Abstract
Antibiotics are refractory pollutants that have been widely found in various environmental media such as soil and surface water. Existing sewage treatments perform poorly at preventing antibiotics in urban sewage from polluting natural environments. In this study, we designed a bioelectrically enhanced bioretention cell system (bioretention cell-microbial fuel cell, BRC-MFC) that utilizes the unique structure of the BRC system to improve the removal of sewage antibiotics. This new system can efficiently remove antibiotics by using a synergy of plant absorption, filler adsorption, filler filtration and microbial degradation. To study the influences of multiple-antibiotics stress on the decontamination performance of BRC-MFC, ofloxacin (OFLX) and tetracycline (TC) were selected as target antibiotics, and five BRC-MFCs were built to treat sewage containing antibiotics of different concentrations. The concentrations of pollutant in the influent and effluent were measured and the pollutant removal performance of BRC-MFC was studied. The diversity of rhizosphere microorganisms and the abundance of denitrifying functional genes were analyzed. Experimental results showed that over 90% of OFLX and TC in each BRC-MFC were removed, with the removal rates positively correlating with the concentration of antibiotics. In addition, the removal rates of chemical oxygen demand (COD) in BRC-MFC were both over 90%, while the removal rate of total nitrogen (TN) was around 70%. Meanwhile, antibiotics could significantly improve the removal of ammonia nitrogen (NH4+-N, p < 0.01). The microbial richness decreased, and we found that combined antibiotic stress on microorganisms was stronger than single antibiotic stress. The abundance of denitrifying functional genes was reduced by antibiotic stress. The results of this study provide reference values for other projects focusing on removing various antibiotics from domestic sewage using BRC-MFC.
1. Introduction
Antibiotics are widely used in human production and life. Most cannot be fully metabolized by humans and animals, and they eventually enter the natural environment through various means, such as hospital sewage and waste, antibiotic users’ excrement and wastewater from aquaculture [1]. Ofloxacin (OFLX), the representative antibiotic of fluoroquinolones (FQs), comes in one of the largest doses of the broad-spectrum antibiotics available and is thus one of the most important contaminants in the aquatic environment [2]. OFLX cannot only be detected in lakes and sewage plants around the world [3,4] but also often can be found in sediments due to its adsorption [5]. As a typical broad–spectrum antibiotic, Tetracycline (TC) is a representative antibiotic of tetracyclines (TCs), which is widely used in aquaculture, animal husbandry, and other industries [6]. However, tetracycline (TC) is largely left in environmental media [7] due to its strong adsorption in the environment, which exerts a negative impact on environmental ecology and human health. Antibiotics were detected in the tailwater of Lanzhou City’s wastewater treatment plant in Northwest China’s Gansu Province, with the highest concentration value reaching 9.78 μg·L−1. It was deduced that the 14 antibiotics continuously discharged from the wastewater plant to the Lanzhou section of the Yellow River combine to total about 5.66 kg per day, and the risk to the aquatic environment from long-cycle antibiotic exposure cannot be ignored [8].
The bioretention cell-microbial fuel cell (BRC-MFC) has been widely used to remove conventional pollutants from wastewater in recent years [9], and our group has been working on its use for a long time [10,11,12,13,14]. BRC-MFC has a good degradation effect on antibiotics as it can remove pollutants through multiple pathways such as filler adsorption, plant uptake, root secretion degradation, and microbial decomposition [15,16,17]. However, when applying BRC-MFC, plants and microorganisms can also be subjected to reverse stress from antibiotics during the treatment of antibiotic-containing wastewater [18], which, in turn, affects the decontamination performance of the BRC-MFC. For example, TC can inhibit the growth of plants and toxify their roots by inhibiting the protein synthesis of Microcystis aeruginosa and green algae [19], as well as by inhibiting the activity of enzymes in chloroplasts [20,21]. The residues of OFLX (>20 μg·mL−1) showed significant negative effects on tomato growth, photosynthesis, fluorescence parameters, antioxidant enzyme activity and transcript-level expression [22]. Antibiotics can also bind to active sites in soil enzyme molecules including sulfhydryl groups and imidazole-containing ligands, forming more stable complexes that inhibit the active centers of enzymes and reduce the activity of soil hydrolytic enzymes, thus affecting the conversion of soil nutrients as well as having indirect effects on plant growth [23]. High concentrations of norfloxacin inhibited the phyla Aspergillus and Actinomycetes [24], with the number of bacteria and fungi in the soil significantly reduced by 10 mg·kg−1 TC, marking reductions of 80.67% and 57.20%, while the maximum reduction rates were 86.91% and 74.01%, respectively [25]. Based on the many effects of antibiotics on plants and microorganisms in BRC-MFC systems, further systematic studies on the effects of multiple antibiotics on the decontamination performance of BRC-MFC and the response characteristics of rhizosphere microorganisms are warranted.
A microbial fuel cell (MFC) is a device that uses microorganisms to convert chemical energy in organic matter directly into electrical energy. The MFC is divided into an anode layer and a cathode layer, with the anode placed in an anaerobic environment in the submerged layer, the cathode in an aerobic environment in the transition layer and the two electrodes connected by external wires [26]. The electron-producing microorganisms in the submerged layer oxidize organic matter to produce electrons, which are transferred from the anode to the cathode via an external wire, creating a current to accelerate pollutant removal [27]. As a biocatalyst, the microbial community plays an important role in the performance of the MFC. When the MFC degrades antibiotics, the biological composition of the electrode surface is divided into two main components: an electrically active extracellular electricity-producing colony and an antibiotic-degrading colony. The complex interactions between the two are critical to antibiotic degradation and electrical energy production in the MFC [28]. Changes in environmental factors, such as the antibiotic concentration, antibiotic type, pH and temperature, can affect the structure of the microbial community in the MFC, which, in turn, affects the performance of the BRC-MFC in removing contaminants [29].
Based on this, this paper explores the response of the BRC-MFC system in decontamination and ecological restoration under multiple-antibiotics stress, with the aim of providing a scientific basis for the practical engineering of BRC-MFC for the removal of various types of antibiotics from domestic wastewater.
2. Materials and Methods
2.1. Experimental Methods
Based on the antibiotic stress response studies conducted by our group on different plants, the plant with the best resistance to antibiotic stress (Phalaris arundinacea) was selected as the subject of the study to investigate the fouling removal performance and inter-root ecological changes of the BRC-MFC system of the plant in response to the combined stress of TC and OFLX. The experiments were conducted in the water supply and drainage laboratory of Lanzhou University of Technology from June to September 2022. The BRC-MFC device is shown in Figure 1. It is a UVPC cylinder (diameter 300 mm × height 900 mm) and the packing in the device follows the Australian FAWB guidelines [30]. We used a total of 5 devices, each planted with 5 Phalaris arundinacea seedlings with good growth potential, planted at a density of 78 plants/m2. Bioparticulate activated carbon (BAC) was used as the electrode material; furthermore, the volume of both the anode and the cathode was 2.12 dm3. Catalytic iron was added below the anode area to provide the electron donor for the anaerobic system, copper wire was used to connect the two electrodes internally, and a 1000 Ω resistor was connected externally to form the whole circuit.
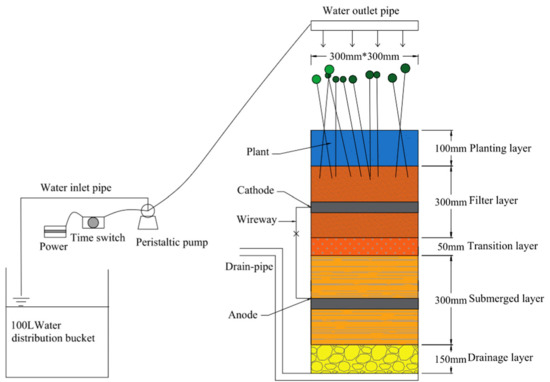
Figure 1.
Bioretention cell–microbial fuel cell (BRC-MFC) device diagram.
2.2. Device Operation
The reactor was operated in 2 stages as follows: OFLX stress stage (I) for 45 day, where no antibiotics were added to R0 as the control group, while OFLX (0.2, 1.2, 2.4 and 3.6 μg·mL−1) was added to R1-4, respectively; OFLX + TC combined stress stage (II) for 45 day, where no antibiotics were added to R0 as the control group, while OFLX (0.2, 1.2, 2.4 and 3.6 μg·mL−1) and TC (0.1, 0.6, 1.2 and 1.8 μg·mL−1) was added to R1-R4, respectively. The test setup used a downflow water intake with a multi-channel peristaltic pump (LEAD15–44, Longer, Hebei, China) for intermittent water intake, a single intake of 1 h, a hydraulic residence time of 7 h and an intake load of 1.0 m3·(m2·day)−1 [31]. The test period was 90 day, the pollutant indices of total nitrogen (TN), ammonia nitrogen (NH4+-N) and chemical oxygen demand (COD) were tested every 3 day and the antibiotic concentrations of TC and OFLX were tested every 7 day. The experimental influent was configured so that each liter of influent contained 0.225 g C6H12O6, 0.222 g NH4Cl, 0.022 g K2HPO4, 0.087 g NaCO3, 0.048 g FeCl2·4H2O, 0.004 g CoCl2·6H2O, 0.002 g NiCl2·6H2O, 0.000058 g MnCl2·4H2O, 0.000058 g CuSO4, 0.000058 g ZnCl2, 0.00043 CaCl2 and 0.0012 g humic acid.
2.3. Test Method
2.3.1. Pollutant Detection
The influent and effluent of the BRC-MFC systems were extracted and enriched using a solid stage extraction column (Infinity Lab Poroshe11 120 EC-C18). TC and OFLX concentrations were determined by the ultra-performance liquid chromatography-ultraviolet and visible spectrophotometry (UPLC-UV) method [32], TN was determined by ultraviolet and visible spectrophotometry using alkaline potassium persulphate digestion [33], NH4+-N was determined by Nessler’s reagent spectrophotometry [34], COD was determined by the digestion colorimetry method. We dropped 2 mL of sewage into the HACH COD prefabrication agent, shook it well and then digested it in the digester (HACH drb200) for 2 h; after cooling it to room temperature, we used a UV-visible spectrophotometer (HACH dr6000) for detection.
2.3.2. High-Throughput Sequencing of Microbial Community
Genomic DNA was extracted using a Rapid Soil DNA Extraction Kit (Qbiogene, Caarlsbad, CA, USA); furthermore, DNA integrity and purity were tested using 1% agarose gel electrophoresis. Polymerase chain reaction (PCR) amplification and product electrophoresis were performed using the genomic DNA as the template. What’s more, primers with barcode and PremixTaq (TaKaRa, Beijing, China) were used for PCR amplification depending on the region selected for sequencing. The PCR products were mixed and sequenced using the high-throughput sequencing platform Hiseq or Miseq. The experiment used 25 μL reaction system: 2 × 12.5 μL SYBR Green Realtime PCR Master Mix, 2.5 μL plus solution, 2 μL sample DNA (diluted 10 times), 0.75 μL upstream and downstream primers (final concentration 0.3 μmol/L), 6.5 μL ddH2O. The cycle conditions were denaturation for 120 s at 95 °C, and annealing for 60 s at 60 °C, for a total of 40 cycles [35,36].
2.3.3. Genetic Testing for Denitrification Function
DNA was extracted from the seed mud samples using a rapid soil DNA extraction kit (Qbiogene, Caarlsbad, CA, USA). Real-time fluorescence quantitative PCR was used for the detection of denitrification genes. Five genes were detected: nitrate reductase functional gene narG, nitrite nitrogen reductase functional gene nirS, NO reductase functional gene norB, N2O reductase functional gene nosZ and total bacteria (Bacteria) 16S rRNA. The primer sequences are shown in Table 1. PCR conditions: initial denaturation at 95 °C for 3 min, followed by holding at 95 °C for 30 s, annealing at the desired annealing temperature for 30 s and final extension at 72 °C for 40 s. All steps were cycled 35 times [37]. The sequencing data were analyzed with the R Programming Language and the National Center for Biotechnology Information database.

Table 1.
The sequences of specific primers in real-time qPCR analysis.
3. Results and Discussion
3.1. Effect of Antibiotics on the Decontamination Performance of BRC-MFC
3.1.1. Nutrient Removal Performance
In stage I, the COD removal effect of the five groups of reactors (R0~R4) was good, and the COD removal rate of BRC-MFC under different concentrations of antibiotic stress remained above 90%. Compared with R0, R1, and R2 showed a slight increase in COD removal rate, while R3 and R4 showed no significant changes. This showed that OFLX stress did not inhibit the COD removal ability of BRC-MFC. There was also a positive effect on COD removal at 0.2 and 1.2 μg·mL−1 OFLX stress. It has been found that antibiotics also have the effect of promoting microbial metabolic activity and stimulating microbial induction of respiration in the low concentration range very commonly [38]. Therefore, microbial activity in the reactor under OFLX loading conditions also increased accordingly, increasing its efficiency in utilizing COD. As shown in Figure 2a, the COD concentration of effluent was less than 30 μg·mL−1, meeting the surface water class IV standard. There was no significant difference (p > 0.05) between the TN removal rate of the test group R1~R4 and the control group R0, and only a slight inhibition effect was observed. From Figure 2b, it can be seen that the microorganisms were in the growth and reproduction period in the early operation period (0–10 days), so their life activities consumed plentiful nutrients. The microorganisms used the nitrogen source to assimilate nitrate-nitrogen reduction, so the TN concentration in the effluent was low. Some scholars point out that the nitrogen source assimilated by the microorganisms themselves can account for up to 80% of the nitrogen removal rate [39], which was the result of the joint action of most microorganisms in the system. During operation (10–45 days), the effluent TN concentrations from R0 to R2 without antibiotics or a low concentration of antibiotic stress were generally lower than those from R3 to R4 under a high concentration of antibiotic stress, which may be due to the biomass advantage of a low antibiotic concentration, which enhances the nitrogen removal effect. In the OFLX stress group, the removal of NH4+-N in R1~R4 was increased by 26.57%, 18.35%, 8.63% and 7.44% compared to the control group R0, respectively. In stage I, the removal of NH4+-N by R1 and R2 was significantly enhanced (p < 0.01), which may be due to the prevalence of resistance genes of fluoroquinolone antibiotics in municipal wastewater [40]. Nitrifying bacteria carrying resistance genes were already present in the reactor during the biofilm formation stage, so the removal rate of NH4+-N was not inhibited by OFLX. Meanwhile, the presence of antibiotics may promote the activity of nitrifying microorganisms [41]. While the nitrification and ammonia oxidation reactions mainly occurred in the surface layer of the BRC-MFC, the denitrification reactions mainly occurred in the submerged layer. Although it was found that the removal of NH4+-N was enhanced by iron filings in the submerged layer under antibiotic stress, the accumulation of more nitrate–nitrogen in the BRC-MFC system was detrimental to the removal of TN. This was consistent with the conclusion of this experiment that TN removal was slightly inhibited by exogenous antibiotics. The approaches consistently taken to enhance TN removal by BRC-MFC in the academia are to increase the submerged layer’s depth and design the submerged layer with an anaerobic environment to enhance the denitrification reaction. When the submerged layer’s depth is increased from 0 cm to 60 cm, the TN removal rate can be increased from −23% to 62% [42], which has a better enhancement of TN removal.

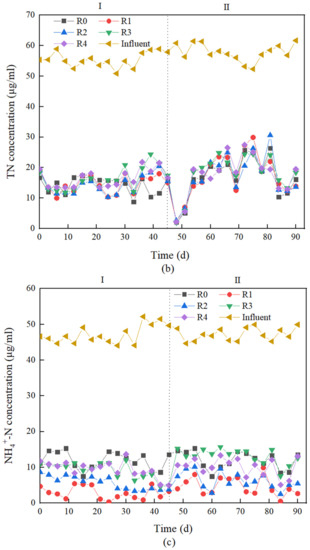
Figure 2.
(a) Removal performance of chemical oxygen demand (COD). (b) Removal performance of total nitrogen (TN). (c) Removal performance of ammonia nitrogen (NH4+-N).
In stage II, the COD removal effects on R0~R4 were good, and the COD removal rates were all maintained above 90%. Compared with the control group R0, there was no significant change in the COD removal rates of R1 and R2, while there was a slight decrease in the COD removal rates of R3 and R4. It can be seen that the COD removal ability of BRC-MFC was not inhibited by the low concentration of OFLX + TC stress. Only 2.4 μg·mL−1 OFLX + 1.2 μg·mL−1 TC and 3.6 μg·mL−1 OFLX + 1.8 μg·mL−1 TC created a slight inhibition of COD removal. This may be due to the prevalence of resistance genes for fluoroquinolone/tetracycline antibiotics in municipal wastewater [40]. The reactor accumulated a certain amount of resistance-gene-carrying bacteria during the start-up period of the hanging film; therefore, there is some resistance to shock under antibiotic stress. Under combined OFLX + TC stress, there was no significant difference (p > 0.05) in the TN removal rate for each reactor from R1 to R4, but there was a slight decrease relative to the control group R0, which reached a high level compared to other domestic and international studies (TN removal rates from −133% to 99%) [43]. In the downflow influent BRC-MFC, pollutants pass through the planting layer, filtration layer, electrode layer cathode, transition layer, submerged layer, electrode layer, and drainage layer from top to bottom. A large amount of root secretions and microorganisms exist in the inter-root microenvironment of the planting layer, and some studies have shown that the organic acid component of root secretions can increase the total number of bacteria in the soil, as well as increase the percentage of effective state nitrogen in the soil [44,45] and promote plant uptake of nitrogen nutrients, which also has a positive effect on TN removal and explains the reason why planting plants in BRC-MFC contributes to TN removal from the perspective of the inter-root microenvironment. The submerged layer was supplemented with an additional carbon source by adding 5% wood chips, and the device has a high outlet to keep the submerged zone anaerobic at all times, which facilitates the denitrification reaction. The external wires between the two electrode layers constitute a complete electron transfer system, and the anode in the submerged layer receives electrons conducted from the cathode, which are used to replenish the denitrifying microorganisms in the submerged layer to ensure the balance of nitrogen conversion [46]. Meanwhile, the activated carbon felt at the anode of the electrode layer and the iron filings in the submerged layer act as an electronics shuttle-like body within the anaerobic environment [47], providing better conditions for denitrification reactions. The drainage layer can also control the extension of hydraulic retention time to increase the microbial concentration in the system, which was an important reason for the high level of TN removal in the effluent. Under the combined stress of OFLX and TC, the removal rates of NH4+-N from R1 to R4 increased by 20.99%, 15.40%, 9.41% and 5.08% compared with the control group R0, especially the removal of NH4+-N by R1 and R2, which increased significantly (p < 0.01). As shown in Figure 2c, the NH4+-N concentrations in the effluent from R1 to R4 were lower than the control group R0. A similar study pointed out that TC has a significant promotion effect on NH4+-N removal, probably because the process of forming complexes between TC and catalytic iron in the BRC-MFC submerged layer consumes some Fe2+, resulting in a weakening of the reduction of nitrate–nitrogen to NH4+-N by catalytic iron [48], leading to a lower effluent concentration of NH4+-N. In addition, MFC connects the surface layer with the deep layer of the BRC system through a wire, forming a closed electronic supply loop, which strengthens the electronic migration within the system and provides additional electrons for the cathode on the surface layer. The nitrification reaction mainly takes place in the surface layer and is strengthened by the influence of additional electrons. The more additional electrons there are, the higher the remove rate of ammonia nitrogen, so NH4+-N was gradually consumed and removed.
The comparison of stage I and stage II data showed that the four antibiotic-stressed reactors did not differ significantly in COD removal, and antibiotics did not have a significant effect on COD removal capacity (p > 0.05), probably due to the presence of antibiotic resistance genes within the BRC-MFC system so that COD removal did not receive much influence from antibiotic stress. In addition, dehydrogenase was thought to play an important role in organic matter degradation. In a study of antibiotic-stressed microorganisms, it was found that single tetracycline antibiotic stress or a combination with other types of antibiotics will have a significant stimulating effect on dehydrogenase activity to enhance the removal of organic matter [49]. The greater decrease in TN removal in stage II compared to stage I throughout the reaction may be due to the fact that the addition of TC inhibited the expression of microbial denitrification functional genes and the inhibition depended on the concentration of TC [50]. However, as shown in Figure 2b, the average TN effluent concentration of each reactor in both stage I and stage II was less than 20 μg·mL−1, which met the primary B standard for urban wastewater treatment plant discharge. The removal rate of NH4+-N was lower in stage II than in stage I. The reason may also be that the absolute abundance of denitrification function genes decreases due to the combined stress of two antibiotics, which weakens the nitrification reaction in the BRC-MFC system. However, the NH4+-N removal rate is always higher than the TN removal rate, which proves that nitrification is always stronger than denitrification in a BRC-MFC system.
3.1.2. OFLX and TC Removal Performance
The concentrations of OFLX in the influent and effluent in stage I and stage II are shown in Figure 3a. In stage I, the average removal rates of OFLX by R1~R4 were 93.91%, 98.98%, 99.52%, and 99.57%. In stage II, the average removal rates of OFLX by R1~R4 were 91.69%, 99.08%, 99.40%, and 99.41%. It can be seen that under OFLX stress of different concentrations, BRC-MFC has a good removal effect on OFLX, which can be maintained at more than 90%, and the removal rate is proportional to the concentration of OFLX. The degradation mechanism of OFLX in BRC-MFC mainly includes electrochemical reduction and biodegradation reaction. Antibiotics become a carbon source and electron donor in an MFC biological anode [51]. Electrogenerated microorganisms and antibiotic-degrading functional bacteria attach to the anode to form a biofilm, which can reduce the overpotential required for the degradation of antibiotics and their metabolites, thus promoting the degradation of antibiotics [52,53]. The degradation of antibiotics at the bioanode relies mainly on anaerobic biodegradation and continuous electrical stimulation, and the anaerobic environment formed at the anode facilitates the mineralization of antibiotics, while continuous electrical stimulation can provide electrons to the environment, and through direct or indirect electron transfer to the bacterial cell, the stimulated microorganisms have enhanced metabolism and are able to rapidly metabolize antibiotics through secreted enzymes [54]. In addition, the cathode of MFC if modified with modified materials (Fe0/TiO2) can promote cathode production of –OH [55], which is extremely oxidizing, thus further enhancing the degradation of antibiotics. In terms of biodegradation, plant roots produce more secretions to decompose antibiotics under OFLX stress [56], but there is a limit to plant resistance to OFLX, and overly high concentrations (30–100 μg·mL−1) of OFLX will cause a decrease in the content of antioxidant enzymes such as CAT, POD and SOD in plants, accompanied by the accumulation of reactive oxygen species such as O2-, H2O2 and MDA, which poison the plant itself [57].
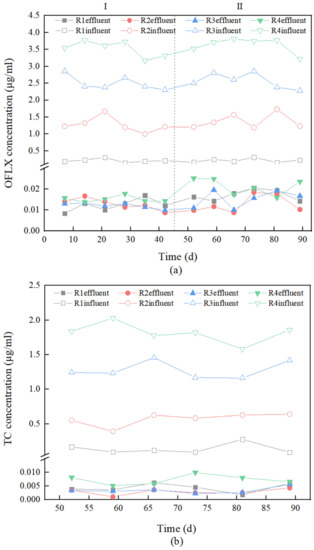
Figure 3.
(a) Removal performance of ofloxacin (OFLX). (b) Removal performance of tetracycline (TC).
The concentrations of TC in the influent and effluent in stage II are shown in Figure 3b. The removal rates of TC by R1~R4 were 95.96%, 99.49%, 99.74%, and 99.60%, respectively. BRC-MFC also maintained more than 90% efficient removal rates for TC of different concentrations. The removal of OFLX and TC from the BRC-MFC reactor mainly depends on the adsorption of fillers and microbial degradation [58]. Since TC has stronger adsorption in the ambient medium [59], it can be seen that the removal rate of TC by BRC-MFC was always higher than that of OFLX. Each layer of fillers in BRC-MFC has a larger adsorption capacity for TC [16], and the maximum adsorption capacity of activated carbon in the electrode layer for TC reached 212.6 mg·g−1 [60]. In addition, the molecular structure of TC contains several hydroxyl groups (-OH) and an amino group (-NH2) [61], and these groups can coordinate with heavy metal ions to generate insoluble salts [62]. For example, if a small amount of iron filings is added to the submerged layer of BRC-MFC, TC will form a red complex with Fe2+ [63]. In terms of microbial degradation, ammonia oxidation and nitrification reactions mainly occur in the surface layer of BRC-MFC, in which ammonia-oxidizing bacteria (AOB) convert ammonia-nitrogen to nitrate-nitrogen by ammonia monooxygenase (AMO), and some studies have found that AMO can also promote antibiotic degradation [64].
3.2. Effect of Antibiotics on Ecological Effects of BRC-MFC
Changes in Microbial Community Characteristics under Combined TC and OFLX Stresses
The microbial diversity indices in different BRC-MFC reactors of stage I and stage II are shown in Table 2. Under antibiotic stress, both richness and chao1 values decreased, indicating that the microbial richness and total number of species decreased. The decrease in stage II was greater than that in stage I, indicating that the inhibitory effect of combined antibiotic stress on microorganisms was stronger than that of single antibiotic stress. The Shannon and Simpson indices did not show significant regular changes, indicating that the homogeneity and diversity of microbial communities did not feel the effect of antibiotic stress.

Table 2.
Microbial diversity indices of different BRC-MFCs.
Due to different classes of antibiotic stress at different concentrations, the distribution of microbial communities in different BRC-MFC reactors also differed. As shown in Figure 4a, the top 12 bacteria in terms of phylum level were Proteobacteria, Bacteroidetes, Acidobacteria, Actinobacteria, Planctomycetes, Chloroflexi, Verrucomicrobia, Patescibacteria, Gemmatimonadetes, Nitrospirae, Firmicutes and Cyanobacteria. It has been shown that Proteobacteria (39.49% to 58.50%), Bacteroidetes (15.33% to 29.67%) and Chloroflexi (0.22% to 3.03%) are the dominant phylum in wetland systems [65,66]. Furthermore, the phylum contains a variety of bacteria that efficiently degrade organic pollutants [67]. In addition, proteobacteria and Chloroflexi are important microorganisms for nitrogen and phosphorus removal [68]. Firmicutes is a nitrifying bacterium that oxidizes nitrite to nitrate, ranked 11th in terms of abundance (0.14–1.43%). Compared to R0, there was a significant increase in the abundance of planctomycetes and a decrease in the abundance of patescibacteria under antibiotic stress. Similar studies by other investigators have shown that when BRC was under two-antibiotics stress, the top 10 bacteria in terms of phylum level are Proteobacteria, Bacteroidetes, Chloroflexi, Patescibacteria, Acidobacteria, Actinobacteria, Firmicutes, Spirochaetes, Verrucomicrobia and Gemmatimonadetes [13]. Among them, nine kinds of bacteria in terms of phylum level were consistent in our study.
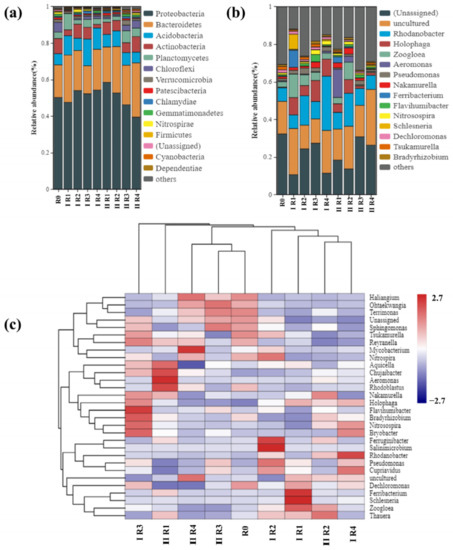
Figure 4.
Microorganism community distribution at the (a) phylum level and (b) genus level in different BRC-MFCs. (c) Clustering heat map of abundance at the genus levels with the 30 highest abundance levels in different BRC-MFCs.3.2.2. Response of Functional Genes for Denitrification under Combined TC and OFLX Stresses.
As shown in Figure 4b, the top 12 bacteria in terms of genus level were Rhodanobacter, Holophaga, Zoogloea, Aeromonas, Pseudomonas, Nakamurella, Ferrobacterium, Flavihumibacter, Nitrosospira, Schlesneria, Dechloromonas and Tsukamurella. The abundance of the dominant genera in the different BRC-MFC reactors varied widely. In the absence of antibiotic stress, the dominant genera were Rhodanobacter (7.20%), Tsukamurella (1.76%), Dechloromonas (1.58%) and Pseudomonas (1.07%). After antibiotic stress, the dominant homogeneous genera changed to Rhodanobacter (7.25–28.87%), Holophaga (1.35–11.30%) and Aeromonas (0.08–15.08%). Only the specific dominant species of Rhodanobacter in BRC-MFC did not change, but its abundance increased to 28.87% after coming under antibiotic stress. Nitrosospira belongs to the nitrifying bacteria that oxidize NH4+-N to NO2– [69]. OFLX stress at 2.4 μg·mL−1 and 3.6 μg·mL−1 increased the abundance of Nitrosospira to 2.47% and 2.09%, respectively, which could enhance the nitrification efficiency. Similar studies by other investigators have shown that when BRC was under two-antibiotics stress, the top 10 bacteria in terms of genus level are Blrii41, denitratisoma, Ferrobacterium, Thiobacillus, Saccharimonadales, Bacteroidetes_vadinHA17, Desulfomicrobium, Sbr1031, Treponema_2 and Subgroup_7 [13]. There were only one kind of bacteria in terms of genus level was consistent in this study.
From the clustering heat map of species abundance at the genus levels, shown in Figure 4c, it can be seen that the microbial community composition structures in different BRC-MFC reactors differed significantly, with the highest similarity between R0, R3 and R4 in stage II. Haliangium, Ohtaekwangia, Terrimonas and Sphingomonas all accounted for a high proportion in R0, II R3 and II R4. In stage Ⅰ, OFLX single-stress caused most growth of Ferruginibacter and Salinimicrobium. In particular, Salinimicrobium was endemic in stage Ⅰ. It might be particularly sensitive to TC, explaining why the species died out in stage Ⅱ with TC stress.
The role of each nitrogen removal functional gene in the denitrification pathway is shown in Figure 5 [70]. Real-time fluorescence quantification of the abundance of total bacterial 16S rRNA genes and denitrifying functional genes narG, nirS, norB, nosZ in the denitrification process showed that the total bacterial 16S rRNA gene copy number was maintained at 108–109 copies·g−1 in the packed soil of the five BRC-MFC reactors at different experimental stages. The absolute copy number of denitrifying functional genes varied at different experimental stages, as shown in Figure 6a, and the corresponding abundance ratio (the ratio of the copy number of denitrifying functional genes to the copy number of total bacterial 16S rRNA genes) is shown in Figure 6b. The absolute abundance levels of denitrifying functional genes in BRC-MFC reactors under OFLX and TC stresses all decreased to some extent, with nosZ decreasing the most, R4 decreasing 95.18% in absolute abundance compared to the control group R0, and R0-R4 narG absolute abundance levels all greater than 106 copies·g−1, thus still possessing a strong nitrate-reduction capacity under various concentrations of antibiotic stress [71]. nirS and nosZ had absolute abundance levels greater than 106 copies·g−1 from R0 to R1, indicating that the BRC-MFC system still had a strong nitrite–nitrogen reduction capacity and N2O reduction capacity under the combined stress of 0.2 μg·mL−1 OFLX + 0.1 μg·mL−1 TC [70].
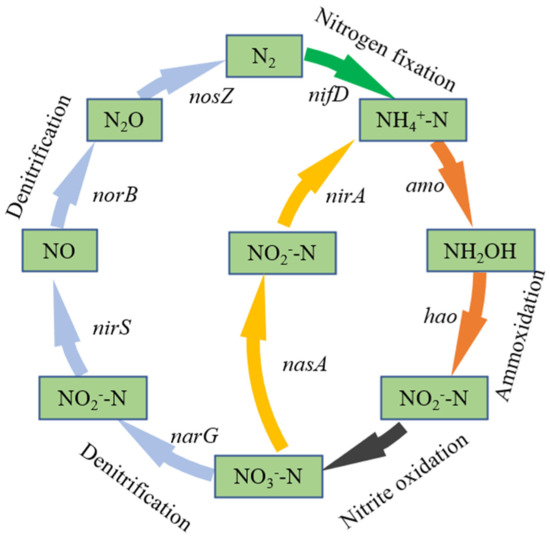
Figure 5.
The role of denitrifying functional genes in the nitrogen transformation pathway.

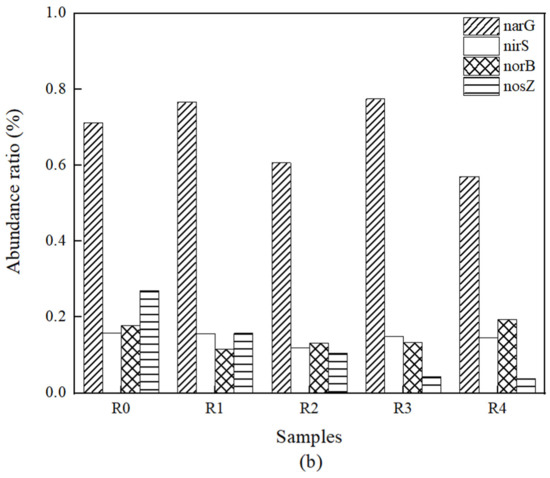
Figure 6.
Effects of antibiotic stress on the abundance of denitrifying functional genes. (a) Absolute abundance. (b) Abundance ratio.
The abundance ratios of three types of denitrifying functional genes, narG, nirS and norB, had little change, and no obvious pattern with the concentration of antibiotics, while the abundance ratio of nosZ showed a significant decrease under the combined stress of two types of antibiotics and showed a negative correlation with antibiotic concentration. The nosZ abundance ratios from R0 to R4 were 0.26859%, 0.15757%, 0.10504%, 0.043% and 0.03865%. This indicates that the antibiotic dosing severely inhibited the conversion of N2O to N2 during denitrification [72], which was detrimental to the denitrification process.
4. Conclusions
The removal rates of COD in BRC-MFC system were all maintained above 90%, and antibiotics had little effect on COD removal (p > 0.05). The removal rate of TN has maintained at 70%, and there was no significant difference among different BRC-MFC reactors (p > 0.05). The NH4+-N removal rate was stable at 74% without antibiotic stress, and the removal rate increased to 77–93% after adding antibiotics. Antibiotics could significantly promote NH4+-N removal (p < 0.01), but the enhancement rate was inversely proportional to the antibiotic concentration. Under different concentrations of antibiotic stress, the removal rates of OFLX and TC of each BRC-MFC system reached more than 90%. The removal rates were proportional to the antibiotic concentrations. The removal rate of OFLX in stage I was higher than that in stage II, indicating that the performance of OFLX removal by the BRC-MFC system was negatively affected by antibiotic complex stress.
Microbial abundance and the total number of species decreased under antibiotic stress, and the decrease was greater in stage II than in stage I, indicating that combined antibiotic stress had a stronger inhibitory effect on microorganisms than single antibiotic stress. Proteobacteria and Bacteroidetes were the dominant phylum within the BRC-MFC system, with the highest abundance levels of 58.50% and 29.67%, respectively. The abundance of denitrifying functional genes in the BRC-MFC reactor under antibiotic stress decreased to some extent; furthermore, the absolute and abundance ratio of nosZ decreased the most, and the absolute abundance of R4 decreased by 95.18% more compared with that of R0 in the control group.
Author Contributions
Writing—original draft preparation, Y.Y.; writing—review and editing, L.C.; formal analysis, T.C.; data curation, H.S.; project administration and funding acquisition Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
Supported by the National Natural Science Foundation of China (41967043 and 52160003) and the Gansu Province Natural Science Foundation (20JR5RA461).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hou, L.; Zhang, L.; Li, F.; Huang, S.; Hu, A. Urban ponds as hotspots of antibiotic resistome in the urban environment. J. Hazard. Mater. 2020, 56, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Q.; Ying, G.G.; Pan, C.G.; Liu, Y.S.; Zhao, J.L. Comprehensive Evaluation of Antibiotics Emission and Fate in the River Basins of China: Source Analysis, Multimedia Modeling, and Linkage to Bacterial Resistance. Environ. Sci. Technol. 2015, 49, 6772–6782. [Google Scholar] [CrossRef] [PubMed]
- Grenni, P.; Ancona, V.; Caracciolo, A.B. Ecological effects of antibiotics on natural ecosystems: A review. Microchem. J. 2017, 262, 1108. [Google Scholar] [CrossRef]
- Yya, B.; Ws, C.; Hui, L.D.; Ww, A.; Ld, E.; Wei, X.A. Antibiotics and antibiotic resistance genes in global lakes: A review and meta-analysis—ScienceDirect. Environ. Int. 2018, 116, 60–73. [Google Scholar]
- Li, S.; Shi, W.Z.; Liu, W.; Li, H.M.; Zhang, W.; Hu, J.R.; Ke, Y.C.; Sun, W.L.; Ni, J.R. A duodecennial national synthesis of antibiotics in China’s major rivers and seas (2005–2016). Sci. Total Environ. 2018, 615, 906–917. [Google Scholar] [CrossRef]
- Zhang, Y.; Boyd, S.A.; Teppen, B.J.; Tiedje, J.M.; Zhang, W.; Zhu, D.; Li, H. Bioavailability of tetracycline to antibiotic resistant Escherichia coli in water-clay systems. Environ. Pollut. 2018, 243, 1078–1086. [Google Scholar] [CrossRef]
- Chen, C.Q.; Zheng, L.; Zhou, J.L.; Zhao, H. Persistence and risk of antibiotic residues and antibiotic resistance genes in major mariculture sites in Southeast China. Sci. Total Environ. 2017, 580, 1175–1184. [Google Scholar] [CrossRef]
- Gao, J.; Wang, Z.; Zhang, H.; Zhu, J.; Xie, C.; Xie, X. Occurrence and the fate of typical antibiotics in sewage treatment plants in Lanzhou. Acta Sci. Circumstantiae 2016, 53, 773–782. [Google Scholar]
- Mo, W.; Zhang, D.; Yong, L.; Hou, Q.; Yu, Y. Effect of a Submerged Zone and Carbon Source on Nutrient and Metal Removal for Stormwater by Bioretention Cells. Water Res. 2018, 241, 145–150. [Google Scholar]
- Wang, Y.J.; Singh, R.P.; Geng, C.; Fu, D. Carbon-to-nitrogen ratio influence on the performance of bioretention for wastewater treatment. Environ. Sci. Pollut. Res. 2020, 27, 17652–17660. [Google Scholar] [CrossRef]
- Wang, Y.J.; Singh, R.P.; Zhang, J.; Xu, Y.; Fu, D. Nitrogen removal performance of microbial fuel cell enhanced bioretention system. J. Water Supply: Res. Technol. 2019, 78, 68. [Google Scholar] [CrossRef]
- Wang, Y.J.; Li, J.; Yan, Y.; Si, Y.; Xu, J.; Li, M.; Peng, X. La(OH)3 loaded magnetic nanocomposites derived from sugarcane bagasse cellulose for phosphate adsorption: Characterization, performance and mechanism. Colloids Surf. A Physicochem. Eng. Asp. 2021, 626, 127060. [Google Scholar] [CrossRef]
- Wang, Y.J.; Geng, C.; Chen, T.; Li, J.; Xu, Y.; Fu, D. Adaptability of enhanced bioretention cell for nitrogen and phosphorus removal under two antibiotics stress. Ecotoxicol. Environ. Saf. 2022, 230, 113114. [Google Scholar]
- Wang, Y.J.; Chen, T.J.; Li, J.S.; Si, Y.M.; Wang, Z.Y. The influence of electrode spacing on the performance of bioretention cell coupled with MFC. R. Soc. Open Sci. 2021, 68, 46–53. [Google Scholar]
- Wang, Y.J.; Si, Y.; Yang, S.; Singh, R. Study on Flow Distribution Pattern and Conductivity of Porous Media in Bioretention Cells. Bioengineered 2021, 452, 351–359. [Google Scholar] [CrossRef]
- Chen, J.; Ying, G.G.; Wei, X.D.; Liu, Y.S.; Liu, S.S.; Hu, L.X.; He, L.Y.; Chen, Z.F.; Chen, F.R.; Yang, Y.Q. Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: Effect of flow configuration and plant species. Sci. Total Environ. 2016, 571, 974–982. [Google Scholar] [CrossRef]
- Smyth, K.; Drake, J.; Li, Y.; Rochman, C.; Passeport, E. Bioretention cells remove microplastics from urban stormwater. Water Res. 2020, 191, 116785. [Google Scholar] [CrossRef]
- Zhang, C.; Li, C.N.; Yao, T.; He, L.; Chen, X.; Zhang, L.; Xi, B.A. Effects of compound microorganisms on growth and physiological characteristics of Lolium multiflorum seedlings under tetracycline stress. J. Grassl. Sci. 2019, 32, 1–10. [Google Scholar]
- Ferreira, C.; Nunes, B.A.; Henriques-Almeida, J.M.; Guilhermino, L. Acute toxicity of oxytetracycline and florfenicol to the microalgae Tetraselmis chuii and to the crustacean Artemia parthenogenetica. Ecotoxicol. Environ. Saf. 2007, 67, 452–458. [Google Scholar] [CrossRef]
- Zhou, J.N.; Yang, C.; Song, Z.Y.; He, C.F.; He, J.H.; Huang, W.L.; Dang, Z. Influences of tetracycline and cadmium on rice roots: Growth and root exudates. Acta Sci. Circumstantiae 2021, 41, 1518–1528. [Google Scholar] [CrossRef]
- Liu, T.; Lu, L.L.; Luo, W.J.; Tian, S.K.; Yang, X.E. Degradation of tetracyclines in soils and their effects on root growth of Chinese cabbages (Brassica campestris L.). Acta Sci. Circumstantiae 2017, 37, 1957–1966. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X.; Lv, Y.; Li, N.; Xu, K. Grafting resulting in alleviating tomato plant oxidative damage caused by high levels of ofloxacin. Environ. Pollut. 2021, 286, 117331. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Sun, S.; Zhou, T.; Du, Z.; Wang, J.; Li, B.; Wang, J.; Zhu, L. Effects of pyroxsulam on soil enzyme activity, nitrogen and carbon cycle-related gene expression, and bacterial community structure. J. Clean. Prod. 2022, 25, 355. [Google Scholar] [CrossRef]
- Xiong, W.G.; Sun, Y.X.; Zhang, T.; Ding, X.Y.; Li, Y.F. Antibiotics, Antibiotic Resistance Genes, and Bacterial Community Composition in Fresh Water Aquaculture Environment in China. Microb. Ecol. 2015, 70, 425–432. [Google Scholar] [CrossRef]
- Chen, M.J.; Qian, Y.H.; Yu, Q.Y.; Guo, Q.; Cheng, H.Z.; Xu, D.M. Effects of Typical Tetracycline Antibiotics on Soil Microorganisms and Plant Growth. Asian J. Ecotoxicol. 2019, 14, 276–283. [Google Scholar]
- Li, H.; Tian, Y.; Qu, Y.; Qiu, Y.; Liu, J.; Feng, Y. A Pilot-scale Benthic Microbial Electrochemical System (BMES) for Enhanced Organic Removal in Sediment Restoration. Sci. Rep. 2017, 7, 39802. [Google Scholar] [CrossRef]
- Peixoto, L.; Parpot, P.; Martins, G. Assessment of Electron Transfer Mechanisms during a Long-Term Sediment Microbial Fuel Cell Operation. Energies 2019, 12, 3–15. [Google Scholar] [CrossRef]
- Zhou, L.A.; Jiang, Q.; Sun, S.Q.; Zhang, W.; Gao, Y.; Wang, X. Removal of antibiotics from water by bioelectrochemical system:A review. J. Civ. Environ. Eng. 2021, 43, 113–123. [Google Scholar]
- Yan, W.; Guo, Y.; Xiao, Y.; Wang, S.; Ding, R.; Jiang, J.; Gang, H.; Wang, H.; Yang, J.; Zhao, F. The changes of bacterial communities and antibiotic resistance genes in microbial fuel cells during long-term oxytetracycline processing. Water Res. 2018, 142, 105–114. [Google Scholar] [CrossRef]
- Hatt, B.; Morison, P.; Fletcher, T.; Deletic, A. (Eds.) Stormwater Biofiltration Systems—Adoption Guidelines; Image & Signal Processing-International Conference; Monash University Publishing: Melbourne, Australia, 2009. [Google Scholar]
- Wang, Y.J.; Chen, T.J.; Li, J.S. Permeability characteristics test on wastewater in unplanted bioretention cell under continous operation. Environ. Eng. 2022, 40, 27–31. [Google Scholar] [CrossRef]
- Sun, H.Q.; Cui, D.N. Determination of Aldicarb,Aldicarb Sulfoxide,Aldicarb Sulfone in Water by Solid Phase Extraction and UPLC⁃MS/MS with Isotope Dilution. Chin. J. Appl. Chem. 2022, 39, 470–479. [Google Scholar]
- HJ 636-2012; Water Quality: Determination of Total Nitrogen—Alkaline Potassium Persulfate Digestion UV Spectrophotometric Method; Ministry of Ecology and Environment of the People’s Republic of China: Beijing, China, 2012.
- HJ 535-2009; Water Quality: Determination of Ammonia Nitrogen—Nessler’s Reagent Spectrophotometry; Ministry of Ecology and Environment of the People’s Republic of China: Beijing, China, 2009.
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Walker, N.J. Real-time and quantitative PCR: Applications to mechanism-based toxicology. J. Biochem. Mol. Toxicol. 2010, 15, 325. [Google Scholar] [CrossRef]
- Li, Y.; Liu, B.; Wang, J.; Gao, M.; Zhang, X. Effects of chlortetracycline on soil microbial communities: Comparisons of enzyme activities to the functional diversity via Biolog EcoPlates (TM). Eur. J. Soil Biol. 2015, 68, 69–76. [Google Scholar] [CrossRef]
- Ray, S.; Scholz, M.; Haritash, A.K. Kinetics of carbon and nitrogen assimilation by heterotrophic microorganisms during wastewater treatment. Environ. Monit. Assess. 2019, 191, 71–79. [Google Scholar] [CrossRef]
- Wu, M.; Que, C.; Tang, L.; Xu, H.; Xiang, J.; Wang, J.; Shi, W.; Xu, G. Distribution, fate, and risk assessment of antibiotics in five wastewater treatment plants in Shanghai, China. Environ. Sci. Pollut. Res. 2016, 23, 18055–18063. [Google Scholar] [CrossRef]
- Zhang, Y.; Geng, J.; Ma, H.; Ren, H.; Ke, X.; Ding, L. Characterization of microbial community and antibiotic resistance genes in activated sludge under tetracycline and sulfamethoxazole selection pressure. Sci. Total Environ. 2016, 571, 479–486. [Google Scholar] [CrossRef]
- Yan, Z.Q.; Li-Qing, L.I.; Liu, Y.Q.; Sheng-Liang, T.U. Removal of Dissolved Nitrogen and Phosphorus in Urban Stormwater by Bioretention Cell Incorporated a Submerged Zone. China Water Wastewater 2017, 33, 33–38. [Google Scholar]
- Xu, Y.; Xi, M.H.; Geng, C.C.; Xu, L.; Liu, Z.M.; Fu, D.F.; Wang, Y.J. Removals of typical antibiotics in sewage by unplanted bioretention cells:Efficiency and its enhancement. J. Southeast Univ. 2020, 50, 12. [Google Scholar]
- Gong, F.F.; Fan, W.G. Effects of Exogenous Citric Acids on Nutrient Activation of Calcareous Yellow Soil and Promotion Effects of Nutrient Absorption and Growth of Rosa roxburghii Seedlings. Sci. Agric. Sin. 2018, 43, 552–564. [Google Scholar]
- Gza, B.; Wca, C.; Jb, A.; Cx, D.; Nz, A.; Sga, B.; Rmr, E.; Fd, F. Co-incorporation of rice straw and leguminous green manure can increase soil available nitrogen (N) and reduce carbon and N losses: An incubation study—ScienceDirect. Pedosphere 2020, 30, 661–670. [Google Scholar]
- Wang, Y.J.; Geng, C.C.; Xu, Y.; Xi, M.H.; Wang, J.X. Effect of Different Enhanced Methods on Efficiency of Denitrification and Phosphorus Removal in Bioretention Cell. China Water Wastewater 2020, 36, 6. [Google Scholar]
- He, Q.; Yu, L.; Li, J.; He, D.; Zhou, S. Electron shuttles enhance anaerobic oxidation of methane coupled to iron(III) reduction. Sci. Total Environ. 2019, 688, 321–330. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, Y.; Zhang, B.; Bu, C.; Wang, Y.; Zhang, D.; Xi, M.; Qin, Q. Enhanced removal of sulfamethoxazole and tetracycline in bioretention cells amended with activated carbon and zero-valent iron: System performance and microbial community. Sci. Total Environ. 2021, 797, 148992. [Google Scholar] [CrossRef]
- Fang, H.; Han, Y.; Yin, Y.; Pan, X.; Yu, Y. Variations in dissipation rate, microbial function and antibiotic resistance due to repeated introductions of manure containing sulfadiazine and chlortetracycline to soil. Chemosphere Environ. Toxicol. Risk Assess. 2014, 96, 51–56. [Google Scholar] [CrossRef]
- Liao, Q.; Rong, H.; Zhao, M.; Luo, H.; Wang, R. Interaction between tetracycline and microorganisms during wastewater treatment: A review. Sci. Total Environ. 2021, 757, 143981. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.; Ma, J.; Zhao, F. Rapid degradation of sulphamethoxazole and the further transformation of 3-amino-5-methylisoxazole in a microbial fuel cell. Water Res. 2016, 88, 322–328. [Google Scholar] [CrossRef]
- Hou, Y.; Yuan, G.; Qin, S.; Tu, L.; Wang, S. Photocathode optimization and microbial community in the solar-illuminated bio-photoelectrochemical system for nitrofurazone degradation. Bioresour. Technol. 2020, 302, 122761. [Google Scholar] [CrossRef]
- Kong, D.; Yun, H.; Cui, D.; Qi, M.; Shao, C.; Cui, D.; Ren, N.; Liang, B.; Wang, A. Response of antimicrobial nitrofurazone-degrading biocathode communities to different cathode potentials. Bioresour. Technol. 2017, 241, 951. [Google Scholar] [CrossRef]
- Dominguez-Benetton, X.; Varia, J.C.; Pozo, G.; Modin, O.; Heijne, A.T.; Fransaer, J.; Rabaey, K. Metal recovery by microbial electro-metallurgy. Prog. Mater. Sci. 2018, 94, 435–461. [Google Scholar] [CrossRef]
- Jiang, C.; Liu, L.; Crittenden, J.C. An electrochemical process that uses an Fe0/TiO2 cathode to degrade typical dyes and antibiotics and a bio-anode that produces electricity. Front. Environ. Sci. Eng. 2016, 10, 15. [Google Scholar] [CrossRef]
- Lv, Y.; Xu, J.; Xu, K.; Liu, X.; Xi, B. Accumulation characteristics and biological response of ginger to sulfamethoxazole and ofloxacin. Environ. Pollut. 2020, 262, 114203. [Google Scholar] [CrossRef]
- Riaz, L.; Mahmood, T.; Coyne, M.S.; Khalid, A.; Rashid, A.; Hayat, M.T.; Gulzar, A.; Amjad, M. Physiological and antioxidant response of wheat (Triticum aestivum) seedlings to fluoroquinolone antibiotics. Chemosphere 2017, 177, 250. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, H.; Xiong, P.; Zhu, Q.; Jiang, G. Occurrence, fate, and risk assessment of typical tetracycline antibiotics in the aquatic environment: A review. Sci. Total Environ. 2020, 753, 141975. [Google Scholar] [CrossRef]
- Xuan, D.; Hao, H.; Bi, J.; Sun, S.; Huang, X. Surface Complexation Enhanced Adsorption of Tetracycline by ALK-MXene. Ind. Eng. Chem. Res. 2022, 223, 321–333. [Google Scholar]
- Li, Z.X.; Zhu, X.Y.; Shen, H. Preparation of Activated Carbon from Corn Straw and Its Adsorption Properties of Tetracycline. Chem. World 2020, 61, 640–643. [Google Scholar] [CrossRef]
- Hu, P.; Shao, J.; Qian, G.; Adeleye, A.S.; Hao, T. Removal of tetracycline by aerobic granular sludge from marine aquaculture wastewater: A molecular dynamics investigation. Bioresour. Technol. Biomass Bioenergy Biowastes Convers. Technol. Biotransform. Prod. Technol. 2022, 96, 355. [Google Scholar] [CrossRef]
- Ma, X.; Yang, C.; Jiang, Y.; Zhang, X.; Wang, Q.; Dang, Z. Desorption of heavy metals and tetracycline from goethite-coated sands: The role of complexation. Colloids Surf. A Physicochem. Eng. Asp. 2019, 134, 1452–1458. [Google Scholar] [CrossRef]
- Kurup, L. Isothermal and Kinetic Study of the Adsorptionof Tetracycline Hydrochloride andSulphamethoxazole by Bottom Ash and Alkalitreated Bottom Ash. Asian J. Pharm. Technol. Innov. 2018, 214, 563–571. [Google Scholar]
- Fernandez-Fontaina, E.; Gomes, I.B.; Aga, D.S.; Omil, F.; Lema, J.M.; Carballa, M. Biotransformation of pharmaceuticals under nitrification, nitratation and heterotrophic conditions. Sci. Total Environ. 2016, 26, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, Z.J.; Huang, J.C.; Sun, S.; He, S. Salinity-driven nitrogen removal and its quantitative molecular mechanisms in artificial tidal wetlands. Water Res. 2021, 202, 351. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.F.; Hou, L.N.; Li, H. Effects of pollution load and salinity shock on nitrogen removal and bacterial community in two-stage vertical flow constructed wetlands. Bioresour. Technol. 2021, 342, 126031. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Yang, Y.L.; Liu, Y.W.; Li, X.; Zhu, W.B. Quantitative ecology associations between heterotrophic nitrification-aerobic denitrification, nitrogen-metabolism genes, and key bacteria in a tidal flow constructed wetland. Bioresour. Technol. 2021, 337, 125449. [Google Scholar] [CrossRef]
- Zhu, Y.; Cui, L.; Li, J.; Wang, R.; Wei, J. Long-term performance of nutrient removal in an integrated constructed wetland. Sci. Total Environ. 2021, 779, 146268. [Google Scholar] [CrossRef]
- Lei, C.A.; Yang, Y.; Jiang, D.P.; Li, X.Q.; Wang, Y.Y. Characteristics of Anaerobic Digestion Solution Nitrification Process Under Low Dissolved Oxygen for a Long Time. China Biogas 2022, 40, 8–17. [Google Scholar] [CrossRef]
- Peng, Y.Z.; Qian, W.T.; Wang, Q.; Li, X.Y.; Zhang, Q.; Wu, L.; Ma, B. Unraveling Microbial Structure of Activated Sludge in a Full-scale Nitrogen Removal Plant Using Metagenomic Sequencing. J. Beijing Univ. Technol. 2019, 45, 8. [Google Scholar]
- Yang, Z.; Sun, H.; Wu, W. Intensified simultaneous nitrification and denitrification performance in integrated packed bed bioreactors using PHBV with different dosing methods. Environ. Sci. Pollut. Res. 2020, 27, 21560–21569. [Google Scholar] [CrossRef]
- Du, R.; Peng, Y.; Cao, S.; Li, B.; Wang, S. Mechanisms and microbial structure of partial denitrification with high nitrite accumulation. Appl. Microbiol. Biotechnol. 2016, 100, 2011–2021. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).