Abstract
Ultrasound (US) is a flexible imaging modality used globally as a first-line medical exam procedure in many different clinical cases. It benefits from the continued evolution of ultrasonic technologies and a well-established US-based digital health system. Nevertheless, its diagnostic performance still presents challenges due to the inherent characteristics of US imaging, such as manual operation and significant operator dependence. Artificial intelligence (AI) has proven to recognize complicated scan patterns and provide quantitative assessments for imaging data. Therefore, AI technology has the potential to help physicians get more accurate and repeatable outcomes in the US. In this article, we review the recent advances in AI-assisted US scanning. We have identified the main areas where AI is being used to facilitate US scanning, such as standard plane recognition and organ identification, the extraction of standard clinical planes from 3D US volumes, and the scanning guidance of US acquisitions performed by humans or robots. In general, the lack of standardization and reference datasets in this field makes it difficult to perform comparative studies among the different proposed methods. More open-access repositories of large US datasets with detailed information about the acquisition are needed to facilitate the development of this very active research field, which is expected to have a very positive impact on US imaging.
1. Introduction
The connection between medicine and computer science started in the early 1960s, when scientists developed a digital computer-based monitoring system [1] that laid the foundations of the current standard in intensive care units across the world. Since then, the relationship between clinical practice and computing advances has been constant for the last half century [2]. Computer capabilities have been evolving at an exponential rate thanks to Moore’s Law and significant improvements in processor design, and now the recent developments in artificial intelligence (AI) and deep learning (DL) methods are starting to fulfill some of the promises of computer science pioneers [3].
Software systems in medicine have taken advantage of the increasing capabilities of modern computers to provide real-time solutions to intricate challenges. Complex medical imaging problems, such as computer-assisted ultrasonography, are now being tackled with AI with great results by exploiting the collaborative work of physicians and engineers, the capabilities of modern computing hardware [4,5], and the access to digital medical images.
Nevertheless, the introduction of deep learning-based strategies into clinical practice has just started, and much work lies ahead. Many leading AI experts agree that there is a huge gap between proof-of-concept research papers and actually deploying a machine learning (ML) system in the real world [6], especially in clinical devices that have to pass regulatory processes. In fact, there is undoubtedly huge potential for ML to transform healthcare, but going ‘from code to clinic’ is the hardest part. The list of challenges to overcome in this field [6] includes the difficulties in obtaining datasets with sufficiently large, curated, high-quality data; the lack of robust clinical validations; the need for interdisciplinary collaborations of computational scientists, signal processing engineers, physics experts, and medical experts [7]; and some technical limitations of AI algorithms.
1.1. Current Challenges of Ultrasound Imaging
Portable ultrasound has the potential to become the new stethoscope. The ultrasound imaging modality stands out for its safety, portability, non-invasiveness, and relatively low cost, which support its use in many different clinical scenarios. In fact, ultrasound imaging has become an important diagnostic tool for an increasing number and range of clinical conditions, enabling a greater variety of treatment options. This situation has led to the design of multiple complex diagnostic procedures and the atomization of their medical use.
However, there are several challenges currently associated with ultrasound imaging:
- Operator dependency. As US imaging depends on the ability of the operator to position the transducer and interpret the images correctly, it can be difficult to obtain consistent and reliable results from different operators, requiring long and specialized training.
- Subjectivity in image interpretation. The interpretation of the US images is quite subjective. Unlike other imaging modalities, which produce objective, quantitative data, the interpretation of US images depends heavily on the experience and skills of the person performing the scan. This can lead to variability in the accuracy and reliability of the images, particularly when they are used for diagnostic purposes.
- Limited penetration depth, which limits its use for imaging deep structures in the body, and an inability to penetrate certain tissues or structures, such as bone and air-filled cavities.
- Limited quality of US images. Ultrasound images are typically less detailed than those produced by other modalities. This can make it difficult to accurately diagnose certain conditions, particularly when the structures being imaged are small or located deep within the body.
While advances in technology are helping to address some of these challenges, they still remain an important consideration in the use of ultrasound imaging.
1.2. The Deep-Learning Revolution in Ultrasound Imaging
The use of deep learning in medical imaging is one of the fields that has reached the most significant advances in the field of AI [8]. It has already achieved many meaningful results in US imaging [9,10]. The tremendous potential of the combination of both clinical and commercial technologies is widely recognized, making computer-assisted ultrasound a hot research field.
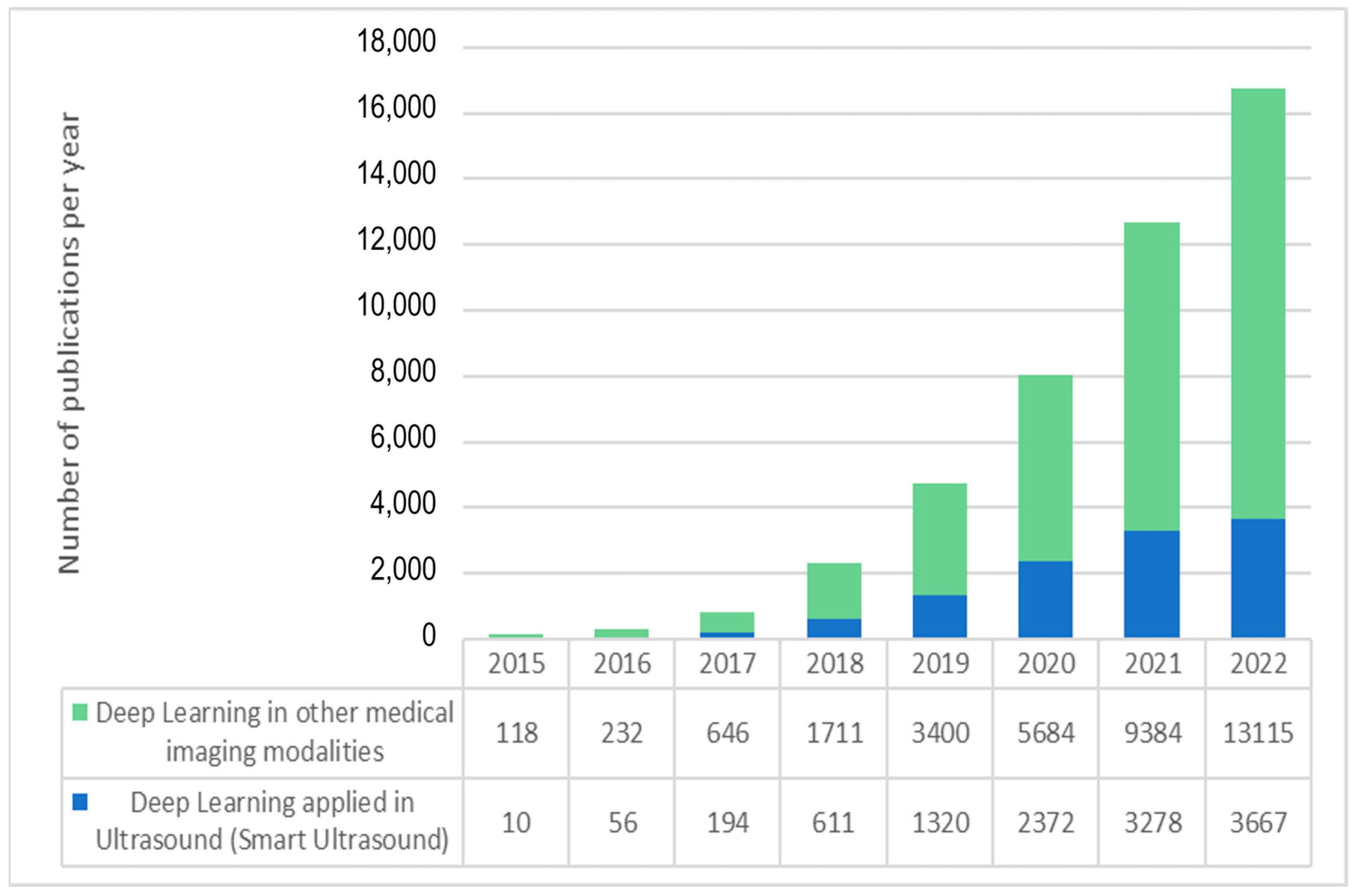
As shown in Figure 1, the number of academic publications on “deep learning” in Pubmed has soared from less than 1000 in 2017 to more than 16,000 in 2022, and the number of deep learning applications in ultrasound imaging has also followed the same exponential trend. Furthermore, if we consider that, despite their high impact and consideration within the community, conference proceedings are usually not listed under PubMed and other databases may include some additional publications not included in PubMed, the number of recent scientific works on ultrasound and artificial intelligence is even higher than shown in Figure 1. These numbers demonstrate the current enthusiasm for this new technology.
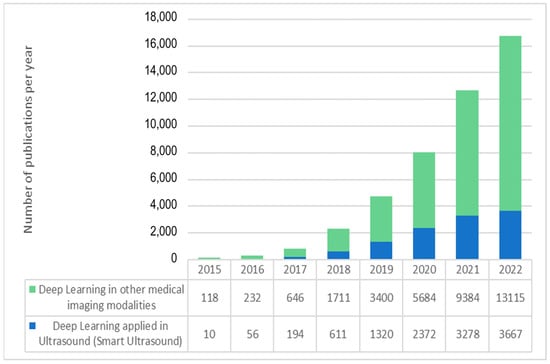
Figure 1.
Exponential growth in the number of articles indexed in PubMed with “deep learning” and “ultrasound” (blue) and “deep learning” applied to other data and medical image modalities.
There are many applications of AI in US imaging that, in general, address the main challenges of US. This review paper aims to provide an overview of the most recent and representative research works in computer-assisted medical ultrasound scanning, which is receiving considerable attention from the academic and medical image industry communities.
1.3. Main Applications of AI in US Imaging
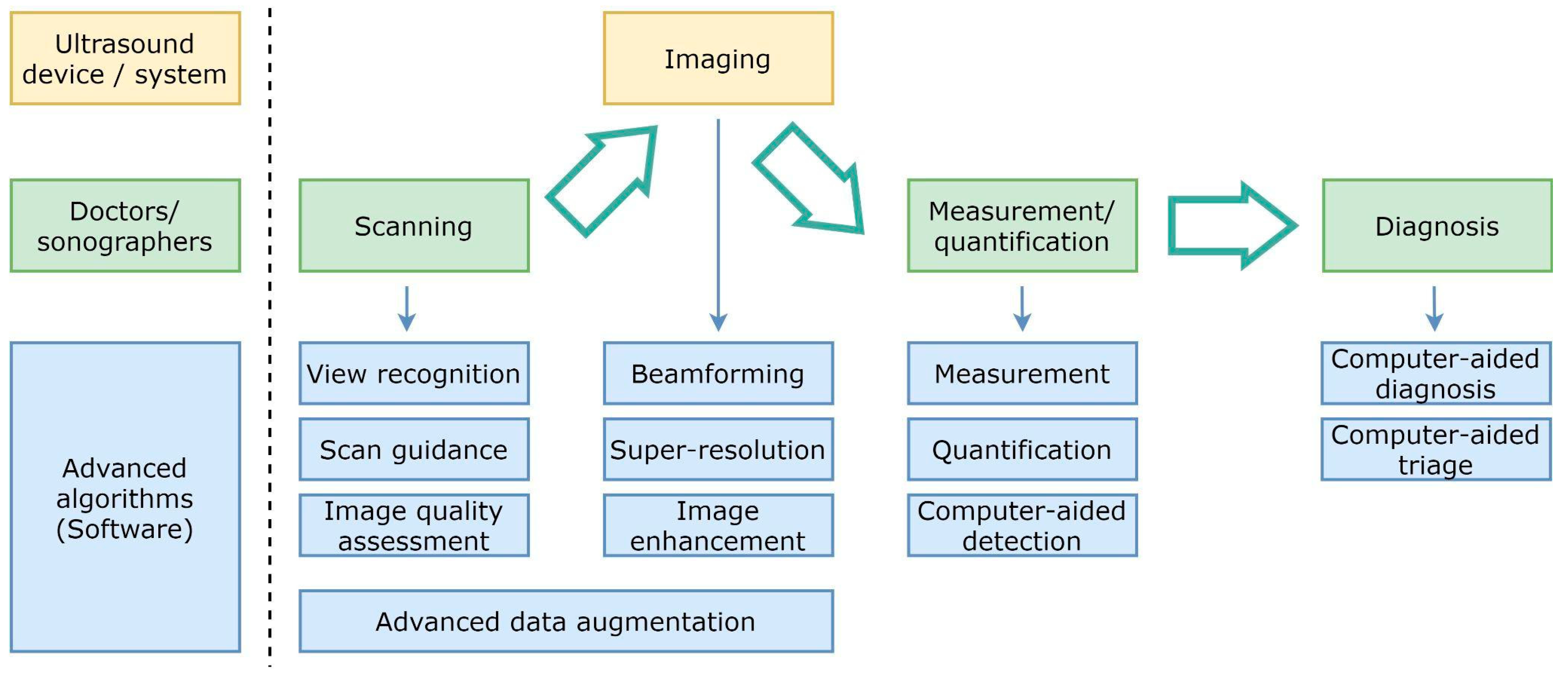
Figure 2 shows a diagram of the diagnostic workflow with computer-assisted US devices, where the main tasks performed during the examination are outlined. Some of them are done by practitioners (and technicians), others are in charge of the hardware, but a significant and growing portion of them are evolving as advanced software pieces that try to improve the efficiency of the two previous actors. The illustrated workflow starts by placing the probe on the patient’s body surface, which will be traversed by pressure waves whose echo is accurately measured. US imaging systems process the recorded signals to generate 2D images, which are taken to perform measurements and diagnoses. Assistance can be provided in the examination procedure by providing indications on how to perform the scan, recognizing the scanned organ, and making additional assessments about how to improve the image quality in the acquired frames.
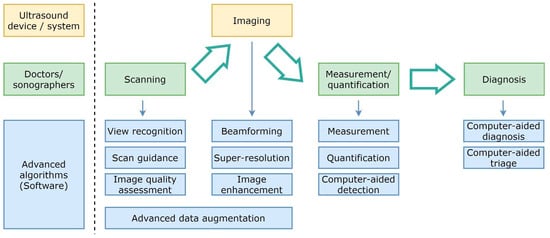
Figure 2.
Simplified diagram of the diagnostic workflow with computer-assisted ultrasound devices.
Beamforming, higher resolution, and image enhancement are three areas in which conventional signal processing can be improved. Computer assistance tools could also replace repetitive measurement and quantification tasks. Additionally, physicians will be able to consider additional feedback from computer-aided diagnosis and computer-aided triage tools.
There are four main research areas where artificial intelligence and computer-assisted solutions may help standardize medical practice, reduce training and examination times, and improve ultrasound image quality.
- The application of machine learning techniques to ultrasound image formation has a large margin of improvement to provide less blurry and more significant images, even by creating new ultrasound medical imaging modalities [11]. New approaches in beamforming, super-resolution, and image enhancement require modifications to hardware elements, which are usually more ambitious than more straightforward software post-processing approaches. In spite of the more difficult adoption, many research advances are overcoming traditional and simple reconstruction algorithms, which translate physical measurements of ultrasound waves into images in the display. The more computational power available in medical devices, the more sophisticated real-time solutions may enrich the different ultrasound imaging modalities.
- AI algorithms can help to reduce the steep learning curve in ultrasound scanning by assisting physicians, nurses, and technicians to perform full examinations, as is shown in detail in the following sections.
- Image processing algorithms for measurement, quantification, and computer-aided detection have left behind traditional feature engineering, and the most competitive solutions in ultrasound imaging take advantage of deep learning-based approaches.
- Finally, computer-assisted diagnosis, triage, detection, and quantification have also attracted the research community’s attention because of their significant impact on reducing physicians’ workload.
There are a large number of applications and publications in this field. Therefore, this state-of-the-art review focuses on the latest advances in computer-assisted ultrasound scanning. The search strategy followed sought to obtain a broad overall view of the state-of-the-art. Specific terms on Pubmed, the most common free database of publications on medical imaging, such as “ultrasound”, “AI”, “computer-assisted” and “AI-guided,” were used to obtain the references considered in this review.
1.4. Computer-Assisted Ultrasound Scanning
As indicated in Section 1.1, ultrasound imaging is highly operator-dependent. Therefore, sonographers must be properly trained to be able to acquire clinically useful images. Their training process has a steep learning curve, which is mainly caused by these factors:
- The operator must place the transducer on the patient’s body surface at precise locations to allow the signal to reach the desired organ and bounce back to the sensor. Bones (such as the ribs) and certain organs (such as the lungs) are opaque to pressure waves, so they must be avoided to reach the target organ.
- Both the exact location of the acoustic windows and the force to be exerted with the probe slightly depend on the patient’s morphology.
- In conventional US imaging, the operator manually moves the transducer, obtaining a series of two-dimensional (2D) images of the target region. Then, he/she performs a mental 3D reconstruction of the underlying anatomy based on the content and the movement patterns used. Physicians have defined a set of protocols to obtain standard views that contain most of the relevant features in the 2D cutting plane of the organ. However, such views are not easy to locate for a person with limited training.
- Ultrasound images are noisy and are affected by signal dropout, attenuation, speckle, and acoustic shadows. So, the operator does not always perceive the anatomy as clearly as one would like. Moreover, a soft-textured image free of noise due to excessive filtering or incorrect gain settings is useless from a clinical point of view.
Because of these unique challenges, computer-assisted scanning is a promising set of technologies that may democratize ultrasound medical imaging. Medical practitioners will benefit from any improvements in the effectiveness and autonomy of the acquisition workflow of this imaging modality. The smart guidance for assisted scanning will simplify their work, increase their productivity, decrease their expenses, and optimize their workflow.
Its impact on developing countries can be even bigger. Because there are not enough skilled operators in developing countries, many women do not get any US exams during their pregnancy [12]. Creating a system that can help reduce the level of expertise needed during scanning could have a very significant impact. With this kind of system, any person with a basic anatomical background in a remote area would be able to do an ultrasound exam. Only images with clinical value would be sent to radiologist experts to be evaluated, no matter where the physician lives.
We distinguish two main subfields in computer-assisted ultrasound scanning: view recognition and scan guidance, which are described in the following subsections. Table 1, Table 2, Table 3 and Table 4 summarize the references analyzed in this work with information on the explored organ, the ultrasound scanner used, the number of cases, the type of AI model used, and some performance metrics. In any case, it is important to note that the variety of cases, US probes, and the significant lack of standardization in US imaging make it hard to compare the results from different publications.
2. Standard Planes Recognition with AI
Physicians have established over the years specific protocols to identify and acquire the most relevant ultrasound planes for each organ of interest. The ultrasound probe has to be posed on the available acoustic windows with the right location, orientation, and pressure. Those indications are far from trivial for an inexperienced user, as they depend on the patient’s morphology, and the images typically have a poor signal-to-noise ratio. Furthermore, they are one of the first and largest steps in the steep learning curve. Consequently, one of the main goals of computer-assisted ultrasound scanning systems is the real-time recognition of standard planes with clinical value as the first step in assisted guidance. Many studies have used AI to make it easier to recognize the standard planes of the target organs during a scanning session.
2.1. Scan Plane Detection
Non-expert physicians may have difficulties determining which organ or part of the body is being scanned with US, as only a limited cross-sectional 2D image is usually available. The lack of information on the specific location of each plane reduces the diagnostic accuracy and makes it quite dependent on the experience and skills of the physician.
Automatic plane detection methods can be used in practice in two different ways: in real-time scanning to assist inexperienced sonographers and, as a postprocessing step, retrieving standard scan planes from acquired videos that could be further analyzed afterward. Each use case has its specific challenges, as fast computation is more critical than accuracy in real-time applications, while in the case of offline image analysis it is the opposite.
The first developments in automatic scan plane detection date back almost 20 years, with several works based on support vector machines (SVM) [13,14] and conventional ML [15,16]. Recently, US view recognition has been further improved with deep-learning techniques. Most of these works are limited to specific solutions for individual organs or organ regions.

Table 1.
Summary of the selected publications on scanning, plane detection, plane classification, and organ identification.
Table 1.
Summary of the selected publications on scanning, plane detection, plane classification, and organ identification.
| Scan Plane Detection, Plane Classification, and Organ Identification | |||||
|---|---|---|---|---|---|
| Reference | Target Organ | # Cases | US System | AI Model | Results |
| R.-F. Chang et al. [14] | Breast | 250 images | ATL HDI 3000 | SVM | Accuracy 95.4% |
| C. P. Bridge et al. [15] | Heart | 91 videos from 12 subjects | GE Voluson E8 | RF | Classification error <20% |
| M. W. Attia et al. [16] | Kidney | 66 images | Several (Databases) | Multilayer ANN | Accuracy 97%. |
| H. Chen et al. [17] | Hrain, Stomach, Spine, Heart | 300 training 331 evaluation | Siemens Acuson Sequoia 512 US | RNN | AUC 0.94 |
| H. Chen et al. [18] | Fetal Abdomen | 20,660 images | Unknown | CNN | Accuracy 90.4% |
| D. Ni et al. [19] | Fetal Abdomen | 1664 images 100 videos | Siemens Acuson Sequoia 512 | AdaBoost, Selective Search (SSD/SSSD) | Accuracy 81% (SSSD) |
| D. Ni et al. [20] | Fetal Abdomen | 1995 images | Siemens Acuson Sequoia 512 | RF + SVM | Accuracy 85.6% AUC >0.98 |
| B. Rahmatullah et al. [21] | Fetal Abdomen | 2384 images | Philips HD9 | AdaBoost | Accuracy 80.7% |
| B. Rahmatullah et al. [22] | Fetal Abdomen | 8749 images | Philips HD9 | Local Phase Feature | AUC 0.88 |
| H. Wu et al. [23] | Heart | 270 videos | Philips CX50 | SVM | Accuracy 98.5% |
| D. Agarwal et al. [24] | Heart | 703 images | GE Vivid series | SVM | Accuracy 98% |
| R. Kumar et al. [25] | Heart | 113 videos (2470 frames) | Several (Database) | SVM | Recognition rate 81% |
| W.-C. Ye et al. [26] | Liver | 20 livers | frame grabber | SVM | Accuracy 91% |
| B. Lei et al. [27] | Fetal Brain | 1.735 images | Siemens Acuson Sequoia 512 | SVM Classifier | Accuracy 93.27% Precision 99.19% |
| R. Qu et al. [28] | Fetal Brain | 31,200 images | Hitachi ARIETTA 70 | CNN | AUC 0.90 |
| Y. Cai et al. [29] | Fetal Abdomen | 1292 images | Philips HD9 (V7-3) | CNN | Precision 96.5% |
| C. F Baumgartner et al. [30] | Fetal Anatomy | 1003 videos | GE Voluson E8 | CNN | Accuracy 66–90% |
| C. F. Baumgartner et al. [31] | Fetal Anatomy | 2694 videos | GE Voluson E8 | CNN | Accuracy 90.1% |
| Y. Chen et al. [32] | Lung and Liver | 23,000 images | Several (Database) | CNN | Accuracy 94.2% |
| Y. Gao et al. [33] | Fetal head | >30,000 frames | Konted GEN 1 C10R | CNN | Precision 86% |
| M. Yaqub et al. [34] | Fetal brain | 19,838 images (10,595 scans) | Several | CNN | Agreement 94.1% |
| X. P. Burgos-Artizzu et al. [35] | Fetus and Mother’s Cervix | 12,400 images (1792 patients) | Several (Database) | CNN (19 models) | Accuracy Classification 93.6% |
| C. P Bridge et al. [15] | Fetal Heart | 91 videos | GE Voluson E8 | RF | ClassificationError 18% |
| C. P Bridge et al. [36] | Fetal Heart | 63 videos | - | Fourier and SVM | Accuracy 88% |
For instance, fully automatic left ventricle (LV) detection in cardiology is a challenging task. This is due to the noise and artifacts in the moving cardiac ultrasound image and the variety of LV shapes. A faster R-CNN and an active shape model (ASM) were combined to come up with a new way to find and track the LV in imaging sequences [17]. In addition, a fast R-CNN [18] was proposed to identify the ROI, and an ASM [19] obtained the parameters that most precisely characterized the LV’s shape.
In obstetrics, scans for the detection of possible abnormal fetal development typically involve the acquisition of several standard planes, which are used to obtain biometric measurements and identify possible abnormalities. The earliest works on automated fetal standard plane detection were able to identify only 1–3 standard planes in short videos of 2D fetal US sweeps [17,18,19,20]. Some of these early works tried to figure out where standard planes were by looking for major anatomical structures in 2D US images. Zhang et al. [24] proposed detecting these standard planes using local context information and cascading AdaBoost, similar to other early works based on Haar-like features extracted from the data and AdaBoost or Random Forest classifiers [22,23].
Rahmatullah et al. [21,22] proposed detecting anatomical structures from abdominal US images by the integration of their global and local features. However, the method was semi-automatic, and the detection accuracy was not good enough for clinical use.
Other pre-deep-learning works were based on object classification methods. An 8-way echocardiogram view classification was obtained in [23] with a Gabor filter and a kernel-based SVM classifier. The work in [24] utilized the Histograms of Oriented Gradients (HOG) feature and SVM classifier. In [25], 2D echocardiogram view classification was obtained based on a set of novel salient features obtained from local spatial, textural, and kinetic information. An SVM classifier with a pyramid-matching kernel was used to learn a hierarchical feature dictionary for view classification. Yeh et al. [26] also used an SVM classifier for the features extracted using wavelet decomposition and a gray-level co-occurrence matrix, and Lei et al. [27] applied it for the automatic recognition of fetal facial standard planes (FFSPs).
The results obtained with “traditional” methods before the deep-learning revolution were generally limited due to the following aspects:
- Manual feature extraction settings depend on subjective human choices.
- The number and type of extracted features are usually very limited.
- Feature extraction methods cannot be optimized in real-time, which may be helpful when there are changes in the datasets.
Chen et al. [17] were one of the first research teams to exploit deep learning in ultrasound imaging. They employed a CNN with five convolutional and two fully connected layers to detect the standard abdominal plane. In this method, each input video frame is analyzed multiple times with overlapping image patches, which improves the accuracy but increases the computational cost. This work was extended to three scan planes and a recurrent architecture that considers temporal information, although it did not reach real-time performance [18].
A very challenging problem in standard plane recognition is the detection of the six standard planes in the fetal brain, which is required for fetal brain abnormality detection. The large diversity of fetal postures, similarities between the standard planes, and reduced amount of data cause these difficulties. Qu et al. [28] approached this problem using a domain transfer learning approach based on a deep CNN and data augmentation, outperforming other deep learning methods.
The automatic recognition of the standard abdominal circumference (AC) planes on fetal ultrasonography was tackled by Cai et al. [29]. They developed SonoEyeNet, a CNN that used the information from experienced sonographers’ eye movements. The collected eye movement data were used to generate heat maps with the visual fixation information of each frame, and then that data was used for plane identification.
Baumgartner et al. [30] proposed a fast system for automatically detecting twelve fetal standard planes from real clinical fetal US scans, allowing real-time inference. They employed a CNN architecture inspired by AlexNet and freehand data (instead of data acquired in single sweeps), which provided much larger datasets. One of the best features of that work is a novel method to obtain localized saliency maps. This is useful, as localized annotations are not routinely recorded, and creating them for large datasets would require too much time. The saliency maps were applied to both guide the user into the neighboring volume of the desired view and build an approximate bounding box of the fetal anatomy. Baumgartner et al. extended this work with more data and a deeper neural network architecture [31] and transferred learning from natural to US images to speed up the training process. Recent works, such as [32], have also proposed this pretraining concept in several contexts where not enough ultrasound images are available.
In general, supervised deep learning methods for ultrasound plane detection face the same problem: manual annotation of US videos is time-consuming and requires clinical expertise. Some recent works have explored new strategies to reduce the need for labeled data and associated annotations significantly. Guo et al. [33] proposed a semi-supervised learning framework for fetal head images. They formulated the learning as a multi-way classification problem, and the trained model can be used to label unseen video frames densely and automatically.
2.2. Scan Plane Classification
Scan plane classification differs from scan plane detection as, in this case, every image is already known to be a standard plane. Although this is not a common situation in clinical practice, it has received significant attention as ultrasound databases from hospitals (typically within a PACS system) only store frames corresponding to standard planes. Therefore, algorithms developed on these datasets have focused on the classification and categorization of each acquired plane.
A representative example is given by the guided random forest method proposed by Yaqub et al. [34] for the classification of fetal 2D ultrasound images into seven standard planes. The authors included an additional class (“other”) in order to consider possible non-modeled standard views and make the model more general. A more recent work with CNN was developed by Burgos-Artizzu et al. [35] using six classes: four fetal anatomical planes, the mother’s cervix, and an additional class (“other”). Fetal brain images were also classified into the three most common fetal brain planes. Their results indicate that the performance of these CNN-classifying standard planes in fetal examinations is comparable to that achieved by humans.
2.3. Organ Identification
Organ identification in ultrasound images is a similar problem to object detection in natural images in computer vision. Therefore, some algorithms from computer vision have been successfully translated to detect and identify organs in ultrasound imaging [13], and they are also used as the first step in organ segmentation.
Traditional ML methods performed organ classification with multi-way linear classifiers (e.g., SVM) and preselected features extracted from US images. However, these methods were susceptible to spurious factors such as image deformation and specific imaging processing steps. Therefore, deep neural networks (DNNs) were proposed for these tasks, as they can learn abstract features and provide individual prediction labels for each US image. They are particularly useful for identifying pathological organs, tumors, lesions, and nodules.
In practice, sonographers know which organs are being scanned, as the protocols define different acoustic window areas for each organ in adult patients. However, there are some fields where organ identification may be very useful, such as obstetrics, as the fetus may be in many possible positions and can move during the ultrasound scan. For instance, Bridge and Noble [36] proposed using rotation-invariant features and SVM for the localization of the fetal heart in short videos. They later extended the method for localizing three cardiac views, taking into account the temporal structure of the data [15].
2.4. Standard Clinical Plane Extraction from Volumetric (3D) Ultrasound Images
As an alternative to assisting operators in acquiring freehand 2D standard planes, it is also possible to acquire 3D US volumes covering the whole area of interest and then extract the standard planes automatically from them. This approach may simplify the acquisition of specific planes, but it currently faces significant challenges. On the one hand, 3D US is an emerging imaging modality, but it is significantly more expensive and does not provide as good a sampling quality as 2D scans. On the other hand, 3D DL is still a challenging task. It is computationally very expensive, and given the typically limited training data [33] and the large number of parameters of these neural networks, there is a high risk of overfitting. Furthermore, as there is no real-time feedback during the 3D acquisition, if it is performed by a less experienced technician, unexpected situations, such as bad acoustic contact or artifacts, may ruin the 3D acquisition and prevent the system from obtaining the desired planes.
However, there is a very intense line of research on plane extraction from 3D volumetric ultrasound images. Once a 3D scan has been acquired with a 3D US probe or by densely scanning an area with a tracked 2D probe, the standard plane can be extracted as a postprocessing step. This approach does not require real-time computation, as long as all the 3D data has been acquired.
For instance, the localization of cardiac planes in 3D ultrasound was achieved by Chykeyuk et al. [37] with a random forests-based method, and the automatic localization of fetal planes was achieved by landmark alignment [38], regression [39,40], and classification [41] methods.
Dou et al. [42] proposed a reinforcement learning (RL) method to localize standard planes in noisy US volumes. Despite its good performance, this RL-based method had some limitations, as agents learned separately from specific planes. As agents do not communicate with each other, they cannot learn the valuable spatial relationship among planes. Yang et al. [43] went a step further and devised a multi-agent RL framework to localize multiple uterine standard planes in 3D US simultaneously by establishing communication among agents and a collaborative strategy to learn the spatial relationship among standard planes effectively. Besides, they adopted a one-shot neural architecture search [44] to obtain the optimal agent for each plane. This architecture uses gradient-based search using a Differentiable Architecture Sampler [45], which may make the training more stable and faster.
Li et al. [40] used a CNN to regress a rigid transformation iteratively for localizing TC and TV planes in 3D fetal ultrasound. Jiang et al. [46] evolved this idea and combined it with the deep RL approach in Huang’s work to build a deep Q-network-based agent that explores the 3D volume with 6DOF to locate the desired plane. Results are only validated with a simple phantom.
Yu et al. [47] proposed a fully connected NN with seven layers trained to map 2D slice images to their 3D coordinate parameters from an ultrasound volume obtained by a GE ABUS (Automated Breast Ultrasound System). Each slice is described by a pair of translation and rotation parameters that label every possible image in the dataset. However, the model does not generalize the mapping and must be trained from scratch for each new patient. Therefore, it can be seen as image-based indexing that can be used to check slice position and orientation.
Skelton et al. [48] also proposed a DL method to extract standard fetal head planes from 3D-head volumes with similar quality to operator-selected planes. In [49], Baum et al. propose a novel deep neural network architecture for data-driven, non-rigid point-set registration, which they call Free Point Transformer (FPT). Their system is designed to allow continual real-time MRI to transrectal ultrasound (TRUS) image fusion in prostate biopsy using sparse data, which may be generated from automatically segmented sagittal and transverse slices available from existing bi-plane TRUS probes. A significant feature of FPT is that it allows unsupervised learning and does not require ground-truth deformation data to learn non-rigid transformations between multimodal images without any prior constraints.
We should highlight that the described strategies assume that the full 3D volume is available, and therefore, they would not be directly applicable to a real-time system with progressive and exploratory scanning. Furthermore, image quality in 3D is lower than in corresponding classic 2D planes due to limitations in 3D technology and acquisition techniques.

Table 2.
Summary of publications on standard clinical plane extraction from volumes.
Table 2.
Summary of publications on standard clinical plane extraction from volumes.
| Standard Clinical Plane Extraction from 3D Volume | |||||
|---|---|---|---|---|---|
| Reference | Target Organ | # Cases | US System | AI Model | Results |
| K. Chykeyuk et al. [37] | Heart | 25 3D cases | Philips iE33 | RF | Distance <6 mm, Angle <3.5° |
| C. Lorenz et al. [38] | Fetal Abdomen | 126 training 42 evaluation | Several | RF | Plane Offset Error 5.8 mm Circumference error 4.9% |
| Y.i Li et al. [39,40] | Fetal Brain | 72 volumes | - | CNN (ITN) | Translation error 3.5 mm Angular error 12.4 deg |
| H. Ryou et al. [41] | Fetal Brain | 19,838 images (10,595 scans) | Several | CNN | 96.9% Dice coefficient (Head Region Detection) |
| H. Dou et al. [42] | Fetal Brain | 430 | Mindray DC-9 US system | RNN recurrent neural network | |
| X. Yang et al. [43] | Uterus and Fetal Brain | 683 (Uterus) 432 (Fetal Brain) | - | RL, RNN, and DQN | Localization Accuracy Fetal Brain 1.19 mm |
| B. Jiang et al. [46] | Phantom | 6 volumes | Ultrasonix SonixTouch | RL, RNN, and DQN | Translation error 5.5 mm Angular error 2.2 deg |
| T.-F. Yu et al. [47] | Breast | 22,982,656 images (1 volume) | GE ABUS | CNN | Translation Prediction error (x/y) (0.14 mm/0.25 mm) |
| E Skelton et al. [48] | Fetal Brain | 551 images | Philips Healthcare EpiQ | CNN (ITN) | Translation error 3.4 mm Angular error 12.5 deg |
| Z. Baum et al. [49] | Prostate | 108 image pairs | Intraoperative TRUS | PointNet NN | Registration Error 4.4 mm |
2.5. Image Quality Assessment
One important aspect of selecting or identifying good standard planes in an ultrasound acquisition is the evaluation of the image quality of each frame. There are multiple criteria for evaluating the quality of the images in the 2D ultrasound-captured frames, such as the presence of artifacts or blurring effects.
Since the local anatomical structure is important in the US, standard quality measures of US imaging are based on contrast statistics [50]. As standard contrast recovery (CR) does not take into account the noise-to-signal ratio, contrast-to-noise ratio (CNR) is a preferred metric. However, recent research has demonstrated that dynamic range adjustments can arbitrarily create high CNR values without having any discernible impact on the likelihood of lesion identification [51]. In order to solve this problem, other metrics, such as the generalized contrast-to-noise ratio (gCNR) in ultrasound, have been proposed [52]. gCNR is supposed to prevent possible bias from dynamic range alterations.
Other recent works have used task-based metrics for automatic quality assessment of standard planes in 2D fetal US. For instance, Zhang et al. [53] proposed a multi-task approach combining three CNNs. As essential anatomical structures should appear full and remarkable with a clear boundary in each standard plane, the identification (or lack thereof) of these structures can be used to determine whether a fetal ultrasound image is the standard plane.
Another relevant aspect of ultrasound images is the speckle patterns, which could be exploited to track the relative movement and orientation of the transducer [54,55]. Although commonly referred to as speckle noise, it is not an actual noise. The name was given as it is considered an unwanted signal. However, it produces deterministic patterns that depend on the local tissue composition and can be used to locate them. Therefore, it may be important that the image processing used in the US preserves that information so that AI algorithms can retrieve and use it.
3. Scanning Guidance with AI
US is used more frequently in clinical practice than other medical modalities such as magnetic resonance imaging (MRI) or computed tomography (CT). Furthermore, it is carried out by a wider variety of medical professionals with different levels of expertise [56]. However, maneuvering a US transducer requires hand-eye coordination that takes years to master [12]. The US examiner has to guide the transducer to specific scan planes through a highly variable anatomy and, at the same time, interpret the findings and make decisions.
Scanning guidance goes beyond the tasks of standard plane identification. The potential for utilizing AI strategies in ultrasound scanning guidance to improve the clinical workflow is high, as such features could increase diagnostic confidence and productivity as well as shorten the learning curve for new users. Many early studies indicate that integrating AI into this task could improve the performance of a less-experienced radiologist to nearly the level of an expert. Besides, this kind of development might reduce scan times in traditionally lengthy exams so that it can improve the scan according to ALARA considerations. For inexperienced users, information from real-time tools might provide support on how to position the transducer correctly and how to reach the target organ.
Most published solutions for scanning guidance with AI are still in the early stages of development. It is a challenging task due to the images’ complexity and the large variability in patients’ anatomy and operators. Here we cover the most prominent research advances in the last few years and the open challenges and opportunities in this field.
3.1. Human-Operated Systems
As previously mentioned, reinforcement learning (RL) is one of the most successful AI methods. RL maximizes the rewards from the action the computer performs (called an agent in this context). In 2019, Jarosik and Lewandowski [57] developed one of the first applications of deep reinforcement learning (DRL) to ultrasound guidance with a phantom containing only a single cuboid object of interest. The agent receives a 2D image corresponding to the current position and orientation of the transducer, and it can decide to change its lateral position, orientation, and focal-point depth. When the environment changes, the agent receives a new image and a reward according to the specific goal. They demonstrated the feasibility of using DRL for ultrasound guidance, although only in a simple case.
Recently, Milletari et al. [58] have proposed a visual DRL architecture to suggest actions to guide inexperienced medical personnel to obtain clinically relevant cardiac ultrasound images of the parasternal long-axis view. The authors created RL environments by projecting a grid on subjects’ chests and populating them with US frames collected from a set of volunteers. At inference time, the user receives motion recommendations from the RL policy.
Droste et al. [59] proposed a system that employed an NN that received the US video signal and the motion signal from an inertial measurement unit (IMU) attached to the probe. The goal was to provide guidance for fetal standard plane acquisition in freehand obstetric US scanning. The network is based partly on SonoNet [35], MobileNet V2, and a custom recurrent NN mechanism that tries to predict the move that an expert sonographer would make toward the standard plane position. Their behavioral cloning approach [60] is not based on DRL, although they have several points in common. The scenario used is more realistic than in other previous works. However, the indications in this work only refer to probe orientation, as the IMU does not provide accurate position or translation information. Their results showed that the algorithm is robust for guiding the examiner toward the target orientation from distant initial points. However, it does not provide an accurately predicted guidance signal when the probe is close to the desired standard plane due to inter- and intra-sonographer variations and sensor uncertainty.
In a recent study, the DL-based software Caption Guidance [61] has shown its capacity to guide novices (in that case, nurses without prior ultrasonography experience) to acquire cardiac US images. The software provides real-time guidance during scanning to obtain anatomically correct images. Their results showed 92.5% to 98.8% agreement in the diagnostic assessments obtained from the scans performed by experienced cardiac sonographers and nurses guided by this AI tool.

Table 3.
Summary of publications on scanning guidance by human-operated systems.
Table 3.
Summary of publications on scanning guidance by human-operated systems.
| Scanning Guidance Human-Operated Systems | |||||
|---|---|---|---|---|---|
| Reference | Target Organ | # Cases | US System | AI Model | Results |
| P. Jarosik et al. [57] | Phantom | 4 runs 16 time steps | Simulation | Deep RL | Distance from the object’s center 0.2 cm |
| F. Milletari et al. [58] | Heart | 200k images | Unknown | RL + DQN | Correct guidance 86.1% |
| R. Droste et al. [59] | Fetus | 464 scans by 17 experts | GE Voluson E8 scanner | MNet CNN | Prediction accuracy: 88.8% goal, 90.9% action |
| A. Narang et al. [61] | Heart | 240 scans | Terason uSmart 3200t Plus | DL | Diagnostic Quality 98.8% |
3.2. Robot-Operated Systems
A significant amount of the published articles on ultrasound imaging scanning assistance discuss how to control the movement of a robot with an US transducer. Toporek et al. [62], from Philips Research, proposed a system composed of a high-end ultrasound device, a robotic arm, and a fetal US phantom used to mimic patient anatomy. They claim that initial attempts to develop control systems for robots that screen certain organs were traditionally not fully automated or sufficiently accurate [63,64]. They obtained relatively good results in an ideal scenario with phantom data and a simple DNN architecture inside their control system.
Toporek et al. [65] used a transfer learning approach using an 18-layer CNN pre-trained using the ImageNet dataset, and they modified the two latest layers to build two fully connected regressors that are trained from scratch to learn the relative position and orientation with image clues. They modified the loss function considering 6 degrees of freedom [66]. This approach has several limitations, but the system was able to indicate how to move the probe from an arbitrary position to reach a target image in most phantom locations.

Table 4.
Summary of publications on scanning guidance robot-operated systems.
Table 4.
Summary of publications on scanning guidance robot-operated systems.
| Scanning Guidance Robot-Operated Systems | |||||
|---|---|---|---|---|---|
| Reference | Target Organ | # Cases | US System | AI Model | Results |
| G. Toporek et al. [62] | Fetal Phantom | 38,900 train 5400 test | Philips X5-1 EPIQ | CNN | Translational accuracy < 3 mm |
| A.S.B. Mustafa et al. [63] | Liver | Validation on 14 male subjects | Mitsubishi Electric MELFA RV-1 | Image Processing Methods | Localization Initial Posit. Success 94% |
| K Liang et al. [64] | Breast | 5 cyst biopsies | Duke/VMI 3-D scanner | Image Processing Methods | Distance error 1.15mm |
| H. Hase et al. [67] | Backbone | 34 volunteers | Zonare z.one ultra | RL + DQN | Correct Navigation 82.91% |
| Q. Huang et al. [68] | Phantom | - | Sonix RP Ultrasonic Medical Corp. | Point Cloud from Kinect | - |
Hase et al. [67] follow a similar approach to Milletari [58]. However, they used a robotic arm to sample the patients on a grid for scanning the lower back (instead of manually sampling the gridded surface as Milletari did) and, ideally, navigate on a flat surface to locate the sacrum area. Although this work also assumed an ideal simulated scenario, it showed several interesting contributions in RL and NN architecture.
Huang et al. [68] proposed a robotic device that could achieve dense 3D ultrasound scanning over a flat surface in the 2D plane. Even in ideal laboratory conditions, the study presented several weak points, such as the fact that the depth camera (a Kinect-based sensor camera) does not work properly when the acoustic gel is applied on the surface of the phantom due to the specular reflections and refractive optical properties of the acoustic conductor gel.
In summary, these robot-related works show that fully automated ultrasound scanning with robots is promising but still far from being integrated into clinical practice.
4. Discussion
There is a shortage of skilled and experienced US practitioners, and this is particularly significant in developing countries, where the World Health Organization estimates that many US scans are acquired by personnel with little or no formal training. As US imaging is particularly subject to errors, poor image quality, and incorrect imaging, the misinterpretation of images is a significant risk in US-based diagnosis. Therefore, advances in this field are expected to impact clinical practice significantly. AI algorithms can help reduce the steep learning curve in ultrasound scanning by assisting physicians, nurses, and technicians to perform full examinations. AI-assisted US scanning is also of great interest for military medical scenarios where US imaging needs to be used by non-skilled personnel for rapid triaging in remote environments. Even in the future, chronic patients could be able to scan themselves and send the images to specialists thanks to these technologies.
There has been a significant improvement in the field of AI-assisted US scanning in the last few years, especially with the use of deep-learning methods and larger datasets. As shown in this review, for instance in Table 1, works based on deep-learning methods have used typically one or two orders of magnitude more images to train the model than previous works based on more traditional machine-learning approaches such as SVM. The use of large training datasets is a typical requirement of deep-learning methods, as they allow for the generation of more robust and generalizable models.
It is interesting to note that, as shown, for instance, in Table 1, the accuracy of the published results obtained with SVM, RF, and DL methods seems to be similarly good in many cases. In fact, the accuracy achieved in the published results would be good enough for practical purposes if they showed the same performance in other real-case scenarios.
However, the comparison between different publications in this field is very difficult, even when they are focused on imaging the same organ or region, as each one is applied to a different dataset with different US systems and different image quality. A detailed and fair comparison of the best implementations of each proposed AI method applied to a specific study would be very beneficial to the field.
The development of these AI-based technologies has many important challenges that are expected to be addressed in the following years:
- Deep learning systems are only as good as the curated datasets applied in their training. The dependence on the operator creates a large variability in US-acquired images, which may limit the generalizability of DL-based systems [69]. AI results depend on how the target structure is represented and defined by the examiner in the captured image [70] and, furthermore, on whether the target is correctly identified and captured. Therefore, the data used for training AI systems must be acquired by examiners with some degree of competency and experience [70].
- Training data are usually taken by a group of physicians that share the same protocols and criteria to choose the right standard views. This may create considerable differences between the imaging data collected to train AI algorithms and the data from real-world practice. Normalization of scanning and image acquisition may be required to mitigate this problem, and it could be critical for the successful future application of AI in the US. In fact, this requirement of AI tools may promote the standardization and image quality verification of US examinations performed by humans.
- A lot of expert sonographer time is required to create datasets by sampling the body surface in all needed probe positions and orientations. Even in a simplified case, as the one presented in [71] for the parasternal long axis (Plax) acoustic window, it required more than 20 min per patient.
- Most of the proposed approaches for AI-based ultrasound guidance in this review paper require labeled data to train the algorithms. Labels may be obtained from external devices such as an IMU [59], an external optical tracking system in the probe [58], or by expert US practitioners. In both cases, labels are prone to errors, which limit the best achievable performance of the DL models trained with them.
- The development of these technologies requires collaboration between experts from multiple disciplines. Most of the successful ML applications in medicine arose from collaborations of computational scientists, signal-processing engineers, physics experts, and medical experts [7]. However, sometimes it is not straightforward to combine different perspectives, different data models, and different approaches to data analysis.
In the next few years, it is expected that large US datasets will become available to the scientific community, and this will facilitate the development of better algorithms. In this field, the robustness of the implemented method, i.e., its capability to assist the scanning of any patient, is probably more important than its final accuracy (as long as it is within a certain range). This is an important difference with respect to other medical imaging fields where AI is being used, such as CT, where the variability between scans is low and accuracy and other related metrics are sometimes the only required consideration. Therefore, special efforts should be devoted in AI-assisted US scanning to the robustness of the algorithms to make them useful for the physicians.
The approval of these assistance tools by agencies such as the FDA in the USA and the CE mark in Europe has already started [72], and it is expected to continue in the following years with other tools. Their availability will provide very useful feedback from the users, which is expected to improve how the information from the AI tools is used to guide the scanning.
We acknowledge that this review may not have considered all publications in this field. Our goal has been to provide an overview of the state of the art, and the fact that this is a very active field makes it difficult to include all works and methods that are being published and proposed. Therefore, if any related work has not been mentioned, the omission has been totally unintentional.
5. Conclusions
Ultrasound imaging stands out for its safety, portability, non-invasiveness, and relatively low cost. It can dynamically display 2D images of the region of interest (ROI) in real-time with lightweight, portable devices. However, ultrasound imaging is highly operator-dependent, making it essential that the sonographer is properly trained in order to be able to implement the full diagnostic capabilities of the technique. Computer-assisted tools can help to reduce the steep learning curve in ultrasound scanning by assisting physicians, nurses, and technicians to perform full examinations. In this work, we have reviewed the most recent advances in AI-based methods toward facilitating the acquisition of high-quality US images. The results obtained in recent years have shown that AI-based assisted US is becoming a mature technology that will facilitate the widespread use of the technique, starting a new era in US scanning.
Author Contributions
Conceptualization and methodology (J.L.H. and D.M.); writing—original draft preparation (all authors); funding acquisition (C.I.I.). All authors have read and agreed to the published version of the manuscript.
Funding
Funded by the Spanish Ministry of Economic Affairs and Digital Transformation (Project MIA.2021.M02.0005 TARTAGLIA, from the Recovery, Resilience, and Transformation Plan financed by the European Union through Next Generation EU funds). TARTAGLIA takes place under the R&D Missions in Artificial Intelligence program, which is part of the Spain Digital 2025 Agenda and the Spanish National Artificial Intelligence Strategy.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Warner, H.R.; Gardner, R.M.; Toronto, A.F. Computer-Based Monitoring of Cardiovascular Functions in Postoperative Patients. Circulation 1968, 37, II68–II74. [Google Scholar] [CrossRef] [PubMed]
- Ambinder, E.P. A History of the Shift Toward Full Computerization of Medicine. J. Oncol. Pract. 2005, 1, 54–56. [Google Scholar] [CrossRef] [PubMed]
- Turing, A.M. Intelligent Machinery, 1948, 4. Reprinted in Mechanical Intelligence (Collected Works of AM Turing); North-Holland Publishing Co.: Amsterdam, The Netherlands, 1992. [Google Scholar]
- Keane, P.A.; Topol, E.J. With an Eye to AI and Autonomous Diagnosis. NPJ Digit. Med. 2018, 1, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Do, S.; Song, K.D.; Chung, J.W. Basics of Deep Learning: A Radiologist’s Guide to Understanding Published Radiology Articles on Deep Learning. Korean J. Radiol. 2020, 21, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Topol, E.J. High-Performance Medicine: The Convergence of Human and Artificial Intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Littmann, M.; Selig, K.; Cohen-Lavi, L.; Frank, Y.; Hönigschmid, P.; Kataka, E.; Mösch, A.; Qian, K.; Ron, A.; Schmid, S. Validity of Machine Learning in Biology and Medicine Increased through Collaborations across Fields of Expertise. Nat. Mach. Intell. 2020, 2, 18–24. [Google Scholar] [CrossRef]
- Zhou, S.K.; Greenspan, H.; Davatzikos, C.; Duncan, J.S.; Van Ginneken, B.; Madabhushi, A.; Prince, J.L.; Rueckert, D.; Summers, R.M. A Review of Deep Learning in Medical Imaging: Imaging Traits, Technology Trends, Case Studies With Progress Highlights, and Future Promises. Proc. IEEE 2021, 109, 820–838. [Google Scholar] [CrossRef]
- Wang, Y.; Ge, X.; Ma, H.; Qi, S.; Zhang, G.; Yao, Y. Deep Learning in Medical Ultrasound Image Analysis: A Review. IEEE Access 2021, 9, 54310–54324. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Y.; Yang, X.; Lei, B.; Liu, L.; Li, S.X.; Ni, D.; Wang, T. Deep Learning in Medical Ultrasound Analysis: A Review. Engineering 2019, 5, 261–275. [Google Scholar] [CrossRef]
- Deffieux, T.; Demené, C.; Tanter, M. Functional Ultrasound Imaging: A New Imaging Modality for Neuroscience. Neuroscience 2021, 474, 110–121. [Google Scholar] [CrossRef]
- Shah, S.; Bellows, B.A.; Adedipe, A.A.; Totten, J.E.; Backlund, B.H.; Sajed, D. Perceived Barriers in the Use of Ultrasound in Developing Countries. Crit. Ultrasound J. 2015, 7, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Maraci, M.A.; Napolitano, R.; Papageorghiou, A.; Noble, J.A. Searching for Structures of Interest in an Ultrasound Video Sequence; Springer: Berlin/Heidelberg, Germany, 2014; pp. 133–140. [Google Scholar]
- Chang, R.-F.; Wu, W.-J.; Moon, W.K.; Chen, D.-R. Improvement in Breast Tumor Discrimination by Support Vector Machines and Speckle-Emphasis Texture Analysis. Ultrasound Med. Biol. 2003, 29, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Bridge, C.P.; Ioannou, C.; Noble, J.A. Automated Annotation and Quantitative Description of Ultrasound Videos of the Fetal Heart. Med. Image Anal. 2017, 36, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Attia, M.W.; Abou-Chadi, F.; Moustafa, H.E.-D.; Mekky, N. Classification of Ultrasound Kidney Images Using PCA and Neural Networks. Int. J. Adv. Comput. Sci. Appl. 2015, 6, 53–57. [Google Scholar]
- Chen, H.; Dou, Q.; Ni, D.; Cheng, J.-Z.; Qin, J.; Li, S.; Heng, P.-A. Automatic Fetal Ultrasound Standard Plane Detection Using Knowledge Transferred Recurrent Neural Networks; Springer: Berlin/Heidelberg, Germany, 2015; pp. 507–514. [Google Scholar]
- Chen, H.; Ni, D.; Qin, J.; Li, S.; Yang, X.; Wang, T.; Heng, P.A. Standard Plane Localization in Fetal Ultrasound via Domain Transferred Deep Neural Networks. IEEE J. Biomed. Health Inform. 2015, 19, 1627–1636. [Google Scholar] [CrossRef]
- Ni, D.; Li, T.; Yang, X.; Qin, J.; Li, S.; Chin, C.-T.; Ouyang, S.; Wang, T.; Chen, S. Selective Search and Sequential Detection for Standard Plane Localization in Ultrasound; Springer: Berlin/Heidelberg, Germany, 2013; pp. 203–211. [Google Scholar]
- Ni, D.; Yang, X.; Chen, X.; Chin, C.-T.; Chen, S.; Heng, P.A.; Li, S.; Qin, J.; Wang, T. Standard Plane Localization in Ultrasound by Radial Component Model and Selective Search. Ultrasound Med. Biol. 2014, 40, 2728–2742. [Google Scholar] [CrossRef]
- Rahmatullah, B.; Sarris, I.; Papageorghiou, A.; Noble, J.A. Quality Control of Fetal Ultrasound Images: Detection of Abdomen Anatomical Landmarks Using AdaBoost; IEEE: Piscataway, NJ, USA, 2011; pp. 6–9. [Google Scholar]
- Rahmatullah, B.; Papageorghiou, A.T.; Noble, J.A. Integration of Local and Global Features for Anatomical Object Detection in Ultrasound; Springer: Berlin/Heidelberg, Germany, 2012; pp. 402–409. [Google Scholar]
- Wu, H.; Bowers, D.M.; Huynh, T.T.; Souvenir, R. Echocardiogram View Classification Using Low-Level Features; IEEE: Piscataway, NJ, USA, 2013; pp. 752–755. [Google Scholar]
- Agarwal, D.; Shriram, K.; Subramanian, N. Automatic View Classification of Echocardiograms Using Histogram of Oriented Gradients; IEEE: Piscataway, NJ, USA, 2013; pp. 1368–1371. [Google Scholar]
- Kumar, R.; Wang, F.; Beymer, D.; Syeda-Mahmood, T. Echocardiogram View Classification Using Edge Filtered Scale-Invariant Motion Features. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; IEEE: Miami, FL, USA, 2009; pp. 723–730. [Google Scholar]
- Yeh, W.-C.; Huang, S.-W.; Li, P.-C. Liver Fibrosis Grade Classification with B-Mode Ultrasound. Ultrasound Med. Biol. 2003, 29, 1229–1235. [Google Scholar] [CrossRef]
- Lei, B.; Tan, E.-L.; Chen, S.; Zhuo, L.; Li, S.; Ni, D.; Wang, T. Automatic Recognition of Fetal Facial Standard Plane in Ultrasound Image via Fisher Vector. PloS ONE 2015, 10, e0121838. [Google Scholar] [CrossRef]
- Qu, R.; Xu, G.; Ding, C.; Jia, W.; Sun, M. Deep Learning-Based Methodology for Recognition of Fetal Brain Standard Scan Planes in 2D Ultrasound Images. IEEE Access 2019, 8, 44443–44451. [Google Scholar] [CrossRef]
- Cai, Y.; Sharma, H.; Chatelain, P.; Noble, J.A. SonoEyeNet: Standardized Fetal Ultrasound Plane Detection Informed by Eye Tracking; IEEE: Piscataway, NJ, USA, 2018; pp. 1475–1478. [Google Scholar]
- Baumgartner, C.F.; Kamnitsas, K.; Matthew, J.; Smith, S.; Kainz, B.; Rueckert, D. Real-Time Standard Scan Plane Detection and Localisation in Fetal Ultrasound Using Fully Convolutional Neural Networks; Springer: Berlin/Heidelberg, Germany, 2016; pp. 203–211. [Google Scholar]
- Baumgartner, C.F.; Kamnitsas, K.; Matthew, J.; Fletcher, T.P.; Smith, S.; Koch, L.M.; Kainz, B.; Rueckert, D. SonoNet: Real-Time Detection and Localisation of Fetal Standard Scan Planes in Freehand Ultrasound. IEEE Trans. Med. Imaging 2017, 36, 2204–2215. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, C.; Liu, L.; Feng, C.; Dong, C.; Luo, Y.; Wan, X. Effective Sample Pair Generation for Ultrasound Video Contrastive Representation Learning. arXiv 2020, arXiv:2011.13066. [Google Scholar]
- Gao, Y.; Beriwal, S.; Craik, R.; Papageorghiou, A.T.; Noble, J.A. Label Efficient Localization of Fetal Brain Biometry Planes in Ultrasound through Metric Learning. In Medical Ultrasound, and Preterm, Perinatal and Paediatric Image Analysis; Springer: Berlin/Heidelberg, Germany, 2020; pp. 126–135. [Google Scholar]
- Yaqub, M.; Kelly, B.; Papageorghiou, A.T.; Noble, J.A. A Deep Learning Solution for Automatic Fetal Neurosonographic Diagnostic Plane Verification Using Clinical Standard Constraints. Ultrasound Med. Biol. 2017, 43, 2925–2933. [Google Scholar] [CrossRef]
- Burgos-Artizzu, X.P.; Coronado-Gutiérrez, D.; Valenzuela-Alcaraz, B.; Bonet-Carne, E.; Eixarch, E.; Crispi, F.; Gratacós, E. Evaluation of Deep Convolutional Neural Networks for Automatic Classification of Common Maternal Fetal Ultrasound Planes. Sci. Rep. 2020, 10, 10200. [Google Scholar] [CrossRef]
- Bridge, C.P.; Noble, J.A. Object Localisation in Fetal Ultrasound Images Using Invariant Features; IEEE: Piscataway, NJ, USA, 2015; pp. 156–159. [Google Scholar]
- Chykeyuk, K.; Yaqub, M.; Alison Noble, J. Class-Specific Regression Random Forest for Accurate Extraction of Standard Planes from 3D Echocardiography; Springer: Berlin/Heidelberg, Germany, 2013; pp. 53–62. [Google Scholar]
- Lorenz, C.; Brosch, T.; Ciofolo-Veit, C.; Tobias, K.; Lefevre, T.; Salim, I.; Papageorghiou, A.T.; Raynaud, C.; Roundhill, D.; Rouet, L.; et al. Automated Abdominal Plane and Circumference Estimation in 3D US for Fetal Screening; SPIE: Bellingham, WA, USA, 2018. [Google Scholar]
- Li, Y.; Khanal, B.; Hou, B.; Alansary, A.; Cerrolaza, J.J.; Sinclair, M.; Matthew, J.; Gupta, C.; Knight, C.; Kainz, B. Standard Plane Detection in 3d Fetal Ultrasound Using an Iterative Transformation Network; Springer: Berlin/Heidelberg, Germany, 2018; pp. 392–400. [Google Scholar]
- Li, Y.; Cerrolaza, J.J.; Sinclair, M.; Hou, B.; Alansary, A.; Khanal, B.; Matthew, J.; Kainz, B.; Rueckert, D. Standard Plane Localisation in 3D Fetal Ultrasound Using Network with Geometric and Image Loss. 2018. Available online: https://openreview.net/forum?id=BykcN8siz (accessed on 9 March 2023).
- Ryou, H.; Yaqub, M.; Cavallaro, A.; Roseman, F.; Papageorghiou, A.; Noble, J.A. Automated 3D Ultrasound Biometry Planes Extraction for First Trimester Fetal Assessment; Springer: Berlin/Heidelberg, Germany, 2016; pp. 196–204. [Google Scholar]
- Dou, H.; Yang, X.; Qian, J.; Xue, W.; Qin, H.; Wang, X.; Yu, L.; Wang, S.; Xiong, Y.; Heng, P.-A. Agent with Warm Start and Active Termination for Plane Localization in 3D Ultrasound; Springer: Berlin/Heidelberg, Germany, 2019; pp. 290–298. [Google Scholar]
- Yang, X.; Huang, Y.; Huang, R.; Dou, H.; Li, R.; Qian, J.; Huang, X.; Shi, W.; Chen, C.; Zhang, Y. Searching Collaborative Agents for Multi-Plane Localization in 3d Ultrasound. Med. Image Anal. 2021, 72, 102119. [Google Scholar] [CrossRef] [PubMed]
- Ren, P.; Xiao, Y.; Chang, X.; Huang, P.-Y.; Li, Z.; Chen, X.; Wang, X. A Comprehensive Survey of Neural Architecture Search: Challenges and Solutions. ACM Comput. Surv. CSUR 2021, 54, 1–34. [Google Scholar] [CrossRef]
- Dong, X.; Yang, Y. Searching for a Robust Neural Architecture in Four Gpu Hours. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; 2019; pp. 1761–1770. Available online: https://openaccess.thecvf.com/content_CVPR_2019/papers/Dong_Searching_for_a_Robust_Neural_Architecture_in_Four_GPU_Hours_CVPR_2019_paper.pdf (accessed on 9 March 2023).
- Jiang, B.; Xu, K.; Taylor, R.H.; Graham, E.; Unberath, M.; Boctor, E.M. Standard Plane Extraction From 3D Ultrasound With 6-DOF Deep Reinforcement Learning Agent; IEEE: Piscataway, NJ, USA, 2020; pp. 1–4. [Google Scholar]
- Yu, T.-F.; Liu, P.; Peng, Y.-L.; Liu, J.-Y.; Yin, H.; Liu, D.C. Slice Localization for Three-Dimensional Breast Ultrasound Volume Using Deep Learning. DEStech Trans. Eng. Technol. Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Skelton, E.; Matthew, J.; Li, Y.; Khanal, B.; Martinez, J.C.; Toussaint, N.; Gupta, C.; Knight, C.; Kainz, B.; Hajnal, J. Towards Automated Extraction of 2D Standard Fetal Head Planes from 3D Ultrasound Acquisitions: A Clinical Evaluation and Quality Assessment Comparison. Radiography 2021, 27, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Baum, Z.; Hu, Y.; Barratt, D.C. Multimodality Biomedical Image Registration Using Free Point Transformer Networks. In Medical Ultrasound, and Preterm, Perinatal and Paediatric Image Analysis; Springer: Berlin/Heidelberg, Germany, 2020; pp. 116–125. [Google Scholar]
- Khan, S.; Huh, J.; Ye, J.C. Variational Formulation of Unsupervised Deep Learning for Ultrasound Image Artifact Removal. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2021, 68, 2086–2100. [Google Scholar] [CrossRef]
- Rindal, O.M.H.; Austeng, A.; Fatemi, A.; Rodriguez-Molares, A. The Effect of Dynamic Range Alterations in the Estimation of Contrast. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2019, 66, 1198–1208. [Google Scholar] [CrossRef]
- Rodriguez-Molares, A.; Rindal, O.M.H.; D’hooge, J.; Masoy, S.-E.; Austeng, A.; Lediju Bell, M.A.; Torp, H. The Generalized Contrast-to-Noise Ratio: A Formal Definition for Lesion Detectability. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2020, 67, 745–759. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, H.; Luo, H.; Li, K. Automatic Quality Assessment for 2D Fetal Sonographic Standard Plane Based on Multitask Learning. Medicine 2021, 100, e24427. [Google Scholar] [CrossRef]
- Prevost, R.; Salehi, M.; Jagoda, S.; Kumar, N.; Sprung, J.; Ladikos, A.; Bauer, R.; Zettinig, O.; Wein, W. 3D Freehand Ultrasound without External Tracking Using Deep Learning. Med. Image Anal. 2018, 48, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Xu, S.; Wood, B.; Yan, P. Sensorless Freehand 3D Ultrasound Reconstruction via Deep Contextual Learning; Springer: Berlin/Heidelberg, Germany, 2020; pp. 463–472. [Google Scholar]
- Park, S.H. Artificial Intelligence for Ultrasonography: Unique Opportunities and Challenges. Ultrasonography 2021, 40, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Jarosik, P.; Lewandowski, M. Automatic Ultrasound Guidance Based on Deep Reinforcement Learning; IEEE: Piscataway, NJ, USA, 2019. [Google Scholar]
- Milletari, F.; Birodkar, V.; Sofka, M. Straight to the Point: Reinforcement Learning for User Guidance in Ultrasound. In Smart Ultrasound Imaging and Perinatal, Preterm and Paediatric Image Analysis; Springer: Berlin/Heidelberg, Germany, 2019; pp. 3–10. [Google Scholar]
- Droste, R.; Drukker, L.; Papageorghiou, A.T.; Noble, J.A. Automatic Probe Movement Guidance for Freehand Obstetric Ultrasound; Springer: Berlin/Heidelberg, Germany, 2020; pp. 583–592. [Google Scholar]
- Pan, Y.; Cheng, C.-A.; Saigol, K.; Lee, K.; Yan, X.; Theodorou, E.; Boots, B. Agile Autonomous Driving Using End-to-End Deep Imitation Learning. arXiv 2017, arXiv:1709.07174 2017. [Google Scholar]
- Narang, A.; Bae, R.; Hong, H.; Thomas, Y.; Surette, S.; Cadieu, C.; Chaudhry, A.; Martin, R.P.; McCarthy, P.M.; Rubenson, D.S.; et al. Utility of a Deep-Learning Algorithm to Guide Novices to Acquire Echocardiograms for Limited Diagnostic Use. JAMA Cardiol. 2021, 6, 624. [Google Scholar] [CrossRef] [PubMed]
- Toporek, G.; Wang, H.; Balicki, M.; Xie, H. Autonomous Image-Based Ultrasound Probe Positioning via Deep Learning; EasyChair Preprint no. 119. 2018. Available online: https://easychair.org/publications/preprint/XmZ9 (accessed on 9 March 2023).
- Mustafa, A.S.B.; Ishii, T.; Matsunaga, Y.; Nakadate, R.; Ishii, H.; Ogawa, K.; Saito, A.; Sugawara, M.; Niki, K.; Takanishi, A. Development of Robotic System for Autonomous Liver Screening Using Ultrasound Scanning Device; IEEE: Piscataway, NJ, USA, 2013; pp. 804–809. [Google Scholar]
- Liang, K.; Rogers, A.J.; Light, E.D.; von Allmen, D.; Smith, S.W. Three-Dimensional Ultrasound Guidance of Autonomous Robotic Breast Biopsy: Feasibility Study. Ultrasound Med. Biol. 2010, 36, 173–177. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Kendall, A.; Grimes, M.; Cipolla, R. Posenet: A convolutional network for real-time 6-dof camera relocalization. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 11–18 December 2015; pp. 2938–2946. [Google Scholar]
- Hase, H.; Azampour, M.F.; Tirindelli, M.; Paschali, M.; Simson, W.; Fatemizadeh, E.; Navab, N. Ultrasound-Guided Robotic Navigation with Deep Reinforcement Learning; IEEE: Piscataway, NJ, USA, 2020; pp. 5534–5541. [Google Scholar]
- Huang, Q.; Lan, J.; Li, X. Robotic Arm Based Automatic Ultrasound Scanning for Three-Dimensional Imaging. IEEE Trans. Ind. Inform. 2018, 15, 1173–1182. [Google Scholar] [CrossRef]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key Challenges for Delivering Clinical Impact with Artificial Intelligence. BMC Med. 2019, 17, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jeong, E.Y.; Kim, H.L.; Ha, E.J.; Park, S.Y.; Cho, Y.J.; Han, M. Computer-Aided Diagnosis System for Thyroid Nodules on Ultrasonography: Diagnostic Performance and Reproducibility Based on the Experience Level of Operators. Eur. Radiol. 2019, 29, 1978–1985. [Google Scholar] [CrossRef]
- Kennedy Hall, M.; Coffey, E.; Herbst, M.; Liu, R.; Pare, J.R.; Andrew Taylor, R.; Thomas, S.; Moore, C.L. The “5Es” of Emergency Physician–Performed Focused Cardiac Ultrasound: A Protocol for Rapid Identification of Effusion, Ejection, Equality, Exit, and Entrance. Acad. Emerg. Med. 2015, 22, 583–593. [Google Scholar] [CrossRef]
- FDA Authorizes Marketing of First Cardiac Ultrasound Software That Uses Artificial Intelligence to Guide User. Feb 2020. FDA news. Available online: https://www.fda.gov/news-events/press-announcements/fda-authorizes-marketing-first-cardiac-ultrasound-software-uses-artificial-intelligence-guide-user (accessed on 9 March 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).