Machine Learning for COVID-19 and Influenza Classification during Coexisting Outbreaks
Abstract
:1. Introduction
2. Related Works
3. Materials and Methods
3.1. Data Collection and Preprocessing
3.2. Attribute Selection
3.3. Validation Method and Algorithms
3.4. Attribute Importance
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Monteiro de Oliveira, M.; Fuller, T.L.; Brasil, P.; Gabaglia, C.R.; Nielsen-Saines, K. Controlling the COVID-19 pandemic in Brazil: A challenge of continental proportions. Nat. Med. 2020, 26, 1505–1506. [Google Scholar] [CrossRef] [PubMed]
- Konala, V.M.; Adapa, S.; Gayam, V.; Naramala, S.; Daggubati, S.R.; Kammari, C.B.; Chenna, A. Co-infection with Influenza A and COVID-19. Eur. J. Case Rep. Intern. Med. 2020, 7, 001656. [Google Scholar] [CrossRef] [PubMed]
- Istepanian, R.S.H.; Al-Anzi, T. m-Health 2.0: New perspectives on mobile health, machine learning and big data analytics. Methods 2018, 151, 34–40. [Google Scholar] [CrossRef] [PubMed]
- da Silveira, A.C.M.; Sobrinho, Á.; Dias da Silva, L.; de Barros Costa, E.; Pinheiro, M.E.; Perkusich, A. Exploring Early Prediction of Chronic Kidney Disease Using Machine Learning Algorithms for Small and Imbalanced Datasets. Appl. Sci. 2022, 12, 3673. [Google Scholar] [CrossRef]
- Sobrinho, A.; Queiroz, A.C.M.D.S.; Silva, L.D.D.; Costa, E.D.B.; Pinheiro, M.E.; Perkusich, A. Computer-Aided Diagnosis of Chronic Kidney Disease in Developing Countries: A Comparative Analysis of Machine Learning Techniques. IEEE Access 2020, 8, 25407–25419. [Google Scholar] [CrossRef]
- Kar, K.; Kornblith, S.; Fedorenko, E. Interpretability of artificial neural network models in artificial intelligence versus neuroscience. Nat. Mach. Intell. 2022, 4, 1065–1067. [Google Scholar] [CrossRef]
- Belard, A.; Buchman, T.; Forsberg, J.; Potter, B.K.; Dente, C.J.; Kirk, A.; Elster, E. Precision diagnosis: A view of the clinical decision support systems (CDSS) landscape through the lens of critical care. J. Clin. Monit. Comput. 2017, 31, 261–271. [Google Scholar] [CrossRef]
- Viana Dos Santos Santana, Í.; Cm da Silveira, A.; Sobrinho, Á.; Chaves E Silva, L.; Dias da Silva, L.; Santos, D.F.S.; Gurjão, E.C.; Perkusich, A. Classification Models for COVID-19 Test Prioritization in Brazil: Machine Learning Approach. J. Med. Internet Res. 2021, 8, e27293. [Google Scholar] [CrossRef]
- Son, W.S.; Park, J.E.; Kwon, O. Early detection of influenza outbreak using time derivative of incidence. EPJ Data Sci. 2020, 9, 28. [Google Scholar] [CrossRef]
- Kumar, S. Monitoring Novel Corona Virus (COVID-19) Infections in India by Cluster Analysis. Ann. Data Sci. 2020, 7, 417–425. [Google Scholar] [CrossRef]
- Aftab, M.; Amin, R.; Koundal, D.; Aldabbas, H.; Alouffi, B.; Iqbal, Z. Classification of COVID-19 and Influenza Patients Using Deep Learning. Contrast Media Mol. Imaging 2022, 2022, 8549707. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ma, J.; Shende, N.; Castaneda, G.; Chakladar, J.; Tsai, J.C.; Apostol, L.; Honda, C.O.; Xu, J.; Wong, L.M.; et al. Using machine learning of clinical data to diagnose COVID-19: A systematic review and meta-analysis. BMC Med. Inform. Decis. Mak. 2020, 20, 247. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, Z.; Li, S.; Liu, T.; Wang, X.; Xia, J.; Zhao, Y. Machine Learning-Based Decision Model to Distinguish Between COVID-19 and Influenza: A Retrospective, Two-Centered, Diagnostic Study. Risk Manag. Healthc. Policy 2021, 14, 595–604. [Google Scholar] [CrossRef]
- Elbasi, E.; Zreikat, A.; Mathew, S.; Topcu, A.E. Classification of influenza H1N1 and COVID-19 patient data using machine learning. In Proceedings of the 44th International Conference on Telecommunications and Signal Processing (TSP), Brno, Czech Republic, 26–28 July 2021; pp. 278–282. [Google Scholar]
- Phu, D.N.; Vinh, P.C.; Quoc, N.K. Enhanced Diagnosis of Influenza and COVID-19 Using Machine Learning. EAI Endorsed Trans. Context Aware Syst. App. [Internet] 2023, 9, 1–6. [Google Scholar] [CrossRef]
- Shilaskar, S.; Ghatol, A. Diagnosis system for imbalanced multi-minority medical dataset. Soft Comput. 2018, 23, 4789–4799. [Google Scholar] [CrossRef]
- Chatterjee, A.; Gerdes, M.W.; Martinez, S.G. Identification of Risk Factors Associated with Obesity and Overweight—A Machine Learning Overview. Sensors 2020, 20, 2734. [Google Scholar] [CrossRef]
- Almansour, N.A.; Syed, H.F.; Khayat, N.R.; Altheeb, R.K.; Juri, R.E.; Alhiyafi, J.; Alrashed, S.; Olatunji, S.O. Neural network and support vector machine for the prediction of chronic kidney disease: A comparative study. Comput. Biol. Med. 2019, 109, 101–111. [Google Scholar] [CrossRef]
- Biau, G.; Cadre, B.; Rouvière, L. Accelerated gradient boosting. Mach. Learn. 2019, 108, 971–992. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 5, 1189–1232. [Google Scholar] [CrossRef]
- Xing, W.; Bei, Y. Medical Health Big Data Classification Based on KNN Classification Algorithm. IEEE Access 2020, 8, 28808–28819. [Google Scholar] [CrossRef]
- Valdes, G.; Luna, J.; Eaton, E.; Simone, C.B., II; Ungar, L.H.; Solberg, T.D. MediBoost: A Patient Stratification Tool for Interpretable Decision Making in the Era of Precision Medicine. Sci. Rep. 2016, 6, 37854. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Alam, S.; Shi, P.; Dexter, F.; Kong, N. Interpretable machine learning models for hospital readmission prediction: A two-step extracted regression tree approach. BMC Med. Inform. Decis. Mak. 2023, 23, 104. [Google Scholar] [CrossRef] [PubMed]
- Joyce, D.W.; Kormilitzin, A.; Smith, K.A.; Cipriani, A. Explainable artificial intelligence for mental health through transparency and interpretability for understandability. NPJ Digit. Med. 2023, 6, 6. [Google Scholar] [CrossRef]
- Ahamad, M.M.; Aktar, S.; Rashed-Al-Mahfuz, M.; Uddin, S.; Liò, P.; Xu, H.; Summers, M.A.; Quinn, J.M.W.; Moni, M.A. A machine learning model to identify early stage symptoms of SARS-Cov-2 infected patients. Expert Syst. Appl. 2020, 160, 113661. [Google Scholar] [CrossRef] [PubMed]
- Sarica, A.; Cerasa, A.; Quattrone, A. Random Forest Algorithm for the Classification of Neuroimaging Data in Alzheimer’s Disease: A Systematic Review. Front. Aging Neurosci. 2017, 9, 329. [Google Scholar] [CrossRef] [PubMed]
- Schober, P.; Vetter, T.R. Logistic Regression in Medical Research. Anesth. Analg. 2021, 132, 365–366. [Google Scholar] [CrossRef]
- Demsar, J. Statistical comparisons of classifiers over multiple data sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- Choi, S.; Park, J.; Park, S.; Byon, I.; Choi, H.Y. Establishment of a prediction tool for ocular trauma patients with machine learning algorithm. Int. J. Ophthalmol. 2021, 14, 1941–1949. [Google Scholar] [CrossRef]
- The Lancet Respiratory Medicine. Opening the black box of machine learning. Lancet Respir. Med. 2018, 6, 801. [Google Scholar] [CrossRef]
- Boyton, R.J.; Altmann, D.M. The immunology of asymptomatic SARS-CoV-2 infection: What are the key questions? Nat. Rev. Immunol. 2021, 21, 762–768. [Google Scholar] [CrossRef]
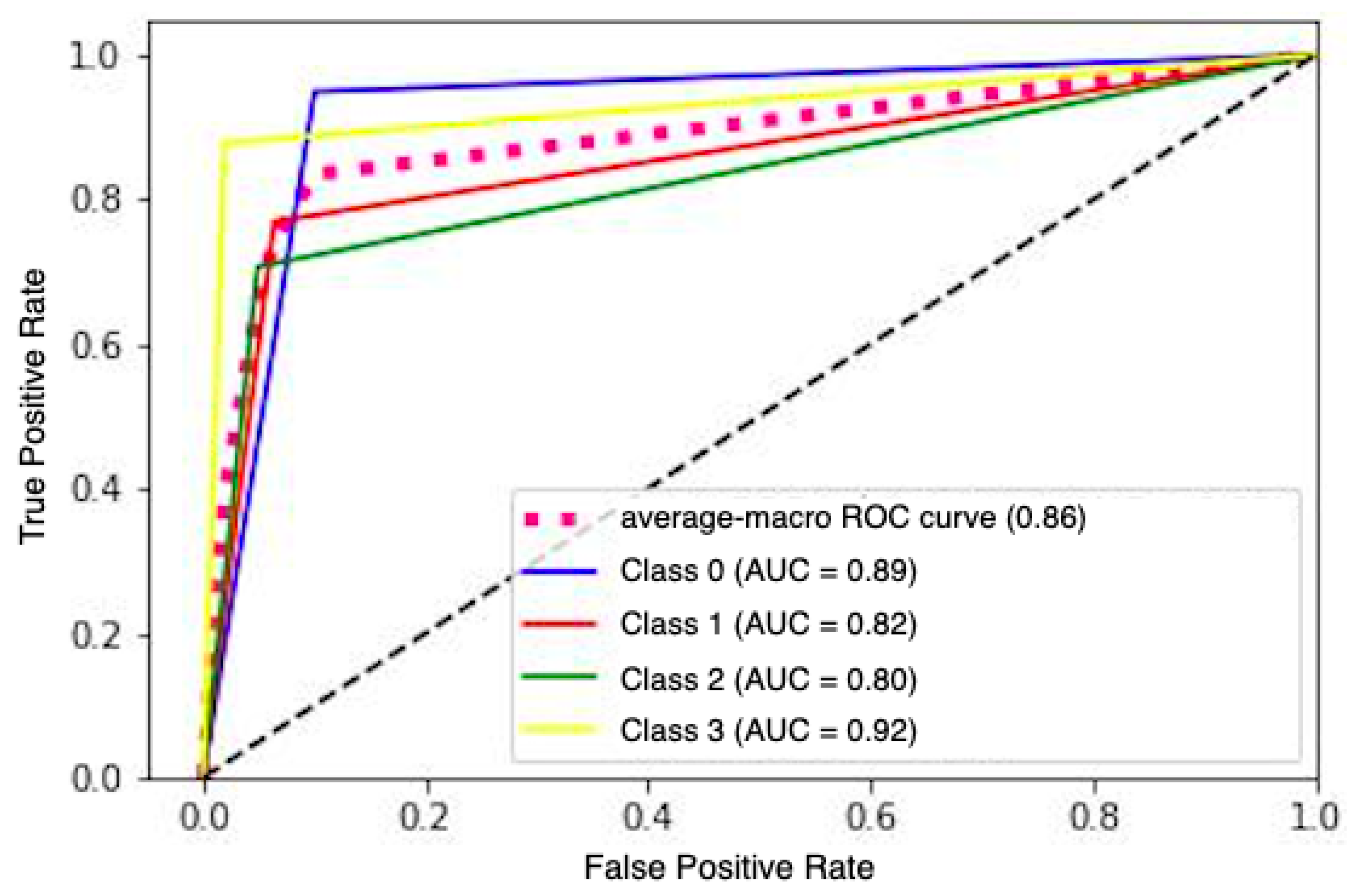
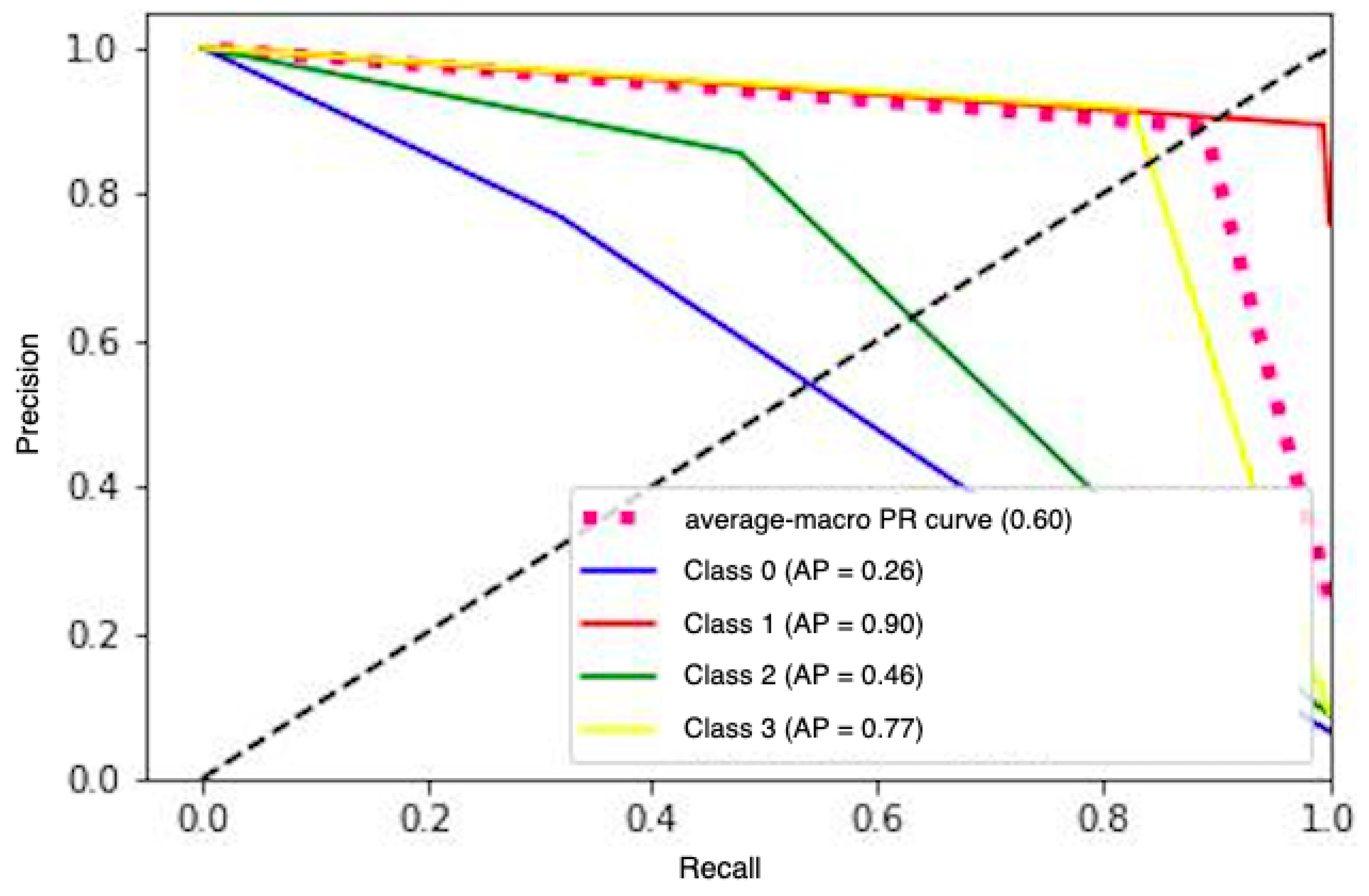

| Database | Model | Precision | Recall | Accuracy | Brier Score |
|---|---|---|---|---|---|
| PCR Imbalanced (Balanced) | MLP | 82.44 (82.72) | 82.72 (82.26) | 82.37 (82.25) | 0.088 (0.088) |
| GBM | 82.59 (82.48) | 82.92 (82.14) | 82.54 (82.14) | 0.08 (0.089) | |
| RF | 83.04 (82.96) | 83.12 (82.47) | 82.84 (82.47) | 0.085 (0.087) | |
| DT | 82.46 (82.37) | 82.60 (81.76) | 82.27 (81.76) | 0.088 (0.091) | |
| XGBoost | 82.73 (82.67) | 82.99 (82.27) | 82.65 (82.27) | 0.086 (0.088) | |
| KNN | 82.16 (82.35) | 82.22 (81.51) | 82.07 (81.61) | 0.089 (0.092) | |
| SVM | 82.23 (81.93) | 82.55 (81.57) | 82.27 (81.57) | 0.088 (0.092) | |
| LRR | 71.76 (71.65) | 70.13 (70.78) | 71.37 (70.78) | 0.143 (0.145) | |
| LR | 72.29 (72.29) | 70.85 (71.38) | 72.02 (71.38) | 0.139 (0.143) | |
| Rapid Imbalanced (Balanced) | MLP | 85.30 (81.40) | 79.41 (81.40) | 96.44 (81.40) | 0.171 (0.092) |
| GBM | 85.61 (81.14) | 79.40 (81.15) | 96.46 (81.15) | 0.017 (0.094) | |
| RF | 86.38 (81.50) | 79.25 (81.56) | 96.53 (81.56) | 0.017 (0.092) | |
| DT | 85.42 (80.99) | 79.61 (80.93) | 96.53 (80.93) | 0.017 (0.095) | |
| XGBoost | 85.76 (80.89) | 79.32 (80.96) | 96.47 (80.96) | 0.017 (0.095) | |
| KNN | 87.81 (80.52) | 78.93 (79.90) | 96.56 (79.90) | 0.017 (0.100) | |
| SVM | 86.12 (80.97) | 79.11 (81.11) | 96.49 (81.11) | 0.017 (0.094) | |
| LRR | 73.32 (68.78) | 55.60 (68.98) | 93.24 (68.98) | 0.033 (0.155) | |
| LR | 72.42 (70.70) | 58.37 (70.24) | 93.50 (70.24) | 0.032 (0.148) | |
| Both imbalanced (Balanced) | MLP | 85.93 (80.95) | 71.11 (80.47) | 90.73 (80.95) | 0.046 (0.097) |
| GBM | 85.89 (81.14) | 71.16 (80.64) | 90.70 (80.64) | 0.046 (0.096) | |
| RF | 86.85 (81.50) | 71.63 (80.95) | 90.96 (80.95) | 0.045 (0.095) | |
| DT | 85.60 (80.74) | 71.66 (80.13) | 90.76 (80.13) | 0.046 (0.099) | |
| XGBoost | 86.07 (81.28) | 71.39 (80.77) | 90.79 (80.77) | 0.046 (0.096) | |
| KNN | 82.23 (80.33) | 66.15 (79.58) | 88.98 (79.58) | 0.055 (0.102) | |
| SVM | 86.43 (80.54) | 70.95 (80.11) | 90.77 (80.11) | 0.046 (0.099) | |
| LRR | 68.87 (70.16) | 52.82 (70.10) | 85.54 (70.10) | 0.072 (0.149) | |
| LR | 71.16 (71.24) | 55.39 (71.22) | 86.29 (71.22) | 0.068 (0.143) |
| Dataset | Feature | GBM | DT | RF | XGBoost |
|---|---|---|---|---|---|
| PCR | Respiratory distress | 0.141 (0.008) | 0.155 (0.008) | 0.156 (0.008) | 0.144 (0.008) |
| Vomit | 0.037 (0.004) | 0.036 (0.004) | 0.035 (0.004) | 0.037 (0.004) | |
| Saturation | 0.149 (0.007) | 0.166 (0.008) | 0.161 (0.007) | 0.154 (0.008) | |
| Fatigue | 0.055 (0.006) | 0.047 (0.005) | 0.057 (0.006) | 0.058 (0.006) | |
| Diarrhea | 0.028 (0.003) | 0.024 (0.003) | 0.027 (0.003) | 0.028 (0.003) | |
| Abdominal pain | 0.013 (0.003) | 0.006 (0.002) | 0.011 (0.002) | 0.013 (0.002) | |
| Gender | 0.138 (0.007) | 0.133 (0.007) | 0.137 (0.007) | 0.137 (0.007) | |
| Health professional | 0.029 (0.003) | 0.026 (0.002) | 0.029 (0.003) | 0.031 (0.003) | |
| Fever | 0.235 (0.009) | 0.230 (0.008) | 0.234 (0.008) | 0.230 (0.008) | |
| Sore throat | 0.077 (0.006) | 0.073 (0.005) | 0.075 (0.005) | 0.079 (0.005) | |
| Dyspnoea | 0.098 (0.007) | 0.095 (0.006) | 0.093 (0.007) | 0.092 (0.007) | |
| Smell disorder | 0.009 (0.002) | 0.017 (0.002) | 0.004 (0.002) | 0.010 (0.002) | |
| Cough | 0.105 (0.007) | 0.101 (0.007) | 0.102 (0.007) | 0.102 (0.007) | |
| Runny nose | 0.007 (0.002) | 0.006 (0.002) | 0.007 (0.002) | 0.005 (0.002) | |
| Taste disorder | 0.019 (0.002) | 0.014 (0.002) | 0.009 (0.002) | 0.010 (0.002) | |
| Headache | 0.012 (0.002) | 0.012 (0.002) | 0.013 (0.002) | 0.008 (0.002) | |
| Rapid | Respiratory distress | 0.109 (0.007) | 0.113 (0.007) | 0.156 (0.008) | 0.121 (0.008) |
| Vomit | 0.019 (0.004) | 0.017 (0.003) | 0.015 (0.004) | 0.020 (0.004) | |
| Saturation | 0.140 (0.008) | 0.194 (0.008) | 0.165 (0.008) | 0.154 (0.007) | |
| Fatigue | 0.056 (0.005) | 0.072 (0.006) | 0.055 (0.005) | 0.055 (0.005) | |
| Diarrhea | 0.013 (0.003) | 0.012 (0.003) | 0.009 (0.002) | 0.015 (0.004) | |
| Abdominal pain | 0.004 (0.002) | 0.008 (0.002) | 0.003 (0.001) | 0.007 (0.002) | |
| Gender | 0.096 (0.009) | 0.096 (0.010) | 0.100 (0.009) | 0.095 (0.008) | |
| Health professional | 0.013 (0.003) | 0.014 (0.002) | 0.015 (0.002) | 0.018 (0.003) | |
| Fever | 0.148 (0.008) | 0.158 (0.009) | 0.148 (0.007) | 0.152 (0.007) | |
| Sore throat | 0.080 (0.006) | 0.073 (0.006) | 0.072 (0.006) | 0.075 (0.006) | |
| Dyspnoea | 0.130 (0.008) | 0.144 (0.009) | 0.136 (0.008) | 0.136 (0.008) | |
| Smell disorder | 0.083 (0.005) | 0.096 (0.006) | 0.087 (0.006) | 0.084 (0.005) | |
| Cough | 0.075 (0.007) | 0.086 (0.008) | 0.072 (0.007) | 0.067 (0.008) | |
| Runny nose | 0.054 (0.004) | 0.046 (0.003) | 0.039 (0.003) | 0.048 (0.003) | |
| Taste disorder | 0.042 (0.004) | 0.036 (0.004) | 0.023 (0.004) | 0.039 (0.004) | |
| Headache | 0.047 (0.004) | 0.053 (0.004) | 0.045 (0.004) | 0.055 (0.004) | |
| Both | Respiratory distress | 0.140 (0.004) | 0.148 (0.004) | 0.147 (0.004) | 0.143 (0.005) |
| Vomit | 0.035 (0.003) | 0.041 (0.004) | 0.039 (0.004) | 0.038 (0.003) | |
| Saturation | 0.171 (0.006) | 0.191 (0.005) | 0.190 (0.006) | 0.184 (0.006) | |
| Fatigue | 0.053 (0.003) | 0.062 (0.003) | 0.057 (0.003) | 0.055 (0.003) | |
| Diarrhea | 0.017 (0.002) | 0.017 (0.002) | 0.015 (0.002) | 0.018 (0.002) | |
| Abdominal pain | 0.018 (0.002) | 0.010 (0.002) | 0.010 (0.002) | 0.018 (0.002) | |
| Gender | 0.123 (0.005) | 0.126 (0.006) | 0.121 (0.005) | 0.119 (0.005) | |
| Health professional | 0.020 (0.002) | 0.016 (0.002) | 0.019 (0.002) | 0.020 (0.002) | |
| Fever | 0.182 (0.006) | 0.187 (0.006) | 0.183 (0.007) | 0.179 (0.007) | |
| Sore throat | 0.084 (0.004) | 0.087 (0.004) | 0.082 (0.004) | 0.083 (0.004) | |
| Dyspnoea | 0.103 (0.005) | 0.108 (0.005) | 0.103 (0.005) | 0.107 (0.005) | |
| Smell disorder | 0.047 (0.003) | 0.053 (0.003) | 0.039 (0.003) | 0.048 (0.003) | |
| Cough | 0.086 (0.004) | 0.088 (0.004) | 0.087 (0.005) | 0.084 (0.004) | |
| Runny nose | 0.017 (0.001) | 0.016 (0.001) | 0.017 (0.002) | 0.016 (0.001) | |
| Taste disorder | 0.024 (0.003) | 0.027 (0.003) | 0.019 (0.003) | 0.024 (0.003) | |
| Headache | 0.035 (0.003) | 0.033 (0.002) | 0.028 (0.003) | 0.031 (0.003) |
| Dataset | Feature | MLP | SVM |
|---|---|---|---|
| RT-PCR | Respiratory distress | 0.139 (0.009) | 0.135 (0.008) |
| Vomit | 0.041 (0.005) | 0.039 (0.004) | |
| Saturation | 0.146 (0.008) | 0.152 (0.007) | |
| Fatigue | 0.056 (0.005) | 0.059 (0.005) | |
| Diarrhea | 0.025 (0.003) | 0.028 (0.003) | |
| Abdominal pain | 0.015 (0.003) | 0.015 (0.003) | |
| Gender | 0.137 (0.007) | 0.125 (0.006) | |
| Health professional | 0.043 (0.003) | 0.021 (0.002) | |
| Fever | 0.230 (0.008) | 0.219 (0.008) | |
| Sore throat | 0.081 (0.007) | 0.069 (0.006) | |
| Dyspnoea | 0.100 (0.006) | 0.080 (0.006) | |
| Smell disorder | 0.013 (0.003) | 0.010 (0.002) | |
| Cough | 0.105 (0.007) | 0.098 (0.007) | |
| Runny nose | 0.010 (0.002) | 0.003 (0.002) | |
| Taste disorder | 0.018 (0.002) | 0.014 (0.002) | |
| Headache | 0.020 (0.002) | 0.007 (0.002) | |
| Rapid | Respiratory distress | 0.090 (0.007) | 0.092 (0.007) |
| Vomit | 0.019 (0.004) | 0.019 (0.004) | |
| Saturation | 0.132 (0.007) | 0.125 (0.007) | |
| Fatigue | 0.067 (0.005) | 0.061 (0.006) | |
| Diarrhea | 0.018 (0.003) | 0.017 (0.004) | |
| Abdominal pain | 0.011 (0.003) | 0.007 (0.002) | |
| Gender | 0.090 (0.009) | 0.076 (0.007) | |
| Health professional | 0.017 (0.003) | 0.016 (0.003) | |
| Fever | 0.141 (0.007) | 0.143 (0.007) | |
| Sore throat | 0.064 (0.006) | 0.062 (0.006) | |
| Dyspnoea | 0.129 (0.008) | 0.118 (0.007) | |
| Smell disorder | 0.074 (0.005) | 0.075 (0.005) | |
| Cough | 0.071 (0.007) | 0.068 (0.008) | |
| Runny nose | 0.041 (0.003) | 0.047 (0.003) | |
| Taste disorder | 0.032 (0.004) | 0.034 (0.003) | |
| Headache | 0.049 (0.005) | 0.044 (0.004) | |
| Both | Respiratory distress | 0.123 (0.005) | 0.123 (0.005) |
| Vomit | 0.038 (0.003) | 0.035 (0.004) | |
| Saturation | 0.146 (0.005) | 0.157 (0.006) | |
| Fatigue | 0.052 (0.003) | 0.047 (0.003) | |
| Diarrhea | 0.017 (0.002) | 0.016 (0.002) | |
| Abdominal pain | 0.020 (0.002) | 0.018 (0.002) | |
| Gender | 0.122 (0.005) | 0.109 (0.005) | |
| Health professional | 0.025 (0.002) | 0.018 (0.002) | |
| Fever | 0.180 (0.006) | 0.173 (0.006) | |
| Sore throat | 0.083 (0.004) | 0.083 (0.004) | |
| Dyspnoea | 0.108 (0.005) | 0.092 (0.005) | |
| Smell disorder | 0.058 (0.003) | 0.040 (0.002) | |
| Cough | 0.089 (0.005) | 0.079 (0.005) | |
| Runny nose | 0.023 (0.02) | 0.017 (0.001) | |
| Taste disorder | 0.030 (0.003) | 0.022 (0.003) | |
| Headache | 0.048 (0.003) | 0.028 (0.003) |
| Dataset | Model | Top 1 | Top 2 | Top 3 | Top 4 | Top 5 |
|---|---|---|---|---|---|---|
| PCR | MLP | Fever | Saturation | Respiratory distress | Gender | Cough |
| GBM | Fever | Saturation | Respiratory distress | Gender | Cough | |
| RF | Fever | Saturation | Gender | Respiratory distress | Cough | |
| DT | Saturation | Fever | Gender | Cough | Respiratory distress | |
| XGBoost | Saturation | Health professional | Respiratory distress | Fever | Fatigue | |
| SVM | Fever | Saturation | Respiratory distress | Gender | Cough | |
| Rapid | MLP | Fever | Saturation | Dyspnoea | Respiratory distress | Gender |
| GBM | Saturation | Dyspnoea | Fever | Smell disorder | Respiratory distress | |
| RF | Saturation | Fever | Dyspnoea | Fatigue | Gender | |
| DT | Fever | Saturation | Gender | Respiratory distress | Smell disorder | |
| XGBoost | Saturation | Smell disorder | Runny nose | Headache | Respiratory distress | |
| SVM | Fever | Saturation | Dyspnoea | Respiratory distress | Gender | |
| Both | MLP | Fever | Saturation | Respiratory distress | Gender | Dyspnoea |
| GBM | Diarrhea | Abdominal pain | Taste disorder | Headache | Runny nose | |
| RF | Abdominal pain | Diarrhea | Runny nose | Headache | Health professional | |
| DT | Health professional | Abdominal pain | Headache | Taste disorder | Runny nose | |
| XGBoost | Taste disorder | Cough | Gender | Dyspnoea | Sore throat | |
| SVM | Fever | Saturation | Respiratory distress | Gender | Dyspnoea |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viana dos Santos Santana, I.; Sobrinho, Á.; Dias da Silva, L.; Perkusich, A. Machine Learning for COVID-19 and Influenza Classification during Coexisting Outbreaks. Appl. Sci. 2023, 13, 11518. https://doi.org/10.3390/app132011518
Viana dos Santos Santana I, Sobrinho Á, Dias da Silva L, Perkusich A. Machine Learning for COVID-19 and Influenza Classification during Coexisting Outbreaks. Applied Sciences. 2023; 13(20):11518. https://doi.org/10.3390/app132011518
Chicago/Turabian StyleViana dos Santos Santana, Iris, Álvaro Sobrinho, Leandro Dias da Silva, and Angelo Perkusich. 2023. "Machine Learning for COVID-19 and Influenza Classification during Coexisting Outbreaks" Applied Sciences 13, no. 20: 11518. https://doi.org/10.3390/app132011518
APA StyleViana dos Santos Santana, I., Sobrinho, Á., Dias da Silva, L., & Perkusich, A. (2023). Machine Learning for COVID-19 and Influenza Classification during Coexisting Outbreaks. Applied Sciences, 13(20), 11518. https://doi.org/10.3390/app132011518










