Deep Learning-Based ECG Arrhythmia Classification: A Systematic Review
Abstract
1. Introduction
- We perform a systematic review for DL-based arrhythmia classification with ECG signals from perspectives of ECG database, preprocessing, DL methodology, evaluation paradigm, and performance metrics in the complete DL workflow as well as the code availability of the reviewed studies;
- The trend of techniques in each perspective in recent years is analyzed to summarize the historical road map and illustrate possible future research directions;
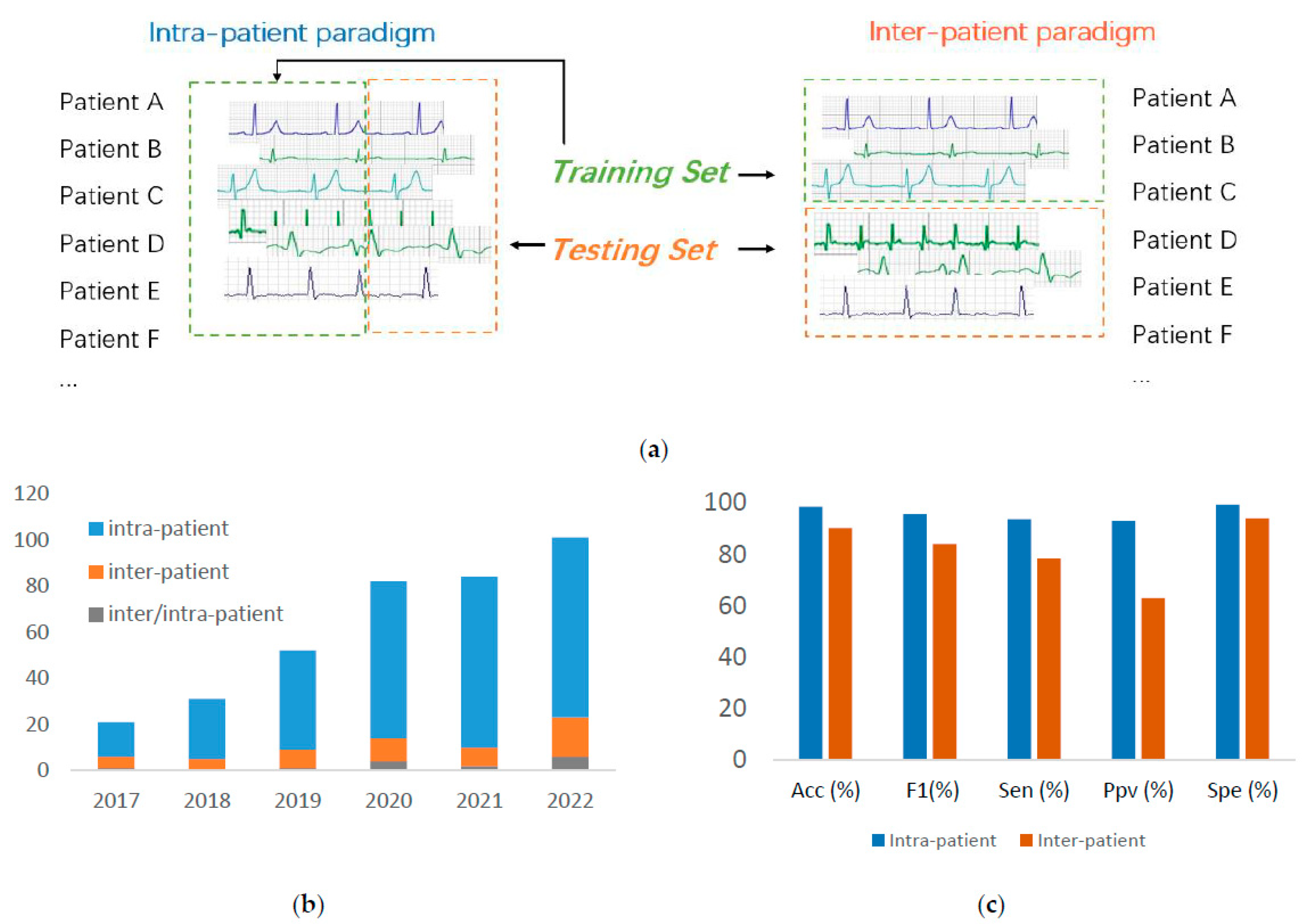
- We present the detailed performance gap between the ECG arrhythmia classification under intra- and inter-patient paradigms.
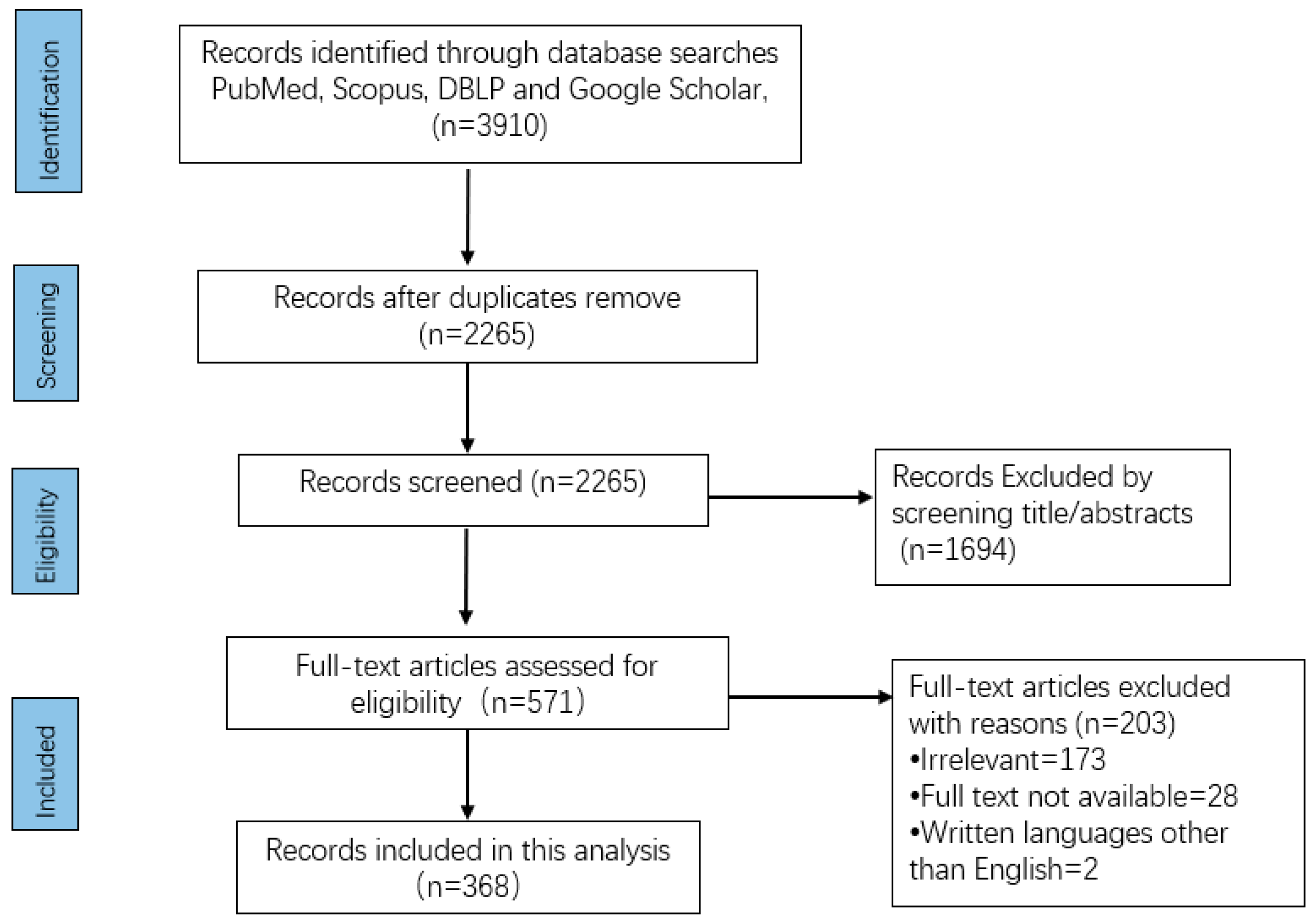
2. Materials and Methods
2.1. Search Strategy
2.2. Data Extraction
- General Information: An overview of the origin of the selected studies, i.e., the conference proceedings or journals in which they are published and their publication years, is provided;
- ECG Database: The publication information, ECG signal information, and demographic information are analyzed for popular ECG databases employed for arrhythmia classification;
- Preprocessing: Two types of commonly used preprocessing techniques, i.e., denoising to remove artifacts and data augmentation to deal with imbalanced datasets, are summarized;
- DL Methodology: The DL algorithms from all the selected studies are investigated and summarized. The information about the types of DL models, optimization techniques, and classification categories for arrhythmia is presented;
- Evaluation Paradigm: The data-driven ECG diagnosis can be categorized into intra- and inter-patient paradigms depending on how the training and testing ECG data from patients are organized;
- Performance Metric: In addition to the widely used performance metrics such as overall accuracy, other metrics such as sensitivity (Sen), positive predictivity (Ppv), false positive rate (FPR) and score of the selected studies is discussed;
- Code Availability: Detailed information about studies that publish their code and the source of the code is listed.
3. Results
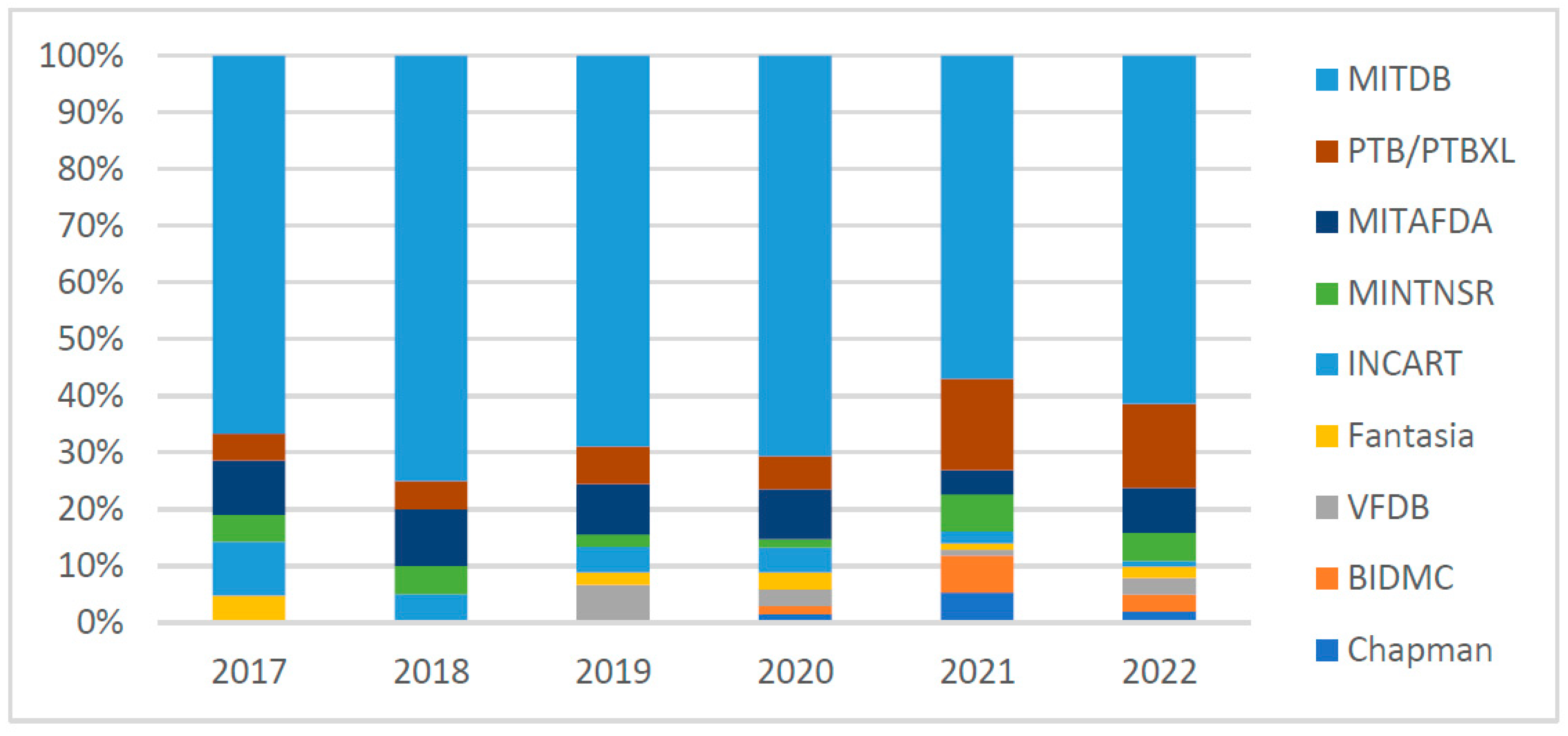
3.1. Database
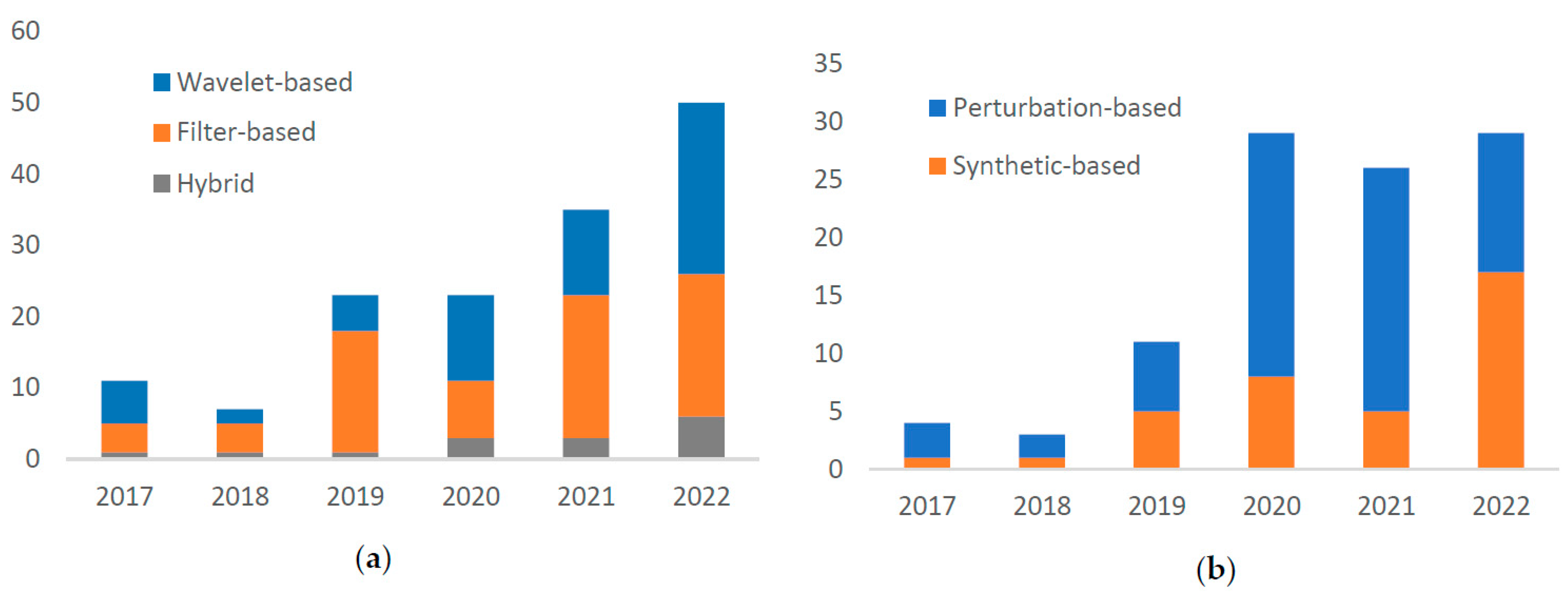
3.2. Preprocessing
3.2.1. Denosing
3.2.2. Data Augmentation
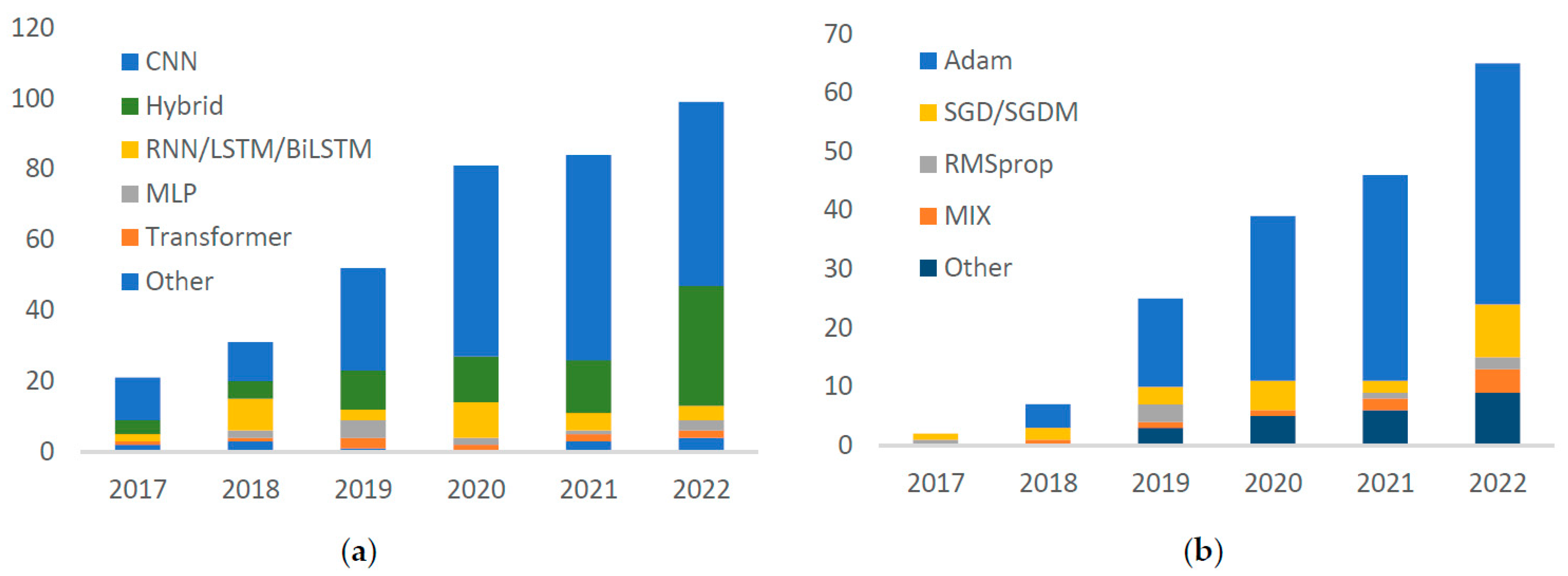
3.3. DL Methodology
3.3.1. Model
- CNN
- RNN/LSTM/BiLSTM
- Transformer
- Hybrid DL model
3.3.2. Optimizer
3.3.3. Classification Categories
3.4. Evaluation Paradigm
3.5. Performance Metrics
3.6. Code Availability
4. Discussion
4.1. ECG Database
4.2. Preprocessing
4.3. DL Methodology
4.4. Evaluation Paradigm
4.5. Performance Metrics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ebrahimi, Z.; Loni, M.; Daneshtalab, M.; Gharehbaghi, A. A review on deep learning methods for ECG arrhythmia classification. Expert Syst. Appl. X 2020, 7, 100033. [Google Scholar] [CrossRef]
- Hong, S.; Zhou, Y.; Shang, J.; Xiao, C.; Sun, J. Opportunities and challenges of deep learning methods for electrocardiogram data: A systematic review. Comput. Biol. Med. 2020, 122, 103801. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Wulan, N.; Wang, K.; Zhang, H. Detecting atrial fibrillation by deep convolutional neural networks. Comput. Biol. Med. 2018, 93, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.S.; Mak, M.-W.; Cheung, C.-C. Towards End-to-End ECG Classification With Raw Signal Extraction and Deep Neural Networks. IEEE J. Biomed. Heal Inform. 2018, 23, 1574–1584. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, H.; Li, Z.; Qin, L. Deep learning in ECG diagnosis: A review. Knowl.-Based Syst. 2021, 227, 107187. [Google Scholar] [CrossRef]
- Egger, J.; Gsaxner, C.; Pepe, A.; Pomykala, K.L.; Jonske, F.; Kurz, M.; Li, J.; Kleesiek, J. Medical deep learning—A systematic meta-review. Comput. Methods Programs Biomed. 2022, 221, 106874. [Google Scholar] [CrossRef]
- Siontis, K.C.; Noseworthy, P.A.; Attia, Z.I.; Friedman, P.A. Artificial intelligence-enhanced electrocardiography in cardiovascular disease management. Nat. Rev. Cardiol. 2021, 18, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Roy, Y.; Banville, H.; Albuquerque, I.; Gramfort, A.; Falk, T.H.; Faubert, J. Deep learning-based electroencephalography analysis: A systematic review. J. Neural Eng. 2019, 16, 051001. [Google Scholar] [CrossRef]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH Arrhythmia Database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Mark, R.G.; Moody, G.B. A new method for detecting atrial fibrillation using R-R intervals. Comput. Cardiol. 1983, 10, 227–230. [Google Scholar]
- Bousseljot, R.; Kreiseler, D.; Schnabel, A. Nutzung der EKG-Signaldatenbank CARDIODAT der PTB über das Internet. Biomed. Eng./Biomed. Tech. 1995, 40, 317–318. [Google Scholar] [CrossRef]
- Wagner, P.; Strodthoff, N.; Bousseljot, R.-D.; Kreiseler, D.; Lunze, F.I.; Samek, W.; Schaeffter, T. PTB-XL, a large publicly available electrocardiography dataset. Sci. Data 2020, 7, 154. [Google Scholar] [CrossRef] [PubMed]
- Berkaya, S.K.; Uysal, A.K.; Gunal, E.S.; Ergin, S.; Gunal, S.; Gulmezoglu, M.B. A survey on ECG analysis. Biomed. Signal Process. Control 2018, 43, 216–235. [Google Scholar] [CrossRef]
- Du, C.; Liu, P.X.; Zheng, M. Classification of Imbalanced Electrocardiosignal Data using Convolutional Neural Network. Comput. Methods Programs Biomed. 2022, 214, 106483. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Feng, X.; Yang, C. A Deep Learning Method to Detect Atrial Fibrillation Based on Continuous Wavelet Transform. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019. [Google Scholar] [CrossRef]
- Nurmaini, S.; Darmawahyuni, A.; Mukti, A.N.S.; Rachmatullah, M.N.; Firdaus, F.; Tutuko, B. Deep Learning-Based Stacked Denoising and Autoencoder for ECG Heartbeat Classification. Electronics 2020, 9, 135. [Google Scholar] [CrossRef]
- Pestana, J.; Belo, D.; Gamboa, H. Detection of Abnormalities in Electrocardiogram (ECG) using Deep Learning. In Proceedings of the BIOSIGNALS 2020—13th International Conference on Bio-Inspired Systems and Signal Processing, Part of 13th International Joint Conference on Biomedical Engineering Systems and Technologies, BIOSTEC. Valletta, Malta, 24–26 February 2020; pp. 236–243. [Google Scholar] [CrossRef]
- Liu, S.; Wang, A.; Deng, X.; Yang, C. MGNN: A multiscale grouped convolutional neural network for efficient atrial fibrillation detection. Comput. Biol. Med. 2022, 148, 105863. [Google Scholar] [CrossRef]
- Liu, Z.; Zhou, B.; Jiang, Z.; Chen, X.; Li, Y.; Tang, M.; Miao, F. Multiclass Arrhythmia Detection and Classification From Photoplethysmography Signals Using a Deep Convolutional Neural Network. J. Am. Hear Assoc. 2022, 11, 023555. [Google Scholar] [CrossRef]
- Zhao, Z.; Murphy, D.; Gifford, H.; Williams, S.; Darlington, A.; Relton, S.D.; Fang, H.; Wong, D.C. Analysis of an adaptive lead weighted ResNet for multiclass classification of 12-lead ECGs. Physiol. Meas. 2022, 43, 034001. [Google Scholar] [CrossRef]
- Mathunjwa, B.M.; Lin, Y.-T.; Lin, C.-H.; Abbod, M.F.; Shieh, J.-S. ECG arrhythmia classification by using a recurrence plot and convolutional neural network. Biomed. Signal Process. Control 2020, 64, 102262. [Google Scholar] [CrossRef]
- Tao, Y.; Li, Z.; Gu, C.; Jiang, B.; Zhang, Y. ECG-based expert-knowledge attention network to tachyarrhythmia recognition. Biomed. Signal Process. Control 2022, 76, 103649. [Google Scholar] [CrossRef]
- Oh, S.; Lee, M. A Shallow Domain Knowledge Injection (SDK-Injection) Method for Improving CNN-Based ECG Pattern Classification. Appl. Sci. 2022, 12, 1307. [Google Scholar] [CrossRef]
- Lee, B.T.; Kong, S.T.; Song, Y.; Lee, Y. Self-Supervised Learning with Electrocardiogram Delineation for Arrhythmia Detection. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, Guadalajara, Mexico, 1–5 November 2021; pp. 591–594. [Google Scholar] [CrossRef]
- Nurmaini, S.; Tondas, A.E.; Darmawahyuni, A.; Rachmatullah, M.N.; Partan, R.U.; Firdaus, F.; Tutuko, B.; Pratiwi, F.; Juliano, A.H.; Khoirani, R. Robust detection of atrial fibrillation from short-term electrocardiogram using convolutional neural networks. Futur. Gener. Comput. Syst. 2020, 113, 304–317. [Google Scholar] [CrossRef]
- Mathunjwa, B.M.; Lin, Y.-T.; Lin, C.-H.; Abbod, M.F.; Sadrawi, M.; Shieh, J.-S. ECG Recurrence Plot-Based Arrhythmia Classification Using Two-Dimensional Deep Residual CNN Features. Sensors 2022, 22, 1660. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Chen, C.; Liu, H.; Zhou, S.; Shu, M. A Deep-Learning Approach to ECG Classification Based on Adversarial Domain Adaptation. Healthcare 2020, 8, 437. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Lv, J.; Kong, D. CNN-FWS: A Model for the Diagnosis of Normal and Abnormal ECG with Feature Adaptive. Entropy 2022, 24, 471. [Google Scholar] [CrossRef]
- Li, X.; Zhang, F.; Sun, Z.; Li, D.; Kong, X.; Zhang, Y. Automatic heartbeat classification using S-shaped reconstruction and a squeeze-and-excitation residual network. Comput. Biol. Med. 2021, 140, 105108. [Google Scholar] [CrossRef]
- Wang, J. A deep learning approach for atrial fibrillation signals classification based on convolutional and modified Elman neural network. Futur. Gener. Comput. Syst. 2019, 102, 670–679. [Google Scholar] [CrossRef]
- Luo, K.; Li, J.; Wang, Z.; Cuschieri, A. Patient-Specific Deep Architectural Model for ECG Classification. J. Healthc. Eng. 2017, 2017, 4108720. [Google Scholar] [CrossRef]
- Alkhodari, M.; Apostolidis, G.; Zisou, C.; Hadjileontiadis, L.J.; Khandoker, A.H. Swarm Decomposition Enhances the Discrimination of Cardiac Arrhythmias in Varied-Lead ECG Using ResNet-BiLSTM Network Activations. In Proceedings of the 2021 Computing in Cardiology (CinC), Brno, Czech Republic, 13–15 September 2021. [Google Scholar] [CrossRef]
- Lui, H.W.; Chow, K.L. Multiclass classification of myocardial infarction with convolutional and recurrent neural networks for portable ECG devices. Inform. Med. Unlocked 2018, 13, 26–33. [Google Scholar] [CrossRef]
- Eltrass, A.S.; Tayel, M.B.; Ammar, A.I. Automated ECG multi-class classification system based on combining deep learning features with HRV and ECG measures. Neural. Comput. Appl. 2022, 34, 8755–8775. [Google Scholar] [CrossRef]
- Eltrass, A.S.; Tayel, M.B.; Ammar, A.I. A new automated CNN deep learning approach for identification of ECG congestive heart failure and arrhythmia using constant-Q non-stationary Gabor transform. Biomed. Signal Process. Control 2020, 65, 102326. [Google Scholar] [CrossRef]
- Zhang, G.; Si, Y.; Yang, W.; Wang, D. A Robust Multilevel DWT Densely Network for Cardiovascular Disease Classification. Sensors 2020, 20, 4777. [Google Scholar] [CrossRef] [PubMed]
- Degirmenci, M.; Ozdemir, M.; Izci, E.; Akan, A. Arrhythmic Heartbeat Classification Using 2D Convolutional Neural Networks. IRBM 2021, 43, 422–433. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, J.; Liu, Y.; Qin, C.; Li, Z.; Xiao, D.; Zhao, L.; Liu, C. A Novel Interpretable Method Based on Dual-Level Attentional Deep Neural Network for Actual Multilabel Arrhythmia Detection. IEEE Trans. Instrum. Meas. 2021, 71, 2500311. [Google Scholar] [CrossRef]
- Zhong, Z.; Zheng, L.; Kang, G.; Li, S.; Yang, Y. Random Erasing Data Augmentation. In Proceedings of the AAAI 2020—34th AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; pp. 13001–13008. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, S.; Yuan, X.; Zhang, P. Interpretable deep learning for automatic diagnosis of 12-lead electrocardiogram. iScience 2021, 24, 102373. [Google Scholar] [CrossRef] [PubMed]
- Prabhakararao, E.; Dandapat, S. Multi-Scale Convolutional Neural Network Ensemble for Multi-Class Arrhythmia Classification. IEEE J. Biomed. Health Inform. 2021, 26, 3802–3812. [Google Scholar] [CrossRef]
- He, J.; Rong, J.; Sun, L.; Wang, H.; Zhang, Y.; Ma, J. A framework for cardiac arrhythmia detection from IoT-based ECGs. World Wide Web 2020, 23, 2835–2850. [Google Scholar] [CrossRef]
- Nankani, D.; Baruah, R.D. Ventricular Arrhythmia Classification and Interpretation Using Residual Neural Network with Guided Backpropagation. In Proceedings of the IEEE Region 10 Annual International Conference, Proceedings/TENCON, Auckland, New Zealand, 7–10 December 2021; pp. 574–579. [Google Scholar] [CrossRef]
- Li, X.; Zhang, J.; Chen, W.; Chen, Y.; Zhang, C.; Xiang, W.; Li, D. Inter-patient automated arrhythmia classification: A new approach of weight capsule and sequence to sequence combination. Comput. Methods Programs Biomed. 2021, 214, 106533. [Google Scholar] [CrossRef]
- Singh, P.; Sharma, A. Interpretation and Classification of Arrhythmia using Deep Convolutional Network. IEEE Trans. Instrum. Meas. 2022, 71, 2518512. [Google Scholar] [CrossRef]
- Gatys, L.A.; Ecker, A.S.; Bethge, M. Image Style Transfer Using Convolutional Neural Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2414–2423. [Google Scholar]
- Shaker, A.M.; Tantawi, M.; Shedeed, H.A.; Tolba, M.F. Generalization of Convolutional Neural Networks for ECG Classification Using Generative Adversarial Networks. IEEE Access 2020, 8, 35592–35605. [Google Scholar] [CrossRef]
- Ganguly, B.; Ghosal, A.; Das, A.; Das, D.; Chatterjee, D.; Rakshit, D. Automated Detection and Classification of Arrhythmia From ECG Signals Using Feature-Induced Long Short-Term Memory Network. IEEE Sens. Lett. 2020, 4, 6001604. [Google Scholar] [CrossRef]
- Pandey, S.K.; Janghel, R.R. Automatic detection of arrhythmia from imbalanced ECG database using CNN model with SMOTE. Australas. Phys. Eng. Sci. Med. 2019, 42, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-Time Patient-Specific ECG Classification by 1-D Convolutional Neural Networks. IEEE Trans. Biomed. Eng. 2015, 63, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Tesfai, H.; Saleh, H.; Al-Qutayri, M.; Mohammad, M.B.; Tekeste, T.; Khandoker, A.; Mohammad, B. Lightweight Shufflenet Based CNN for Arrhythmia Classification. IEEE Access 2022, 10, 111842–111854. [Google Scholar] [CrossRef]
- Mostayed, A.; Luo, J.; Shu, X.; Wee, W. Classification of 12-Lead ECG Signals with Bi-Directional LSTM Network. 2018. Available online: http://arxiv.org/abs/1811.02090 (accessed on 26 December 2022).
- Mashrur, F.R.; Roy, A.D.; Saha, D.K. Automatic Identification of Arrhythmia from ECG Using AlexNet Convolutional Neural Network. In Proceedings of the 2019 4th International Conference on Electrical Information and Communication Technology, EICT 2019, Khulna, Bangladesh, 20–22 December 2019. [Google Scholar] [CrossRef]
- de Santana, J.R.G.; Costa, M.G.F.; Filho, C.F.F.C. A New Approach to Classify Cardiac Arrythmias Using 2D Convolutional Neural Networks. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, Auckland, New Zealand, 7–10 December 2021; pp. 566–570. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, M.; Zhang, Y.; Liao, Y.; Huang, Q.; Chang, S.; Wang, H.; He, J. Real-Time Multilead Convolutional Neural Network for Myocardial Infarction Detection. IEEE J. Biomed. Heal Inform. 2017, 22, 1434–1444. [Google Scholar] [CrossRef]
- Singh, S.; Pandey, S.K.; Pawar, U.; Janghel, R.R. Classification of ECG Arrhythmia using Recurrent Neural Networks. Procedia Comput. Sci. 2018, 132, 1290–1297. [Google Scholar] [CrossRef]
- Khan, M.A.; Kim, Y. Cardiac Arrhythmia Disease Classification Using LSTM Deep Learning Approach. Comput. Mater. Contin. 2021, 67, 427–443. [Google Scholar] [CrossRef]
- Zhou, X.; Zhu, X.; Nakamura, K.; Mahito, N. Premature Ventricular Contraction Detection from Ambulatory ECG Using Recurrent Neural Networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018. [Google Scholar] [CrossRef]
- Xu, W.; Wang, L.; Wang, B.; Cheng, W. Intelligent Recognition Algorithm of Multiple Myocardial Infarction Based on Morphological Feature Extraction. Processes 2022, 10, 2348. [Google Scholar] [CrossRef]
- Rahul, J.; Sharma, L.D. Artificial intelligence-based approach for atrial fibrillation detection using normalised and short-duration time-frequency ECG. Biomed. Signal Process. Control 2021, 71, 103270. [Google Scholar] [CrossRef]
- Faust, O.; Shenfield, A.; Kareem, M.; San, T.R.; Fujita, H.; Acharya, U.R. Automated detection of atrial fibrillation using long short-term memory network with RR interval signals. Comput. Biol. Med. 2018, 102, 327–335. [Google Scholar] [CrossRef]
- Jiang, M.; Gu, J.; Li, Y.; Wei, B.; Zhang, J.; Wang, Z.; Xia, L. HADLN: Hybrid Attention-Based Deep Learning Network for Automated Arrhythmia Classification. Front. Physiol. 2021, 12, 683025. [Google Scholar] [CrossRef]
- Hu, R.; Chen, J.; Zhou, L. A transformer-based deep neural network for arrhythmia detection using continuous ECG signals. Comput. Biol. Med. 2022, 144, 105325. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Gan, C.; Li, Z.; Rekik, I.; Yin, Z.; Ji, W.; Gao, Y.; Wang, Q.; Zhang, J.; Shen, D. Transformers in medical image analysis. Intell. Med. 2023, 3, 59–78. [Google Scholar] [CrossRef]
- Yan, G.; Liang, S.; Zhang, Y.; Liu, F. Fusing Transformer Model with Temporal Features for ECG Heartbeat Classification. In Proceedings of the—2019 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2019, San Diego, CA, USA, 18–21 November 2019; pp. 898–905. [Google Scholar] [CrossRef]
- Natarajan, A.; Chang, Y.; Mariani, S.; Rahman, A.; Boverman, G.; Vij, S.; Rubin, J. A Wide and Deep Transformer Neural Network for 12-Lead ECG Classification. Comput. Cardiol. 2020, 2020, 20350657. [Google Scholar] [CrossRef]
- Natarajan, A.; Boverman, G.; Chang, Y.; Antonescu, C.; Rubin, J. Convolution-Free Waveform Transformers for Multi-Lead ECG Classification. In Proceedings of the 2021 Computing in Cardiology (CinC), Brno, Czech Republic, 13–15 September 2021. [Google Scholar] [CrossRef]
- Mousavi, S.; Afghah, F. Inter-and intra-patient ecg heartbeat classification for arrhythmia detection: A sequence to sequence deep learning approach. In Proceedings of the ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1308–1312. [Google Scholar]
- Swapna, G.; Soman, K.P.; Vinayakumar, R. Automated detection of cardiac arrhythmia using deep learning techniques. Procedia Comput. Sci. 2018, 132, 1192–1201. [Google Scholar] [CrossRef]
- Shoughi, A.; Dowlatshahi, M.B. A practical system based on CNN-BLSTM network for accurate classification of ECG heartbeats of MIT-BIH imbalanced dataset. In Proceedings of the 26th International Computer Conference, Computer Society of Iran, CSICC 2021, Tehran, Iran, 3–4 March 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Xiong, Z.; Stiles, M.K.; Gillis, A.M.; Zhao, J. Enhancing the detection of atrial fibrillation from wearable sensors with neural style transfer and convolutional recurrent networks. Comput. Biol. Med. 2022, 146, 105551. [Google Scholar] [CrossRef]
- Meng, L.; Tan, W.; Ma, J.; Wang, R.; Yin, X.; Zhang, Y. Enhancing dynamic ECG heartbeat classification with lightweight transformer model. Artif. Intell. Med. 2022, 124, 102236. [Google Scholar] [CrossRef]
- Yoo, J.; Jun, T.J.; Kim, Y.-H. xECGNet: Fine-tuning attention map within convolutional neural network to improve detection and explainability of concurrent cardiac arrhythmias. Comput. Methods Programs Biomed. 2021, 208, 106281. [Google Scholar] [CrossRef]
- Li, J.; Pang, S.-P.; Xu, F.; Ji, P.; Zhou, S.; Shu, M. Two-dimensional ECG-based cardiac arrhythmia classification using DSE-ResNet. Sci. Rep. 2022, 12, 216. [Google Scholar] [CrossRef]
- Srivastava, A.; Pratiher, S.; Alam, S.; Hari, A.; Banerjee, N.; Ghosh, N.; Patra, A. A deep residual inception network with channel attention modules for multi-label cardiac abnormality detection from reduced-lead ECG. Physiol. Meas. 2022, 43, 064005. [Google Scholar] [CrossRef]
- Nejedly, P.; Ivora, A.; Smisek, R.; Viscor, I.; Koscova, Z.; Jurak, P.; Plesinger, F. Classification of ECG Using Ensemble of Residual CNNs with Attention Mechanism. Comput. Cardiol. 2021, 48, 9662723. [Google Scholar] [CrossRef]
- de Chazal, P.; O’Dwyer, M.; Reilly, R. Automatic Classification of Heartbeats Using ECG Morphology and Heartbeat Interval Features. IEEE Trans. Biomed. Eng. 2004, 51, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Rong, J.; Sun, L.; Wang, H.; Zhang, Y. An Advanced Two-Step DNN-Based Framework for Arrhythmia Detection. Lect. Notes Comput. Sci. 2020, 12085, 422–434. [Google Scholar] [CrossRef]
- Tutuko, B.; Nurmaini, S.; Tondas, A.E.; Rachmatullah, M.N.; Darmawahyuni, A.; Esafri, R.; Firdaus, F.; Sapitri, A.I. AFibNet: An implementation of atrial fibrillation detection with convolutional neural network. BMC Med. Inform. Decis. Mak. 2021, 21, 216. [Google Scholar] [CrossRef]
- Katsushika, S.; Kodera, S.; Nakamoto, M.; Ninomiya, K.; Inoue, S.; Sawano, S.; Kakuda, N.; Takiguchi, H.; Shinohara, H.; Matsuoka, R.; et al. The Effectiveness of a Deep Learning Model to Detect Left Ventricular Systolic Dysfunction from Electrocardiograms. Int. Hear J. 2021, 62, 1332–1341. [Google Scholar] [CrossRef]
- Andersen, R.S.; Peimankar, A.; Puthusserypady, S. A deep learning approach for real-time detection of atrial fibrillation. Expert Syst. Appl. 2018, 115, 465–473. [Google Scholar] [CrossRef]
- Gan, Y.; Shi, J.-C.; He, W.-M.; Sun, F.-J. Parallel classification model of arrhythmia based on DenseNet-BiLSTM. Biocybern. Biomed. Eng. 2021, 41, 1548–1560. [Google Scholar] [CrossRef]
- Sellami, A.; Hwang, H. A robust deep convolutional neural network with batch-weighted loss for heartbeat classification. Expert Syst. Appl. 2018, 122, 75–84. [Google Scholar] [CrossRef]
- Tan, J.H.; Hagiwara, Y.; Pang, W.; Lim, I.; Oh, S.L.; Adam, M.; Tan, R.S.; Chen, M.; Acharya, U.R. Application of stacked convolutional and long short-term memory network for accurate identification of CAD ECG signals. Comput. Biol. Med. 2018, 94, 19–26. [Google Scholar] [CrossRef]
- Jangra, M.; Dhull, S.K.; Singh, K.K.; Singh, A.; Cheng, X. O-WCNN: An optimized integration of spatial and spectral feature map for arrhythmia classification. Complex Intell. Syst. 2021, 2021, 1–14. [Google Scholar] [CrossRef]
- Chen, A.; Wang, F.; Liu, W.; Chang, S.; Wang, H.; He, J.; Huang, Q. Multi-information fusion neural networks for arrhythmia automatic detection. Comput. Methods Programs Biomed. 2020, 193, 105479. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Lu, B.; Nie, B.; Peng, Z.; Liu, H.; Pi, X. Hybrid Network with Attention Mechanism for Detection and Location of Myocardial Infarction Based on 12-Lead Electrocardiogram Signals. Sensors 2020, 20, 1020. [Google Scholar] [CrossRef] [PubMed]
- Zubair, M.; Yoon, C. Cost-Sensitive Learning for Anomaly Detection in Imbalanced ECG Data Using Convolutional Neural Networks. Sensors 2022, 22, 4075. [Google Scholar] [CrossRef]
- Ran, S.; Yang, X.; Liu, M.; Zhang, Y.; Cheng, C.; Zhu, H.; Yuan, Y. Homecare-Oriented ECG Diagnosis with Large-Scale Deep Neural Network for Continuous Monitoring on Embedded Devices. IEEE Trans. Instrum. Meas. 2022, 71, 1–13. [Google Scholar] [CrossRef]
- Rizqyawan, M.I.; Siradj, Y.; Amri, M.F.; Pratondo, A. Re-implementation of Convolutional Neural Network for Arrhythmia Detection. Int. J. Adv. Sci. Eng. Inf. Technol. 2022, 12, 1319–1326. [Google Scholar] [CrossRef]
- Essa, E.; Xie, X. An Ensemble of Deep Learning-Based Multi-Model for ECG Heartbeats Arrhythmia Classification. IEEE Access 2021, 9, 103452–103464. [Google Scholar] [CrossRef]
- Li, Y.; Qian, R.; Li, K. Inter-patient arrhythmia classification with improved deep residual convolutional neural network. Comput. Methods Programs Biomed. 2022, 214, 106582. [Google Scholar] [CrossRef] [PubMed]
- Labib, M.I.; Nahid, A.-A. OptRPC: A novel and optimized recurrence plot-based system for ECG beat classification. Biomed. Signal Process. Control 2021, 72, 103328. [Google Scholar] [CrossRef]
- Pokaprakarn, T.; Kitzmiller, R.R.; Moorman, J.R.; Lake, D.E.; Krishnamurthy, A.K.; Kosorok, M.R. Sequence to Sequence ECG Cardiac Rhythm Classification Using Convolutional Recurrent Neural Networks. IEEE J. Biomed. Health Inform. 2021, 26, 572–580. [Google Scholar] [CrossRef]
- Sarshar, N.T.; Mirzaei, M. Premature Ventricular Contraction Recognition Based on a Deep Learning Approach. J. Heal Eng. 2022, 2022, 1450723. [Google Scholar] [CrossRef]
- Wang, G.; Chen, M.; Ding, Z.; Li, J.; Yang, H.; Zhang, P. Inter-patient ECG arrhythmia heartbeat classification based on unsupervised domain adaptation. Neurocomputing 2021, 454, 339–349. [Google Scholar] [CrossRef]
- Pandey, S.K.; Janghel, R.R.; Dev, A.V.; Mishra, P.K. Automated arrhythmia detection from electrocardiogram signal using stacked restricted Boltzmann machine model. SN Appl. Sci. 2021, 3, 624. [Google Scholar] [CrossRef]
- Qiu, X.; Liang, S.; Meng, L.; Zhang, Y.; Liu, F. Exploiting feature fusion and long-term context dependencies for simultaneous ECG heartbeat segmentation and classification. Int. J. Data Sci. Anal. 2021, 11, 181–193. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, A.; Liang, D.; Chen, X.; Gao, M. Interpatient ECG Heartbeat Classification with an Adversarial Convolutional Neural Network. J. Health Eng. 2021, 2021, 9946596. [Google Scholar] [CrossRef]
- Cai, J.; Zhou, G.; Dong, M.; Hu, X.; Liu, G.; Ni, W. Real-Time Arrhythmia Classification Algorithm Using Time-Domain ECG Feature Based on FFNN and CNN. Math. Probl. Eng. 2021, 2021, 6648432. [Google Scholar] [CrossRef]
- Wang, T.; Lu, C.; Sun, Y.; Yang, M.; Liu, C.; Ou, C. Automatic ECG Classification Using Continuous Wavelet Transform and Convolutional Neural Network. Entropy 2021, 23, 119. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Zhang, N.; Meng, Y.; Qin, Z.; Lu, Q.; Liu, X. ECG Heartbeat Classification Detection Based on WaveNet-LSTM. In Proceedings of the 2020 IEEE 4th International Conference on Frontiers of Sensors Technologies (ICFST), Shanghai, China, 6–9 November 2020. [Google Scholar] [CrossRef]
- Li, Y.; He, Z.; Wang, H.; Li, B.; Li, F.; Gao, Y.; Ye, X. CraftNet: A deep learning ensemble to diagnose cardiovascular diseases. Biomed. Signal Process. Control 2020, 62, 102091. [Google Scholar] [CrossRef]
- Zhao, W.; Hu, J.; Jia, D.; Wang, H.; Li, Z.; Yan, C.; You, T. Deep Learning Based Patient-Specific Classification of Arrhythmia on ECG signal. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 1500–1503. [Google Scholar] [CrossRef]
- Xie, Q.; Tu, S.; Wang, G.; Lian, Y.; Xu, L. Feature Enrichment Based Convolutional Neural Network for Heartbeat Classification From Electrocardiogram. IEEE Access 2019, 7, 153751–153760. [Google Scholar] [CrossRef]
- Zhou, R.; Li, X.; Yong, B.; Shen, Z.; Wang, C.; Zhou, Q.; Cao, Y.; Li, K.-C. Arrhythmia recognition and classification through deep learning-based approach Arrhythmia recognition and classification through deep learning-based approach 507. Arrhythmias 2019, 19, 506. [Google Scholar]
- Mathews, S.M.; Kambhamettu, C.; Barner, K.E. A novel application of deep learning for single-lead ECG classification. Comput. Biol. Med. 2018, 99, 53–62. [Google Scholar] [CrossRef]
- Hu, J.; Zhao, W.; Jia, D.; Yan, C.; Wang, H.; Li, Z.; Fang, J.; Yang, M. Deep Multi-instance Networks for Bundle Branch Block Detection from Multi-lead ECG. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020. [Google Scholar] [CrossRef]
- Guo, L.; Sim, G.; Matuszewski, B. Inter-patient ECG classification with convolutional and recurrent neural networks. Biocybern. Biomed. Eng. 2019, 39, 868–879. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Hannun, A.Y.; Haghpanahi, M.; Bourn, C.; Ng, A.Y. Cardiologist-Level Arrhythmia Detection with Convolutional Neural Networks. 2017. Available online: http://arxiv.org/abs/1707.01836 (accessed on 6 July 2017).
- Ribeiro, A.H.; Ribeiro, M.H.; Paixão, G.M.M.; Oliveira, D.M.; Gomes, P.R.; Canazart, J.A.; Ferreira, M.P.S.; Andersson, C.R.; Macfarlane, P.W.; Meira, W.; et al. Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat. Commun. 2020, 11, 1760. [Google Scholar] [CrossRef] [PubMed]
- Bollepalli, S.C.; Sevakula, R.K.; Au-Yeung, W.M.; Kassab, M.B.; Merchant, F.M.; Bazoukis, G.; Boyer, R.; Isselbacher, E.M.; Armoundas, A.A. Real-Time Arrhythmia Detection Using Hybrid Convolutional Neural Networks. J. Am. Hear Assoc. 2021, 10, 023222. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liang, L. A regularization method to improve adversarial robustness of neural networks for ECG signal classification. Comput. Biol. Med. 2022, 144, 105345. [Google Scholar] [CrossRef] [PubMed]
- Bazargani, M.H.Z.; Pakrashi, A.; Mac Namee, B. The Deep Radial Basis Function Data Descriptor (D-RBFDD) Network: A One-Class Neural Network for Anomaly Detection. IEEE Access 2022, 10, 70645–70661. [Google Scholar] [CrossRef]
- Huang, Y.; Yen, G.G.; Tseng, V.S. Snippet Policy Network V2: Knee-Guided Neuroevolution for Multi-Lead ECG Early Classification. IEEE Trans. Neural. Netw. Learn. Syst. 2022, 99, 3187741. [Google Scholar] [CrossRef] [PubMed]
- Irfan, S.; Anjum, N.; Althobaiti, T.; Alotaibi, A.A.; Siddiqui, A.B.; Ramzan, N. Heartbeat Classification and Arrhythmia Detection Using a Multi-Model Deep-Learning Technique. Sensors 2022, 22, 5606. [Google Scholar] [CrossRef] [PubMed]
- Murat, F.; Yildirim, O.; Talo, M.; Demir, Y.; Tan, R.-S.; Ciaccio, E.J.; Acharya, U.R. Exploring deep features and ECG attributes to detect cardiac rhythm classes. Knowl.-Based Syst. 2021, 232, 107473. [Google Scholar] [CrossRef]
- Radhakrishnan, T.; Karhade, J.; Ghosh, S.; Muduli, P.; Tripathy, R.; Acharya, U.R. AFCNNet: Automated detection of AF using chirplet transform and deep convolutional bidirectional long short term memory network with ECG signals. Comput. Biol. Med. 2021, 137, 104783. [Google Scholar] [CrossRef]
- Liang, Y.; Yin, S.; Tang, Q.; Zheng, Z.; Elgendi, M.; Chen, Z. Deep Learning Algorithm Classifies Heartbeat Events Based on Electrocardiogram Signals. Front. Physiol. 2020, 11, 569050. [Google Scholar] [CrossRef]
- Chen, T.-M.; Huang, C.-H.; Shih, E.S.; Hu, Y.-F.; Hwang, M.-J. Detection and Classification of Cardiac Arrhythmias by a Challenge-Best Deep Learning Neural Network Model. iScience 2020, 23, 100886. [Google Scholar] [CrossRef] [PubMed]
- Hannun, A.Y.; Rajpurkar, P.; Haghpanahi, M.; Tison, G.H.; Bourn, C.; Turakhia, M.P.; Ng, A.Y. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019, 25, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Plesinger, F.; Nejedly, P.; Viscor, I.; Halamek, J.; Jurak, P. Parallel use of a convolutional neural network and bagged tree ensemble for the classification of Holter ECG. Physiol. Meas. 2018, 39, 094002. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Liang, G.; Yue, M. Deep Learning-Based Arrhythmia Detection in Electrocardiograph. Sci. Program. 2021, 2021, 9926769. [Google Scholar] [CrossRef]
- Ma, S.; Cui, J.; Xiao, W.; Liu, L. Deep Learning-Based Data Augmentation and Model Fusion for Automatic Arrhythmia Identification and Classification Algorithms. Comput. Intell. Neurosci. 2022, 2022, 1577778. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Dharahas, G.; Gite, S.; Kotecha, K.; Koundal, D.; Zaguia, A.; Kaur, M.; Lee, H.-N. ECG Data Analysis with Denoising Approach and Customized CNNs. Sensors 2022, 22, 1928. [Google Scholar] [CrossRef] [PubMed]
- Rath, A.; Mishra, D.; Panda, G.; Satapathy, S.C. Heart disease detection using deep learning methods from imbalanced ECG samples. Biomed. Signal Process. Control 2021, 68, 102820. [Google Scholar] [CrossRef]
- Petmezas, G.; Haris, K.; Stefanopoulos, L.; Kilintzis, V.; Tzavelis, A.; Rogers, J.A.; Katsaggelos, A.K.; Maglaveras, N. Automated Atrial Fibrillation Detection using a Hybrid CNN-LSTM Network on Imbalanced ECG Datasets. Biomed. Signal Process. Control 2020, 63, 102194. [Google Scholar] [CrossRef]
- Obeidat, Y.; Alqudah, A.M. A Hybrid Lightweight 1D CNN-LSTM Architecture for Automated ECG Beat-Wise Classification. Traitement Signal 2021, 38, 1281–1291. [Google Scholar] [CrossRef]
- Chen, C.; Hua, Z.; Zhang, R.; Liu, G.; Wen, W. Automated arrhythmia classification based on a combination network of CNN and LSTM. Biomed. Signal Process. Control 2019, 57, 101819. [Google Scholar] [CrossRef]
- Oh, S.L.; Ng, E.Y.; San Tan, R.; Acharya, U.R. Automated diagnosis of arrhythmia using combination of CNN and LSTM techniques with variable length heart beats. Comput. Biol. Med. 2018, 102, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Wang, Y.; Chen, H.; Chen, X.; Guo, J.; Liu, Z.; Tang, Y.; Xiao, A.; Xu, C.; Xu, Y.; et al. A Survey on Vision Transformer. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 45, 87–110. [Google Scholar] [CrossRef] [PubMed]
- Tolstikhin, I.; Houlsby, N.; Kolesnikov, A.; Beyer, L.; Zhai, X.; Unterthiner, T.; Yung, J.; Steiner, A.; Keysers, D.; Uszkoreit, J.; et al. MLP-Mixer: An all-MLP Architecture for Vision. Adv. Neural. Inf. Process. Syst. 2021, 34, 24261–24272. [Google Scholar]
- He, Z.; Yuan, S.; Zhao, J.; Du, B.; Yuan, Z.; Alhudhaif, A.; Alenezi, F.; Althubiti, S.A. A novel myocardial infarction localization method using multi-branch DenseNet and spatial matching-based active semi-supervised learning. Inf. Sci. 2022, 606, 649–668. [Google Scholar] [CrossRef]
- Khajuria, R.; Sarwar, A. Reinforcement Learning in Medical Diagnosis: An Overview. Lect. Notes Electr. Eng. 2022, 832, 179–188. [Google Scholar] [CrossRef]
| Extracted Items | Definition |
|---|---|
| |
| Origin | Journal/conference where the articles were published. |
| Publication year | Years of selected studies were published in. |
| |
| Publication information | Source, release year, and whether the database is public available or not. |
| Signal information | Number of channels, sampling rate, signal duration, subject, and records. |
| Demographic information | Information about the characteristics of subjects, e.g., gender, age. |
| |
| Denoising | Denoising techniques. |
| Data Augmentation | Methods to balance data categories. |
| |
| Model | Deep learning architecture or framework. |
| Optimization | Techniques to optimize the model learnable weights. |
| Category | Number of categories of the DL models. |
| Whether training and testing datasets contain ECG data from the same patients or not. |
| Metrics to evaluate the classification performance, e.g., F1, Sp. |
| Whether the code is shared online or not. |
| Database | Publicly Available | Release Year | No. Channels | Sampling Rate (Hz) | Duration | Subjects | Collection Place | No. of Records | Demographic Information | Papers |
|---|---|---|---|---|---|---|---|---|---|---|
| MIT-BIH Arrhythmia Database (MITDB) | Yes | 2005 | 2 | 360 | 30 min | 47 | USA | 48 | Male: 25, female: 22 (23–89 years old) | 223 |
| MIT-BIH Atrial Fibrillation Database (AFDB) | Yes | 2000 | 2 | 250 | 10 h | 25 | USA | 23 | Subjects are suffering from atrial fibrillation | 26 |
| PTB Diagnostic ECG Database (PTB) | Yes | 2004 | 15 | 1000 | 2 min | 290 | Germany | 549 | Male: 377, female: 139 | 24 |
| PTB-XL ECG dataset (PTBXL) | Yes | 2020 | 12 | 500 | 10 s | 18,885 | Germany | 21,837 | Male: 9820; female: 9065 | 15 |
| MIT-BIH Normal Sinus Rhythm (NSRDB) | Yes | 1999 | 2 | / | 24 h | 18 | USA | 18 | Male:5 (26–45 years old), female:13 (20–50 years old), both no significant arrhythmia; | 15 |
| St Petersburg INCART 12-lead Arrhythmia (INCART) | Yes | 2008 | 12 | 257 | 30 min | 32 | Russia | 75 | Male: 17, female: 15 (18–80 years old). None of the patients had pacemakers; most had ventricular ectopic beats | 11 |
| BIDMC Congestive Heart Failure (BIDMC) | Yes | 2000 | 2 | 250 | 20 h | 15 | USA | 15 | Male:11 (22–710 years old), female: 4 (54–63 years old); subjects with severe congestive heart failure. | 10 |
| MIT-BIH Malignant Ventricular Ectopy Database (VFDB) | Yes | 1999 | 2 | 250 | 30 min | 16 | USA | 22 | Subjects with episodes of sustained ventricular tachycardia, ventricular flutter, and ventricular fibrillation. | 9 |
| Chapman University and Shaoxing People’s Hospital Dataset (Chapman) | Yes | 2020 | 12 | 500 | 10 s | 10,646 | China | 10,646 | Male: 5956, female 4690 (4–98 years old). 17% of subjects had normal sinus rhythm, and 83% had at least one abnormality. | 8 |
| Fantasia | Yes | 2003 | 12 | 250 | 2 h | 40 | USA | 40 | 20 young (21–34 years old) and 20 elderly (68–86 years old) subjects | 7 |
| UCI Machine Learning Repository Arrhythmia Dataset (UCI) | Yes | 1998 | 12 | / | / | 279 | USA | 452 | 203 instances correspond to male subjects; 249 are from female subjects | 4 |
| MIT-BIH Noise Stress Test Database (NSTDB) | Yes | 1984 | 2 | 360 | 30 min | 12 | USA | 15 | Subjects are physically active volunteers | 4 |
| Creighton University Ventricular Tachyarrhythmia Database (CUDB) | Yes | 2007 | 1 | 250 | 8 min | / | USA | 35 | Subjects who experienced episodes of sustained ventricular tachycardia, ventricular flutter, and ventricular fibrillation | 4 |
| The American Heart Association database short/long (AHA) | No (2 samples available) | 1982 | 2 | 250 | 30/150 min | / | USA | 10/67 | / | 4 |
| Chinese Cardiovascular Disease Database (CCDD) | Yes | 2012 | 12 | 500 | 10 s | / | China | 90 | / | 4 |
| The QT Dataset (QT) | Yes | 1999 | 2 | 250 | 15 min | 15 | USA and Europe | 105 | Chosen primarily from among existing ECG databases | 3 |
| European ST-T | Yes | 2009 | 2 | 250 | 2 h | 79 | Europe | 90 | Male: 70 (30–84 years old), female: 8 (55–71 years old); Myocardial ischemia was diagnosed or suspected for each subject | 3 |
| Paper | Train/Validate Data | Test Data | Algorithm | Class | Performance | ||||
|---|---|---|---|---|---|---|---|---|---|
| Acc (%) | F1 (%) | Sen (%) | Ppv (%) | Spe (%) | |||||
| [82] | MITDB DS1 | MITDB DS2 | DenseNet-BiLSTM | 5: N, S, V, F, Q | 92.37 | 63.49 | 68.29 | 60.35 | 94.51 |
| [83] | MITDB DS1 | MITDB DS2 | CNN | 5: N, S, V, F, Q | 88.34 | / | 90.9 | 48.25 | 88.51 |
| [84] | Fantasia + INCART | Fantasia + INCART | CNN-LSTM | 2: N, CAD | 95.76 | 95.57 | 95.7 | / | 95.76 |
| [85] | MITDB DS1 | MITDB DS2 | O-WCNN | 5: N, S, V, F, Q | 99.43 | 92.05 | 91.06 | 93.5 | 99.69 |
| [81] | AFDB | MITDB | CNN+RNN | 2:AF; NoAF | 89.3 | / | 99.82 | 51.71 | 87.94 |
| AFDB | NSRDB | / | / | / | / | 95.01 | |||
| [86] | MITDB DS1 | MITDB DS2 | CNN+BLSTM | 5: N, S, V, F, Q | 96.77 | 77.84 | 74.89 | 81.24 | 95.16 |
| [28] | PTB-XL (by subject) | PTB-XL (by subject) | CNN-FWS | 2: N; abnormal | 90.05 | 90.2 | 88.9 | 91.5 | / |
| [87] | PTB | PTB | MLA-CNN-BiGRU | 6 | 62.94 | / | 63.97 | 63 | / |
| [88] | MITDB DS1 | MITDB DS2 | CNN | 5: N, S, V, F, Q | 96.36 | / | 70.6 | 48.1 | 96.16 |
| [89] | Tongji Hospital, China | CPSC 2018 | DCNN | 6 | / | 84.2 | 80 | / | 98 |
| [90] | MITDB DS1 | MITDB DS2 | CNN | 5: N, S, V, F, Q | 90.22 | / | 35.64 | 27.71 | 87.87 |
| [78] | MITDB DS1 | MITDB DS2 | DDCNN + CLSM | 5: N, S, V, F, Q | 95.1 | 84 | 87.2 | 82.4 | / |
| MITDB DS1 | MIT-BIH-SUP | 88.2 | / | 77.7 | 64.7 | / | |||
| MITDB DS1 | INCART | 91.6 | / | 88 | 65.3 | / | |||
| [91] | MITDB DS1 | MITDB DS2 | CNN-LSTM | 5: N, S, V, F, Q | 95.81 | 71.06 | 69.20 | 74.94 | 94.56 |
| [92] | MITDB DS1 | MITDB DS2 | DRCNN | 5: N, S, V, F, Q | 88.99 | / | 52.10 | 56.82 | 94.75 |
| [93] | MITDB DS1 | MITDB DS2 | OptRPC | 5: N, S, V, F, Q | 98.48 | / | 98.45 | 98.43 | 98.06 |
| [94] | UVA Holter Recordings | MITDB | CNN+RNN | 4: NSR; AF; Other; noise | / | 75.5 | / | / | / |
| [95] | MITDB DS1 | MITDB DS2 | CNN | 2: PVC; no PVC | / | 98.90 | 99.20 | 98.60 | / |
| [29] | MITDB DS1 | MITDB DS2 | SE-ResNet | 5: N, S, V, F, Q | 99.61 | / | 93.78 | / | / |
| [96] | MITDB DS1 | MITDB DS2 | CNN | 5: N, S, V, F, Q | 84,62 | / | / | / | / |
| MITDB | SVDB | 84.17 | / | / | / | / | |||
| MITDB | INCARTDB | 95.36 | / | / | / | / | |||
| [97] | MITDB DS1 | MITDB DS2 | RBM | 5: N, S, V, F, Q | 98.61 | / | 87.31 | / | 98.76 |
| [98] | MITDB DS1 | MITDB DS2 | Faster R-CNN | 5: N, S, V, F, Q | 95.68 | / | 72.8 | 90 | / |
| [99] | MITDB DS1 | MITDB DS2 | CNN | 5: N, S, V, F, Q | 94.70 | 88.9 | 89 | 93.7 | / |
| [100] | MITDB DS1 | MITDB DS2 | FNN+CNN | 5: N, S, V, F, Q | 94.2 | / | 58.2 | 53.6 | / |
| [101] | MITDB DS1 | MITDB DS2 | CWT+CNN | 5: N, S, V, F, Q | 98.74 | 68.76 | 67.47 | 70.75 | / |
| [102] | MITDB DS1 | MITDB DS2 | WaveNet-LSTM | 5: N, S, V, F, Q | 96.80 | / | / | / | / |
| [42] | MITDB DS1 | MITDB DS2 | DHCAF | 5: N, S, V, F, Q | 93.0 | / | 75.1 | 70.4 | / |
| [103] | MITDB DS1 | MITDB DS2 | CraftNet | 5: N, S, V, F, Q | 89.24 | / | 89.25 | 61.84 | 95.79 |
| [48] | MITDB DS1 | MITDB DS2 | BiLSTM | 5: N, S, V, F, Q | 97.3 | / | 77.9 | / | / |
| [27] | MITDB DS1 | MITDB DS2 | CNN | 5: N, S, V, F, Q | 92.3 | / | 73.50 | 68.33 | / |
| [104] | MITDB DS1 | MITDB DS2 | CNNs | 5: N, S, V, F, Q | 98.6 | / | 93 | / | / |
| [4] | MITDB DS1 | MITDB DS2 | DNN | 2: S, non-S | / | / | 61.4 | 98.3 | |
| MITDB DS1 | MITDB DS2 | 2: V, non-V | / | / | 91.8 | / | 99.5 | ||
| [105] | MITDB DS1 | MITDB DS2 | FE-CNN | 5: N, S, V, F, Q | 98.6 | 88.0 | 84.2 | 92.3 | 99.45 |
| [106] | MITDB DS1 | MITDB DS2 | LSTM | 5: N, S, V, F, Q | / | / | 74.91 | 76.16 | / |
| [107] | MITDB DS1 | MITDB DS2 | RBM | 5: N, S, V, F, Q | 95.20 | / | 83.07 | 51.42 | / |
| [31] | MITDB DS1 | MITDB DS2 | DNN | 5: N, S, V, F, Q | 97.5 | / | 85.9 | 84.4 | / |
| [68] | MITDB DS1 | MITDB DS2 | BiLSTM | 5: N, S, V, F, Q | 99.53 | / | 96.19 | 97.21 | 98.58 |
| [108] | CPSC | MITDB | RBNN | 3: L; R; O | 78.58 | / | 78.57 | / | / |
| [109] | MITDB DS1 | MITDB DS2 | CRNN | 5: N, S, V, F, Q | 93.66 | 75.85 | 76.98 | 74.76 | 95.59 |
| MITDB | SVDB | 5: N, S, V, F, Q | 75.33 | 42.11 | 47.36 | 61.66 | 89.12 | ||
| MITDB DS1; SVDB | MITDB DS2 | 5: N, S, V, F, Q | 93.04 | 75.18 | 80.53 | 70.89 | 94.57 | ||
| Paper | Paradigm | Train/Validate Data | Test Data | Algorithm | Class | Cross-Validation | Performance | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc | F1 | Sen | Ppv | Spe | |||||||
| [82] | Intra- | MITDB | MITDB | DenseNet-BiLSTM | 4: N, S, V, F | 10-fold | 99.44 | 95.89 | 95.69 | 96.11 | 99.32 |
| Inter- | MITDB DS1 | MITDB DS2 | 92.37 | 63.49 | 68.29 | 60.35 | 94.51 | ||||
| [83] | Intra- | MITDB | MITDB | CNN | 5: N, S, V, F, Q | 10-fold | 99.48 | / | 96.97 | 98.83 | 99.87 |
| Inter- | MITDB DS1 | MITDB DS2 | 88.34 | / | 90.90 | 48.25 | 88.51 | ||||
| [84] | Intra- | Fantasia+ INCART | Fantasia+ INCART | CNN-LSTM | 2: N, CAD | / | 99.85 | 99.52 | 99.85 | / | 99.84 |
| Inter- | Fantasia+ INCART (by Subjects) | Fantasia + INCART (by Subjects) | 95.76 | 95.57 | 95.70 | / | 95.76 | ||||
| [85] | Intra- | MITDB | MITDB | O-WCNN | 4: N, S, V, F | 10-fold | 99.58 | 99.28 | 99.2 | / | 99.15 |
| Inter- | MITDB DS1 | MITDB DS2 | 99.43 | 92.05 | 91.06 | 93.50 | 99.69 | ||||
| [81] | Intra- | AFDB | AFDB | CNN+RNN | 2:AF; NoAF | 5-fold | 97.80 | / | 98.98 | 95.76 | 96.95 |
| Inter- | AFDB | MITDB | 89.30 | / | 99.82 | 51.71 | 87.94 | ||||
| Inter- | AFDB | NSRDB | / | / | / | / | 95.01 | ||||
| [86] | Intra- | MITDB | MITDB | CNN+BLSTM | 5: N, S, V, F, Q | 10-fold | 99.56 | 96.40 | 95.90 | 97.14 | 99.47 |
| Inter- | MITDB DS1 | MITDB DS2 | 96.77 | 77.84 | 74.89 | 81.24 | 95.16 | ||||
| [28] | Intra- | PTB-XL | PTB-XL | CNN-FWS | 2: N; abnormal | / | 89.92 | 90.70 | 91.40 | 90.01 | / |
| Inter- | PTB-XL (by subject) | PTB-XL (by subject) | 90.05 | 90.20 | 88.90 | 91.50 | / | ||||
| [87] | Intra- | PTB | PTB | MLA-CNN-BiGRU | 6 | 5-fold | 99.11 | / | 99.02 | 99.10 | / |
| Inter- | PTB | PTB | 62.94 | / | 63.97 | 63.00 | / | ||||
| [88] | Intra- | MITDB | MITDB | CNN | 5: N, S, V, F, Q | / | 99.81 | / | 88.82 | 95.68 | 99.54 |
| Inter- | MITDB DS1 | MITDB DS2 | 96.36 | / | 70.6 | 48.10 | 96.16 | ||||
| [89] | Intra- | Tongji Hospital, Database China | Tongji Hospital, Database China | DCNN | 26 | / | / | 91.3 | 89.1 | / | 99.7 |
| Inter- | Tongji Hospital, China | CPSC 2018 | 6 | / | 84.2 | 80.0 | / | 98.0 | |||
| [90] | Intra- | MITDB | MITDB | CNN | 5: N, S, V, F, Q | / | 99.31 | / | 73.66 | 69.6 | 98.83 |
| Inter- | MITDB DS1 | MITDB DS2 | 90.22 | / | 35.64 | 27.71 | 87.87 | ||||
| Average performance compartment of above studies | Intra-patient | 98.39 | 95.52 | 93.51 | 92.78 | 99.19 | |||||
| Inter-patient | 90.15 | 83.89 | 78.16 | 62.82 | 93.86 | ||||||
| Differences | 8.24 | 11.63 | 15.35 | 29.96 | 5.33 | ||||||
| Paper | Code Platform | Database | Evaluation Paradigm | Class Types | Model |
|---|---|---|---|---|---|
| [23] | Website | UEA; UCR; PhysioNet. AF-D1, AF-D2, and Two Lead | Intra-patient | 2 | SDK-CNN |
| [29] | GitHub | MITDB | Intra-patient | 5 | CNN (ResNet) |
| [113] | GitHub | MITDB; CPSC2018 | Intra-patient | 5 | DNN |
| [114] | GitHub | MNIST and Fashion MNIST; MITBIH | Intra-patient | 2 | D-RBFDD |
| [115] | GitHub | PTBXL; CPSC2018 | Intra-patient | 2 | SPN-V2 |
| [116] | GitHub | UCI Machine-learning repository-based arrhythmia dataset; MITDB | Intra-patient | 13 | CDNN |
| [75] | GitHub | 2021 PhysioNet Challenge data | Intra-patient | 30 | d-RINCA |
| [20] | GitHub | 2020 Physionet Challenge data | Intra-patient | 24 | CNN (ResNet) |
| [117] | GitHub | Chapman University and Shaoxing People’s Hospital Dataset | Intra-patient | 4 | Hybrid DNN |
| [118] | GitHub | Physionet Challenge database; AFDB; AF Termination database | Intra-patient | 2 | CNN+ BiLSTM |
| [40] | GitHub | CPSC2018 | Intra-patient | 9 | DNN |
| [98] | GitHub | MITDB | Intra-/Inter-patient | 5 | Faster R-CNN-DNN |
| [70] | Gitlab | MITDB | Intra-patient | 5 | CNN-BLSTM |
| [101] | GitHub | MITDB | Intra-patient | 5 | CWT+CNN |
| [111] | GitHub | Telehealth Network of Minas Gerais (TNMG) | Intra-patient | 6 | DNN |
| [119] | GitHub | MITDB; CPSC2018 | Intra-patient | 17 | CNN+ BiLSTM |
| [120] | Website | CPSC2018 | Intra-patient | 9 | CNN |
| [121] | Website | 2017 PhysioNet Challenge data | Intra-patient | 12 | DNN |
| [68] | GitHub | MITDB | Intra-patient | 5 | Hybrid CNN |
| [122] | Website | 2017 PhysioNet Challenge data | Intra-patient | 4 | CNN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, Q.; Lee, K.; Mokhtar, S.A.; Ismail, I.; Pauzi, A.L.b.M.; Zhang, Q.; Lim, P.Y. Deep Learning-Based ECG Arrhythmia Classification: A Systematic Review. Appl. Sci. 2023, 13, 4964. https://doi.org/10.3390/app13084964
Xiao Q, Lee K, Mokhtar SA, Ismail I, Pauzi ALbM, Zhang Q, Lim PY. Deep Learning-Based ECG Arrhythmia Classification: A Systematic Review. Applied Sciences. 2023; 13(8):4964. https://doi.org/10.3390/app13084964
Chicago/Turabian StyleXiao, Qiao, Khuan Lee, Siti Aisah Mokhtar, Iskasymar Ismail, Ahmad Luqman bin Md Pauzi, Qiuxia Zhang, and Poh Ying Lim. 2023. "Deep Learning-Based ECG Arrhythmia Classification: A Systematic Review" Applied Sciences 13, no. 8: 4964. https://doi.org/10.3390/app13084964
APA StyleXiao, Q., Lee, K., Mokhtar, S. A., Ismail, I., Pauzi, A. L. b. M., Zhang, Q., & Lim, P. Y. (2023). Deep Learning-Based ECG Arrhythmia Classification: A Systematic Review. Applied Sciences, 13(8), 4964. https://doi.org/10.3390/app13084964






