Mechanical Design of DNA Origami in the Classroom
Abstract
1. Introduction
2. Methods
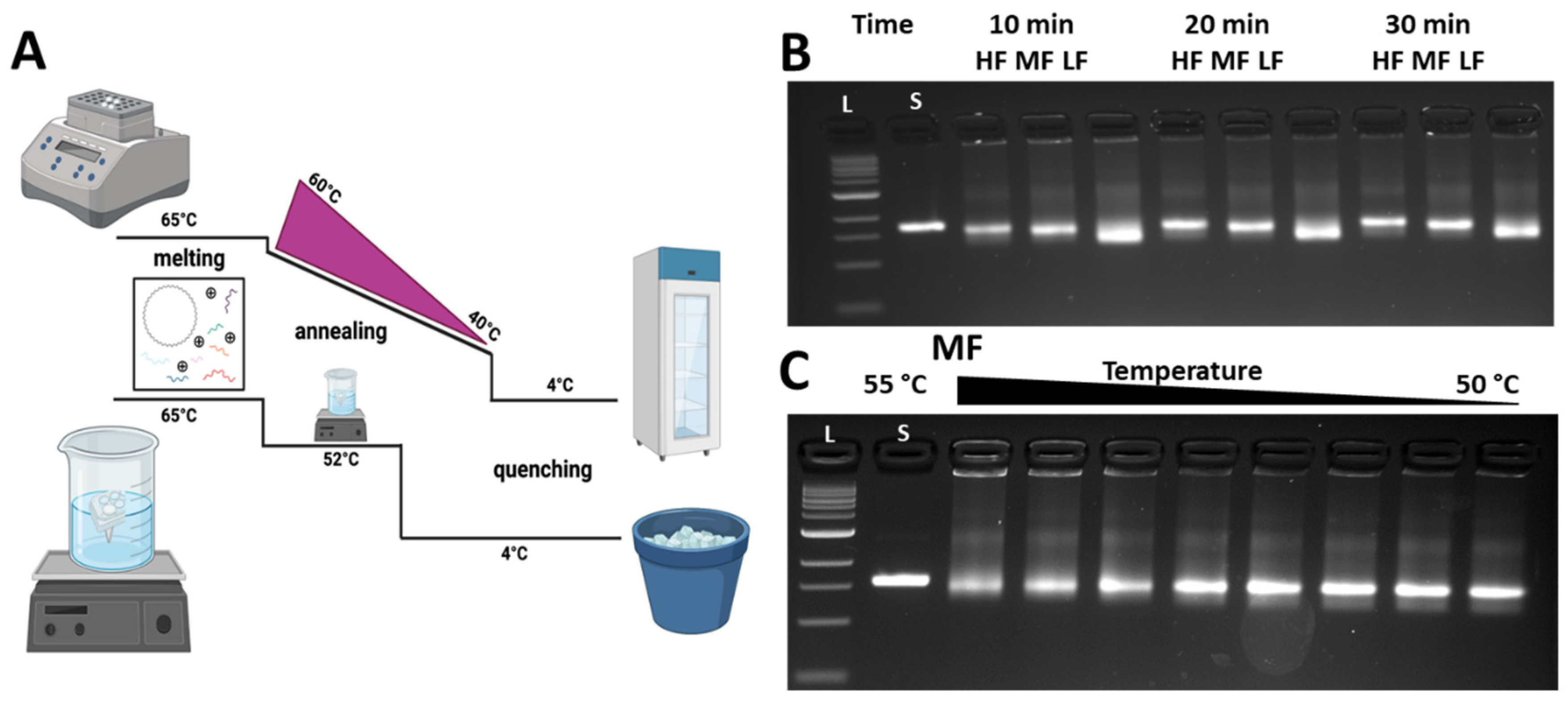
2.1. Fabrication of the DNA Origami Compliant Hinge Joint
2.2. Analysis of CHJ by Agarose Gel Electrophoresis
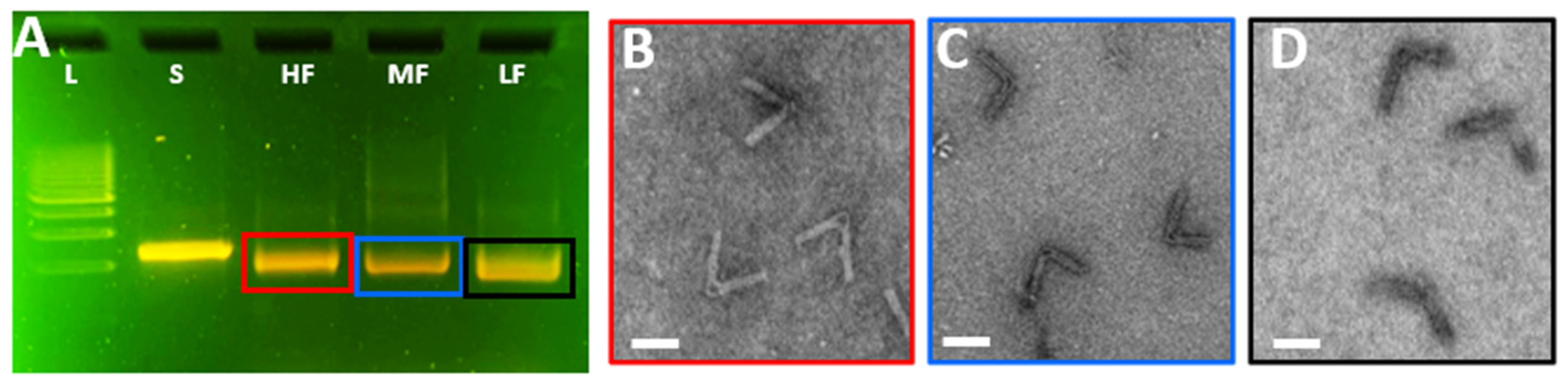
2.3. Imaging of DNA Origami via Transmission Electron Microscopy
2.4. Simulation of DNA Origami CHJ
3. Results
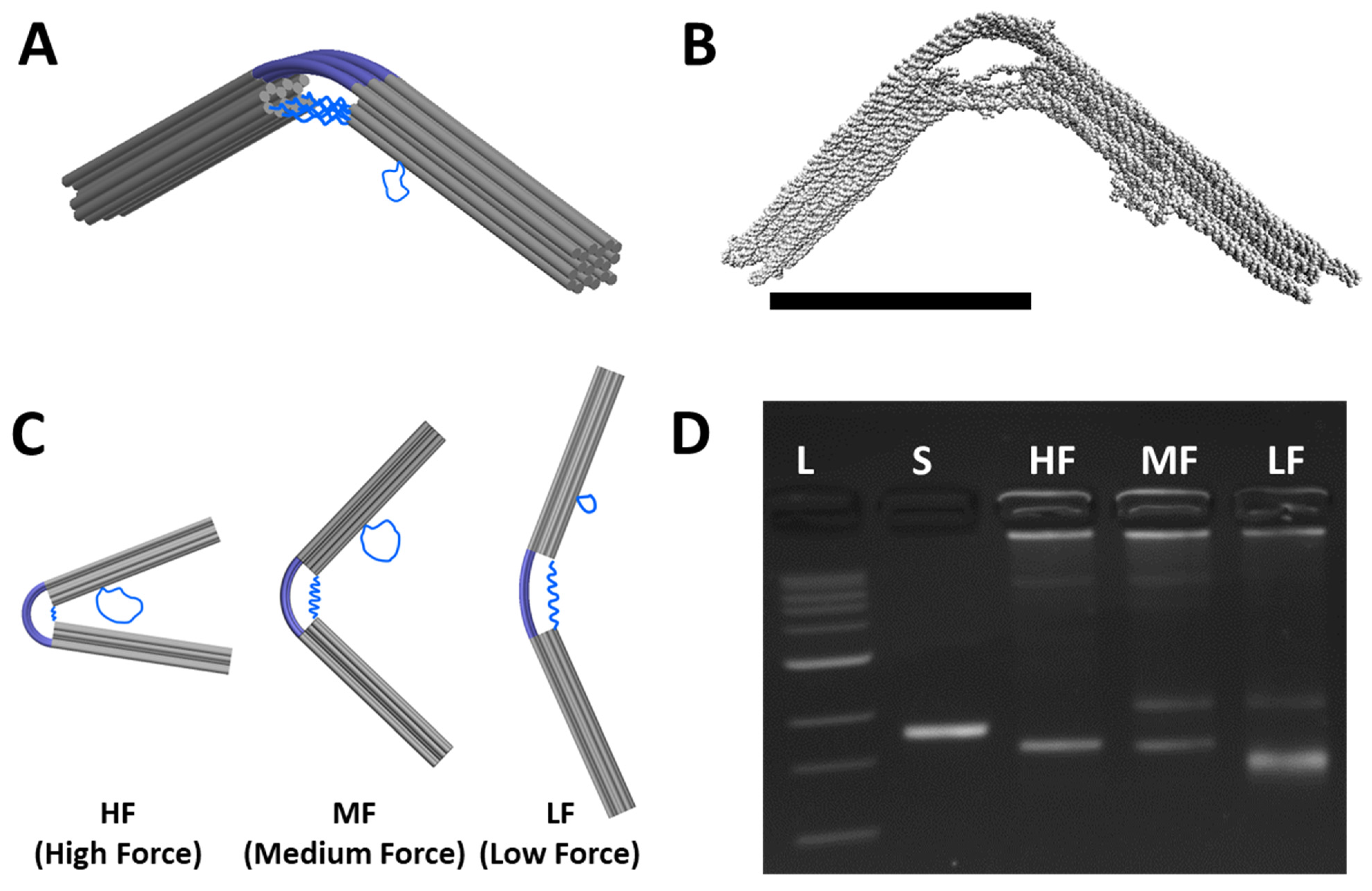
3.1. The Compliant Hinge Joint Design
3.2. Folding the CHJ with Classroom-Friendly Protocols
3.3. Analysis of the CHJ with Classroom Gel Electrophoresis
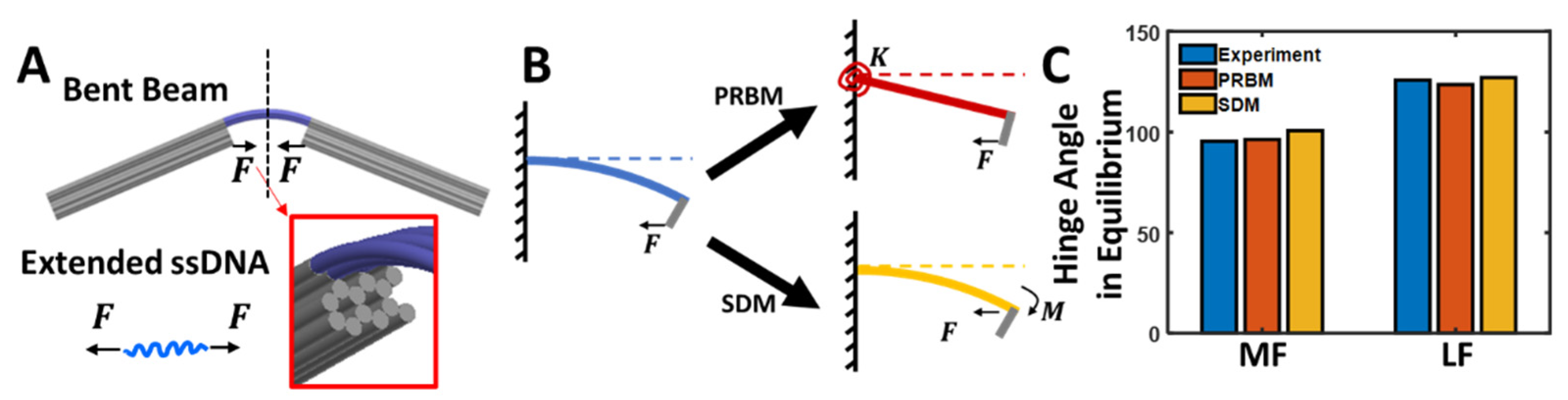
3.4. Mechanical Modeling of the Compliant Hinge Joint Angles
3.5. Pseudo-Rigid-Body Model
3.6. Small Deflection Model
3.7. Predicting the CHJ Joint Angles
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA Origami: Scaffolds for Creating Higher Order Structures. Chem. Rev. 2017, 117, 12584–12640. [Google Scholar] [CrossRef] [PubMed]
- Marras, A.E.; Zhou, L.; Su, H.J.; Castro, C.E. Programmable motion of DNA origami mechanisms. Proc. Natl. Acad. Sci. USA 2015, 112, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-S.; Pierce, N.A. A Synthetic DNA Walker for Molecular Transport. J. Am. Chem. Soc. 2004, 126, 10834–10835. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Wang, J.; Wang, E.; Dong, S. Propelling DNA Computing with Materials’ Power: Recent Advancements in Innovative DNA Logic Computing Systems and Smart Bio-Applications. Adv. Sci. 2020, 7, 2001766. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Chen, H.; Choi, J.H. Auxetic Two-Dimensional Nanostructures from DNA. Angew. Chem. Int. Ed. 2021, 60, 7165–7173. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, Z.; Ma, N.; Yang, S.; Li, K.; Teng, C.; Ke, Y.; Tian, Y. DNA origami-enabled biosensors. Sensors 2020, 20, 6899. [Google Scholar] [CrossRef] [PubMed]
- Udomprasert, A.; Kangsamaksin, T. DNA origami applications in cancer therapy. Cancer Sci. 2017, 108, 1535–1543. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Karna, D.; Mao, H. DNA origami nano-mechanics. Chem. Soc. Rev. 2021, 50, 11966–11978. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Sun, W.; Shen, Z.; Seeman, N.C. A nanomechanical device based on the B–Z transition of DNA. Nature 1999, 397, 144–146. [Google Scholar] [CrossRef] [PubMed]
- Yurke, B.; Turber, A.J.; Mills, A.P., Jr.; Simmel, F.C.; Neumann, J.L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhang, X.; Shen, Z.; Seeman, N.C. A robust DNA mechanical device controlled by hybridization topology. Nature 2002, 415, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, C.M.; Adler, M. Nanomechanical Devices Based on DNA. Angew. Chem. Int. Ed. 2002, 41, 3779–3783. [Google Scholar] [CrossRef]
- Chakraborty, B.; Sha, R.; Seeman, N.C. A DNA-based nanomechanical device with three robust states. Proc. Natl. Acad. Sci. USA 2008, 105, 17245–17249. [Google Scholar] [CrossRef]
- Andersen, E.S.; Dong, M.; Nielsen, M.M.; Jahn, K.; Subramani, R.; Mamdouh, W.; Golas, M.M.; Sander, B.; Stark, H.; Oliveira, C.L.P.; et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 2009, 459, 73–76. [Google Scholar] [CrossRef]
- Marini, M.; Piantanida, L.; Musetti, R.; Bek, A.; Dong, M.; Besenbacher, F.; Lazzarino, M.; Firrao, G. A Revertible, Autonomous, Self-Assembled DNA-Origami Nanoactuator. Nano Lett. 2011, 11, 5449–5454. [Google Scholar] [CrossRef]
- Zhou, L.; Marras, A.E.; Su, H.-J.; Castro, C.E. Direct Design of an Energy Landscape with Bistable DNA Origami Mechanisms. Nano Lett. 2015, 15, 1815–1821. [Google Scholar] [CrossRef]
- Willner, E.M.; Kamada, Y.; Suzuki, Y.; Emura, T.; Hidaka, K.; Dietz, H.; Sugiyama, H.; Endo, M. Single-Molecule Observation of the Photoregulated Conformational Dynamics of DNA Origami Nanoscissors. Angew. Chem. Int. Ed. 2017, 56, 15324–15328. [Google Scholar] [CrossRef]
- Lee, C.; Lee, J.Y.; Kim, D.-N. Polymorphic design of DNA origami structures through mechanical control of modular components. Nat. Commun. 2017, 8, 2067. [Google Scholar] [CrossRef]
- Gür, F.N.; Kempter, S.; Schueder, F.; Sikeler, C.; Urban, M.J.; Jungmann, R.; Nickels, P.C.; Liedl, T. Double- to Single-Strand Transition Induces Forces and Motion in DNA Origami Nanostructures. Adv. Mater. 2021, 33, 2101986. [Google Scholar] [CrossRef] [PubMed]
- DeLuca, M.; Shi, Z.; Castro, C.E.; Arya, G. Dynamic DNA nanotechnology: Toward functional nanoscale devices. Nanoscale Horiz. 2020, 5, 182–201. [Google Scholar] [CrossRef]
- Nummelin, S.; Shen, B.; Piskunen, P.; Liu, Q.; Kostiainen, M.A.; Linko, V. Robotic DNA Nanostructures. ACS Synth. Biol. 2020, 9, 1923–1940. [Google Scholar] [CrossRef] [PubMed]
- Pumm, A.-K.; Engelen, W.; Kopperger, E.; Isensee, J.; Vogt, M.; Kozina, V.; Kube, M.; Honemann, M.N.; Bertosin, E.; Langecker, M.; et al. A DNA origami rotary ratchet motor. Nature 2022, 607, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Beshay, P.E.; Kucinic, A.; Wile, N.; Halley, P.; Rosiers, L.D.; Chowdhury, A.; Hall, J.L.; Castro, C.E.; Hudoba, M.W. Translating DNA origami Nanotechnology to Middle School, High School, and Undergraduate Laboratories. bioRxiv 2022. [Google Scholar] [CrossRef]
- Castro, C.E.; Kilchherr, F.; Kim, D.N.; Shiao, E.L.; Wauer, T.; Wortmann, P.; Bathe, M.; Dietz, H. A primer to scaffolded DNA origami. Nat. Methods 2011, 8, 221–229. [Google Scholar] [CrossRef]
- Seeman, N.C.; Sleiman, H.F. DNA nanotechnology. Nat. Rev. Mater. 2017, 3, 1–23. [Google Scholar] [CrossRef]
- Dey, S.; Fan, C.; Gothelf, K.V.; Li, J.; Lin, C.; Liu, L.; Liu, N.; Nijenhuis, M.A.D.; Saccà, B.; Simmel, F.C.; et al. DNA origami. Nat. Rev. Methods Primer 2021, 1, 1–24. [Google Scholar] [CrossRef]
- Abraham Punnoose, J.; Halvorsen, K.; Chandrasekaran, A.R. DNA Nanotechnology in the Undergraduate Laboratory: Analysis of Molecular Topology Using DNA Nanoswitches. J. Chem. Educ. 2020, 97, 1448–1453. [Google Scholar] [CrossRef]
- Chandrasekaran, A.R. DNA Nanotechnology in the Undergraduate Laboratory: Electrophoretic Analysis of DNA Nanostructure Biostability. J. Chem. Educ. 2023, 100, 316–320. [Google Scholar] [CrossRef]
- Sobczak, J.P.J.; Martin, T.G.; Gerling, T.; Dietz, H. Rapid folding of DNA into nanoscale shapes at constant temperature. Science 2012, 338, 1458–1461. [Google Scholar] [CrossRef] [PubMed]
- Halley, P.D.; Patton, R.A.; Chowdhury, A.; Byrd, J.C.; Castro, C.E. Low-cost, simple, and scalable self-assembly of DNA origami nanostructures. Nano Res. 2019, 12, 1207–1215. [Google Scholar] [CrossRef]
- Castro, C.E.; Su, H.J.; Marras, A.E.; Zhou, L.; Johnson, J. Mechanical design of DNA nanostructures. Nanoscale 2015, 7, 5913–5921. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Marras, A.E.; Su, H.J.; Castro, C.E. DNA origami compliant nanostructures with tunable mechanical properties. ACS Nano 2014, 8, 27–34. [Google Scholar] [CrossRef]
- Douglas, S.M.; Marblestone, A.H.; Teerapittayanon, S.; Vazquez, A.; Church, G.M.; Shih, W.M. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009, 37, 5001–5006. [Google Scholar] [CrossRef]
- Poppleton, E.; Bohlin, J.; Matthies, M.; Sharma, S.; Zhang, F.; Šulc, P. Design, optimization and analysis of large DNA and RNA nanostructures through interactive visualization, editing and molecular simulation. Nucleic Acids Res. 2020, 48, e72. [Google Scholar] [CrossRef]
- Bohlin, J.; Matthies, M.; Poppleton, E.; Procyk, J.; Mallya, A.; Yan, H.; Šulc, P. Design and simulation of DNA, RNA and hybrid protein–nucleic acid nanostructures with oxView. Nat. Protoc. 2022, 17, 1762–1788. [Google Scholar] [CrossRef]
- Veneziano, R.; Ratanalert, S.; Zhang, K.; Zhang, F.; Yan, H.; Chiu, W.; Bathe, M. Designer nanoscale DNA assemblies programmed from the top down. Science 2016, 352, 1534. [Google Scholar] [CrossRef]
- Benson, E.; Mohammed, A.; Gardell, J.; Masich, S.; Czeizler, E.; Orponen, P.; Högberg, B. DNA rendering of polyhedral meshes at the nanoscale. Nature 2015, 523, 441–444. [Google Scholar] [CrossRef]
- Jun, H.; Shepherd, T.R.; Zhang, K.; Bricker, W.P.; Li, S.; Chiu, W.; Bathe, M. Automated Sequence Design of 3D Polyhedral Wireframe DNA Origami with Honeycomb Edges. ACS Nano 2019, 13, 2083–2093. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.M.; Kucinic, A.; Johnson, J.A.; Su, H.J.; Castro, C.E. Integrated computer-aided engineering and design for DNA assemblies. Nat. Mater. 2021, 20, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Pradeep Narayanan, R.; Prasad, A.; Zhang, F.; Williams, D.; Schreck, J.S.; Yan, H.; Reif, J. Automated design of 3D DNA origami with non-rasterized 2D curvature. Sci. Adv. 2022, 8, eade4455. [Google Scholar] [CrossRef] [PubMed]
- Glaser, M.; Deb, S.; Seier, F.; Agrawal, A.; Liedl, T.; Douglas, S.; Gupta, M.K.; Smith, D.M. The Art of Designing DNA Nanostructures with CAD Software. Molecules 2021, 26, 2287. [Google Scholar] [CrossRef]
- Shi, Z.; Castro, C.E.; Arya, G. Conformational Dynamics of Mechanically Compliant DNA Nanostructures from Coarse-Grained Molecular Dynamics Simulations. ACS Nano 2017, 11, 4617–4630. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Zhou, L.; Marras, A.E.; Castro, C.E.; Su, H.-J. Pseudorigid-Body Models of Compliant DNA Origami Mechanisms. J. Mech. Robot. 2016, 8, 051013. [Google Scholar] [CrossRef]
- Bauchau, O.A.; Craig, J.I. Euler-Bernoulli beam theory. In Structural Analysis; Bauchau, O.A., Craig, J.I., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 173–221. [Google Scholar] [CrossRef]
- Marko, J.F.M.; Siggia, E.D.S. Bending and Twisting Elasticity of DNA. Macromolecules 1994, 27, 981–988. [Google Scholar] [CrossRef]
- Howell, L.L. Compliant Mechanisms. In Encyclopedia of Nanotechnology; Bhushan, B., Ed.; Springer: Dordrecht, The Netherlands, 2016; pp. 604–611. ISBN 978-94-017-9780-1. [Google Scholar]
- Hibbeler, R.C. Mechanics of Materials; Prentice Hall: Upper Saddle River, NJ, USA, 2004. [Google Scholar]
- Howell, L.L.; Midha, A. Parametric Deflection Approximations for End-Loaded, Large-Deflection Beams in Compliant Mechanisms. J. Mech. Des. 1995, 117, 156–165. [Google Scholar] [CrossRef]
- Albanesi, A.E.; Fachinotti, V.D.; Pucheta, M.A. A Review on Design Methods for Compliant Mechanisms. Mecánica Comput. 2010, 29, 59–72. [Google Scholar]
- Chepkoech, M.; Alunda, B.O.; Otieno, L.O.; Park, S.J.; Byeon, C.C.; Lee, Y.J. Design and Fabrication of a Low-Cost Teaching Atomic Force Microscope with 3D Printed Parts. New Phys. Sae Mulli 2019, 69, 128–135. [Google Scholar] [CrossRef]
- Kauert, D.J.; Kurth, T.; Liedl, T.; Seidel, R. Direct Mechanical Measurements Reveal the Material Properties of Three-Dimensional DNA Origami. Nano Lett. 2011, 11, 5558–5563. [Google Scholar] [CrossRef] [PubMed]

| Hinge Name | Lower Three ssDNA Springs (Unit: Nucleotides) | Upper Three ssDNA Springs (Unit: Nucleotides) |
|---|---|---|
| HF | 0 | 24 |
| MF | 32 | 56 |
| LF | 74 | 84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Kucinic, A.; Des Rosiers, L.; Beshay, P.E.; Wile, N.; Hudoba, M.W.; Castro, C.E. Mechanical Design of DNA Origami in the Classroom. Appl. Sci. 2023, 13, 3208. https://doi.org/10.3390/app13053208
Wang Y, Kucinic A, Des Rosiers L, Beshay PE, Wile N, Hudoba MW, Castro CE. Mechanical Design of DNA Origami in the Classroom. Applied Sciences. 2023; 13(5):3208. https://doi.org/10.3390/app13053208
Chicago/Turabian StyleWang, Yuchen, Anjelica Kucinic, Lilly Des Rosiers, Peter E. Beshay, Nicholas Wile, Michael W. Hudoba, and Carlos E. Castro. 2023. "Mechanical Design of DNA Origami in the Classroom" Applied Sciences 13, no. 5: 3208. https://doi.org/10.3390/app13053208
APA StyleWang, Y., Kucinic, A., Des Rosiers, L., Beshay, P. E., Wile, N., Hudoba, M. W., & Castro, C. E. (2023). Mechanical Design of DNA Origami in the Classroom. Applied Sciences, 13(5), 3208. https://doi.org/10.3390/app13053208






