.H. Picture indexing and abstraction techniques for pictorial databases. IEEE Trans. Pattern Anal. Mach. Intell. 1984, 4, 475–484. [Google Scholar] [CrossRef]
- Foster, C.C. Information Retrieval: Information Storage and Retrieval Using AVL Trees. In Proceedings of the 1965 20th National Conference, Cleveland, OH, USA, 24–26 August 1965; Association for Computing Machinery: New York, NY, USA, 1965; pp. 192–205. [Google Scholar] [CrossRef]
- Salton, G.; Lesk, M.E. The SMART Automatic Document Retrieval Systems-an Illustration. Commun. ACM 1965, 8, 391–398. [Google Scholar] [CrossRef]
- Salton, G. The SMART Retrieval System-Experiments in Automatic Document Processing; Prentice-Hall, Inc.: Upper Saddle River, NJ, USA, 1971. [Google Scholar]
- Rabitti, F.; Stanchev, P. An Approach to Image Retrieval from Large Image Databases. In Proceedings of the 10th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, New Orleans, LA, USA, 3–5 June 1987; Association for Computing Machinery: New York, NY, USA, 1987; pp. 284–295. [Google Scholar] [CrossRef]
- Wang, X.Y.; Wu, J.F.; Yang, H.Y. Robust image retrieval based on color histogram of local feature regions. Multimed. Tools Appl. 2010, 49, 323–345. [Google Scholar] [CrossRef]
- Lee, Y.B.; Park, U.; Jain, A.K.; Lee, S.W. Pill-ID: Matching and retrieval of drug pill images. Pattern Recognit. Lett. 2012, 33, 904–910. [Google Scholar] [CrossRef]
- Maji, S.; Bose, S. CBIR using features derived by deep learning. ACM/IMS Trans. Data Sci. 2021, 2, 1–24. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Razavian, A.S.; Azizpour, H.; Sullivan, J.; Carlsson, S. CNN Features Off-the-Shelf: An Astounding Baseline for Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Columbus, OH, USA, 23–28 June 2014; pp. 512–519. [Google Scholar] [CrossRef]
- Taigman, Y.; Yang, M.; Ranzato, M.; Wolf, L. DeepFace: Closing the Gap to Human-Level Performance in Face Verification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1701–1708. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Wang, Y.; Ribera, J.; Liu, C.; Yarlagadda, S.; Zhu, F. Pill Recognition Using Minimal Labeled Data. In Proceedings of the IEEE Third International Conference on Multimedia Big Data (BigMM), Laguna Hills, CA, USA, 19–21 April 2017; pp. 346–353. [Google Scholar] [CrossRef]
- Ou, Y.Y.; Tsai, A.C.; Zhou, X.P.; Wang, J.F. Automatic drug pills detection based on enhanced feature pyramid network and convolution neural networks. IET Comput. Vis. 2020, 14, 9–17. [Google Scholar] [CrossRef]
- Zeng, X.; Cao, K.; Zhang, M. MobileDeepPill: A Small-Footprint Mobile Deep Learning System for Recognizing Unconstrained Pill Images. In Proceedings of the 15th Annual International Conference on Mobile Systems, Applications, and Services, Niagara Falls, NY, USA, 19–23 June 2017; Association for Computing Machinery: New York, NY, USA, 2017; pp. 56–67. [Google Scholar] [CrossRef]
- Guo, P.; Stanley, R.; Cole, J.G.; Hagerty, J.; Stoecker, W. Color Feature-based Pillbox Image Color Recognition. In Proceedings of the 12th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications—Volume 4: VISAPP, (VISIGRAPP 2017), Porto, Portugal, 27 February–1 March 2017; pp. 188–194. [Google Scholar] [CrossRef]
- Cordeiro, L.S.; Lima, J.S.; Rocha Ribeiro, A.I.; Bezerra, F.N.; Rebouças Filho, P.P.; Rocha Neto, A.R. Pill Image Classification using Machine Learning. In Proceedings of the 2019 8th Brazilian Conference on Intelligent Systems (BRACIS), Salvador, Brazil, 15–18 October 2019; pp. 556–561. [Google Scholar] [CrossRef]
- Suksawatchon, U.; Srikamdee, S.; Suksawatchon, J.; Werapan, W. Shape Recognition Using Unconstrained Pill Images Based on Deep Convolution Network. In Proceedings of the 2022 6th International Conference on Information Technology (InCIT), Nonthaburi, Thailand, 10–11 November 2022; pp. 309–313. [Google Scholar] [CrossRef]
- Proma, T.P.; Hossan, M.Z.; Amin, M.A. Medicine Recognition from Colors and Text; Association for Computing Machinery: New York, NY, USA, 2019; ICGSP ’19. [Google Scholar] [CrossRef]
- Swastika, W.; Prilianti, K.; Stefanus, A.; Setiawan, H.; Arfianto, A.Z.; Santosa, A.W.B.; Rahmat, M.B.; Setiawan, E. Preliminary Study of Multi Convolution Neural Network-Based Model To Identify Pills Image Using Classification Rules. In Proceedings of the 2019 International Seminar on Intelligent Technology and Its Applications (ISITIA), Surabaya, Indonesia, 28–29 August 2019; pp. 376–380. [Google Scholar] [CrossRef]
- Kwon, H.J.; Kim, H.G.; Lee, S.H. Pill Detection Model for Medicine Inspection Based on Deep Learning. Chemosensors 2022, 10, 4. [Google Scholar] [CrossRef]
- Holtkötter, J.; Amaral, R.; Almeida, R.; Jácome, C.; Cardoso, R.; Pereira, A.; Pereira, M.; Chon, K.H.; Fonseca, J.A. Development and Validation of a Digital Image Processing-Based Pill Detection Tool for an Oral Medication Self-Monitoring System. Sensors 2022, 22, 2958. [Google Scholar] [CrossRef]
- Nguyen, A.D.; Pham, H.H.; Trung, H.T.; Nguyen, Q.V.H.; Truong, T.N.; Nguyen, P.L. High Accurate and Explainable Multi-Pill Detection Framework with Graph Neural Network-Assisted Multimodal Data Fusion. arXiv 2023, arXiv:2303.09782. [Google Scholar]
- Chang, W.J.; Chen, L.B.; Hsu, C.H.; Lin, C.P.; Yang, T.C. A Deep Learning-Based Intelligent Medicine Recognition System for Chronic Patients. IEEE Access 2019, 7, 44441–44458. [Google Scholar] [CrossRef]
- Ting, H.W.; Chung, S.L.; Chen, C.F.; Chiu, H.Y.; Hsieh, Y.W. A drug identification model developed using deep learning technologies: Experience of a medical center in Taiwan. BMC Health Serv. Res. 2019, 20, 312. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.D.; Nguyen, T.D.; Pham, H.H.; Nguyen, T.H.; Nguyen, P.L. Image-based Contextual Pill Recognition with Medical Knowledge Graph Assistance. In Proceedings of the Asian Conference on Intelligent Information and Database Systems, Ho Chi Minh City, Vietnam, 28–30 November 2022. [Google Scholar]
- National Library of Medicine. Pill Identification Challenge. 2016. Available online: https://www.nlm.nih.gov/databases/download/pill_image.html (accessed on 31 July 2022).
- Stanford. CS231n: Convolutional Neural Networks for Visual Recognition. 2022. Available online: http://cs231n.stanford.edu/ (accessed on 31 July 2022).
- Vapnik, V.N. Statistical Learning Theory; Wiley-Interscience: Hoboken, NJ, USA, 1998. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar] [CrossRef]

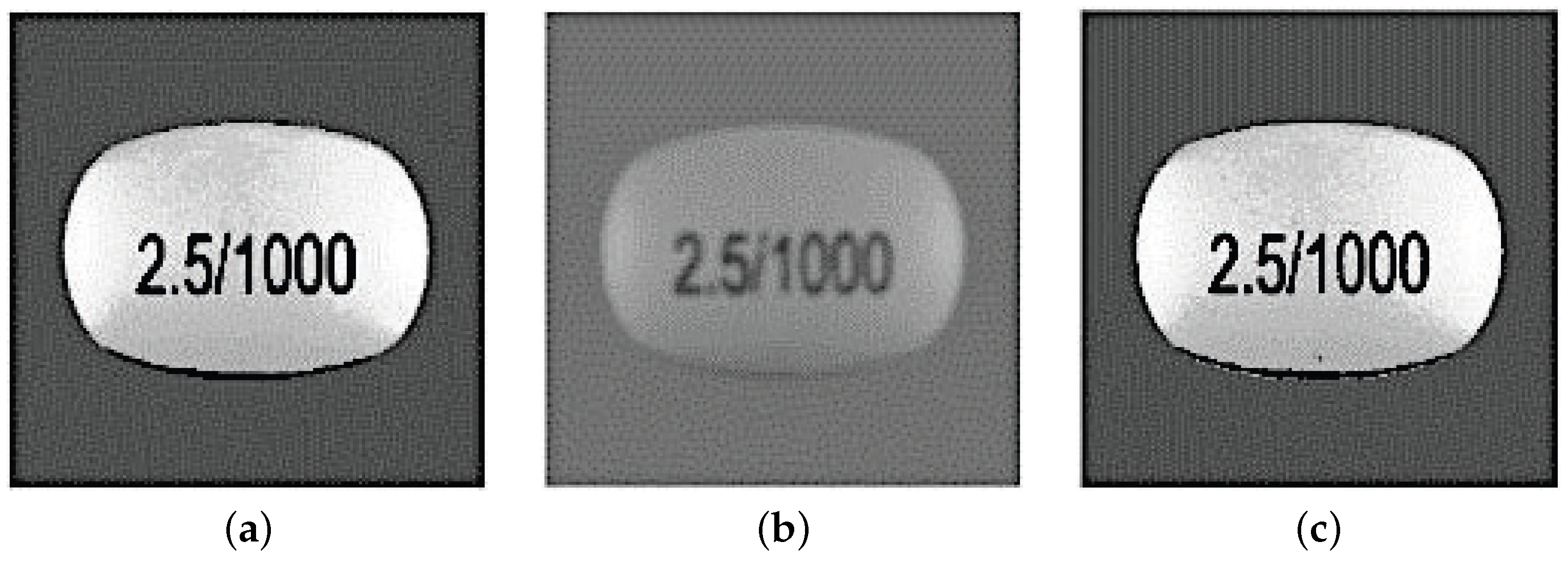
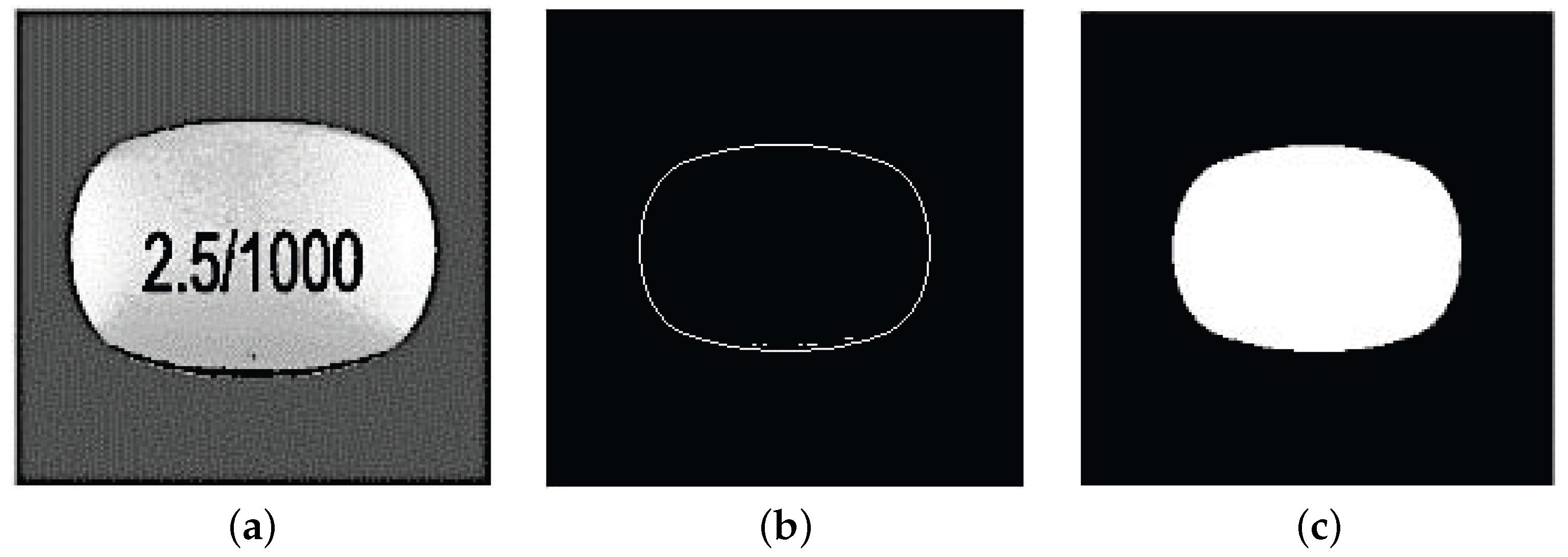


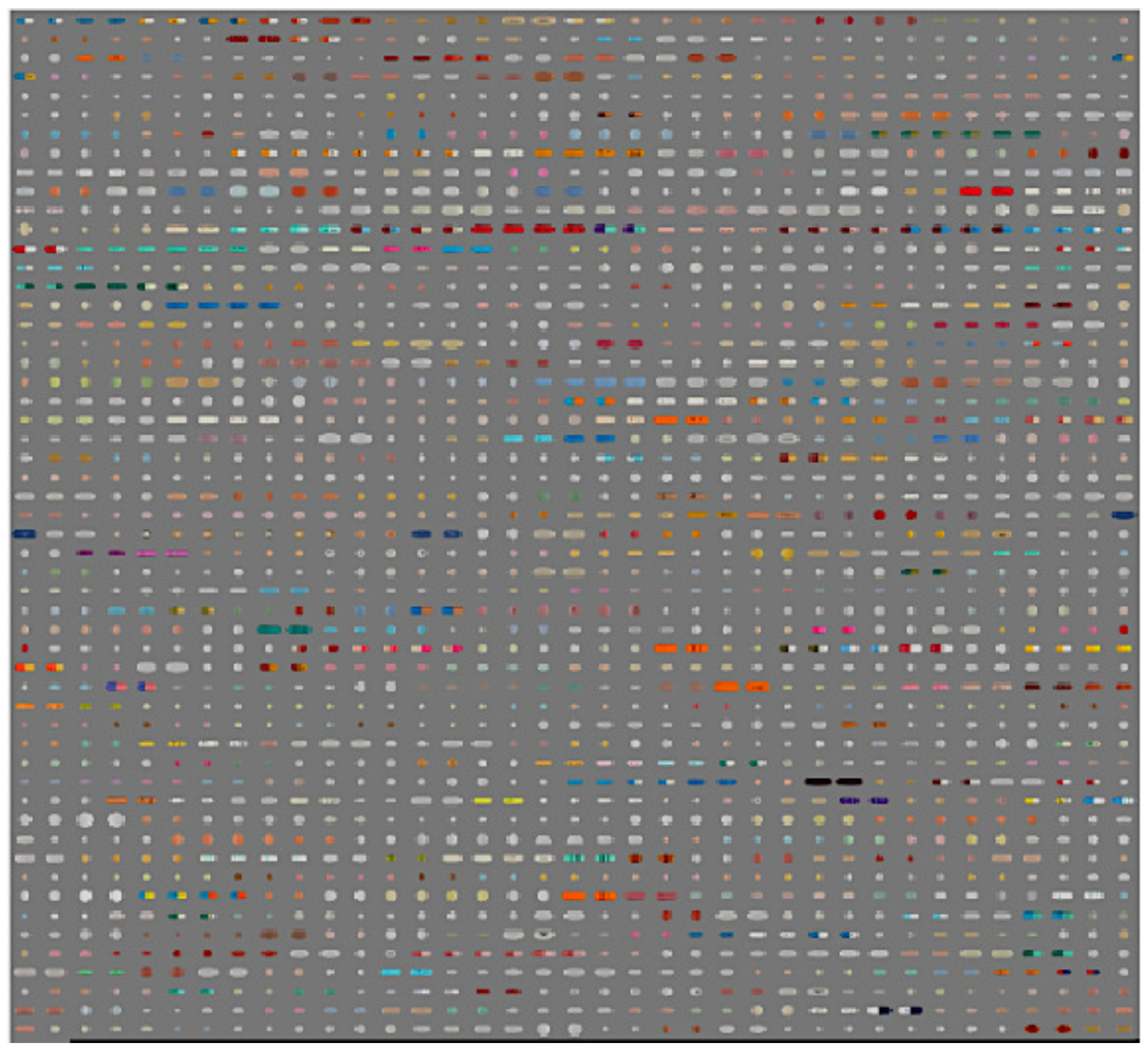
| Features | Reference Image | Consumer Image |
|---|---|---|
| Format | jpeg | jpeg |
| Width | 2400 | 4416 |
| Height | 1600 | 3312 |
| XResolution | 72 | 180 |
| YResolution | 72 | 180 |
| ColorType | TrueColor | TrueColor |
| BitDepth | 24 | 24 |
| YCbCrPositioning | Centered | Co-Sited |
| Models | Mean Average Precision (mAP) |
|---|---|
| ResNet-50 | 80% |
| CNN+SVM | 86.3% |
| CNN+kNN (our proposed model) | 90.8% |
| Method | Top-1 | Top-5 |
|---|---|---|
| SqueezeNet [49] | 49.0% | 76.8% |
| AlexNet [30] | 62.5% | 83.0% |
| ResNet-50 [31] | 71.7% | 85.5% |
| MobileNet [50] | 71.7% | 92.0% |
| MobileDeepPill [34] | 73.7% | 95.6% |
| InceptionV3 [51] | 74.4% | 93.3% |
| CNN+SVM (our suggested model) | 76.5% | 92.0% |
| CNN+kNN (our proposed model) | 80.5% | 96.1% |
| k-Value | ResNet-50 | CNN+SVM | CNN+kNN |
|---|---|---|---|
| k = 1 | 71.7% | 76.5% | 80.5% |
| k = 2 | 74.5% | 82.1% | 86.1% |
| k = 3 | 82.0% | 89.0% | 94.0% |
| k = 4 | 83.0% | 90.5% | 95.5% |
| k = 5 | 85.5% | 92.0% | 96.1% |
| k = 6 | 86.5% | 92.2% | 96.5% |
| k = 7 | 88.0% | 92.5% | 97.0% |
| k = 8 | 89.0% | 92.7% | 97.5% |
| k = 9 | 89.8% | 93.0% | 97.8% |
| k = 10 | 90.5% | 93.0% | 97.9% |
| k = 11 | 90.8% | 93.0% | 98.0% |
| k = 12 | 91.0% | 93.0% | 98.0% |
| k = 13 | 91.0% | 93.0% | 98.0% |
| k = 14 | 91.0% | 93.0% | 98.0% |
| k = 15 | 91.0% | 93.0% | 98.0% |
| ResNet-50 | CNN+SVM | CNN+kNN |
|---|---|---|
| (Best Performing (Table 3) | (Our Proposed Model) | |
| 1.25 ms | 1.05 ms | 1.02 ms |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Hussaeni, K.; Karamitsos, I.; Adewumi, E.; Amawi, R.M. CNN-Based Pill Image Recognition for Retrieval Systems. Appl. Sci. 2023, 13, 5050. https://doi.org/10.3390/app13085050
Al-Hussaeni K, Karamitsos I, Adewumi E, Amawi RM. CNN-Based Pill Image Recognition for Retrieval Systems. Applied Sciences. 2023; 13(8):5050. https://doi.org/10.3390/app13085050
Chicago/Turabian StyleAl-Hussaeni, Khalil, Ioannis Karamitsos, Ezekiel Adewumi, and Rema M. Amawi. 2023. "CNN-Based Pill Image Recognition for Retrieval Systems" Applied Sciences 13, no. 8: 5050. https://doi.org/10.3390/app13085050
APA StyleAl-Hussaeni, K., Karamitsos, I., Adewumi, E., & Amawi, R. M. (2023). CNN-Based Pill Image Recognition for Retrieval Systems. Applied Sciences, 13(8), 5050. https://doi.org/10.3390/app13085050





_Karamitsos.jpg)


