Variability in Phytochemical Contents and Biological Activities among Adenophora triphylla Genotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Extract Preparation
2.2. DNA Extraction and Analysis of DNA Barcoding Regions
2.3. Phytochemical Analysis
2.4. Analysis of the Antioxidant Activities
2.5. Cell Culture
2.6. Determination of Cell Viability, Nitric Oxide (NO), and Melanin Production
2.7. Statistical Analysis
3. Results and Discussion
3.1. Genetic Diversity of A. triphylla Genotypes
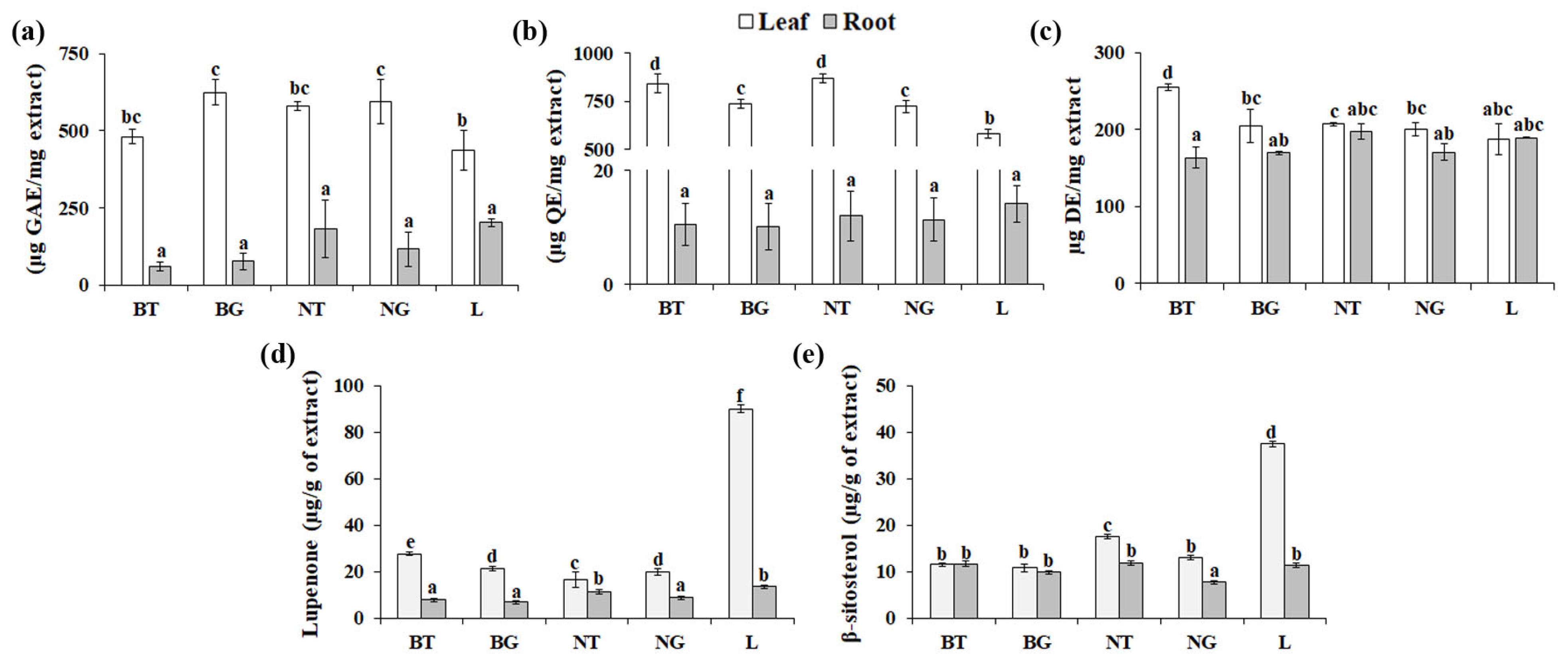
3.2. Variability in Phytochemical Contents among A. triphylla Genotypes
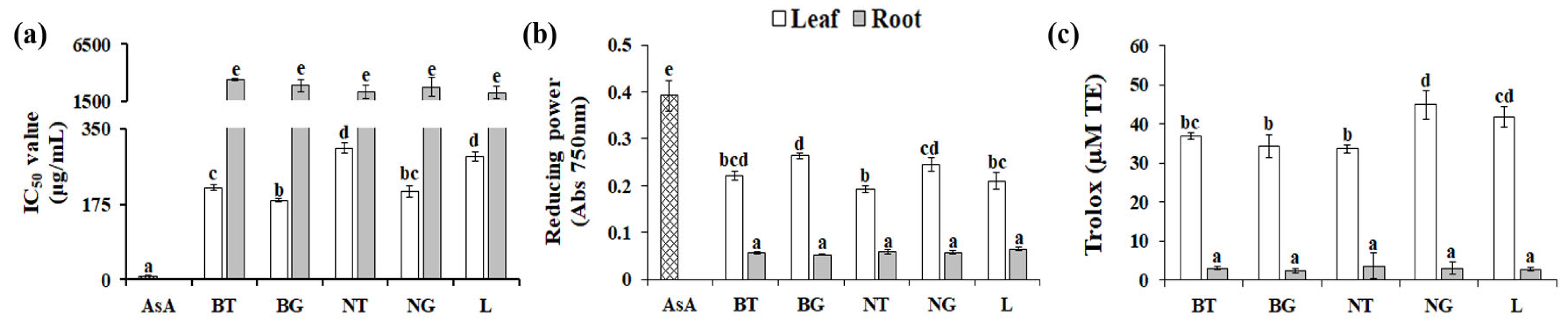
3.3. Effect of Genotype on Antioxidant Activities
3.4. Effect of Genotype on the Anti-Inflammatory and Antimelanogenic Activities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nasim, N.; Sandeep, I.S.; Mohanty, S. Plant-Derived Natural Products for Drug Discovery: Current Approaches and Prospects. Nucleus 2022, 65, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, A. Potential role of bioactive phytochemicals in combination therapies against antimicrobial activity. J. Pharmacopunct. 2022, 25, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Khan, F.; Alshammari, N.; Saeed, A.; Aqil, F.; Saeed, M. Updates on the anticancer potential of garlic organosulfur compounds and their nanoformulations: Plant therapeutics in cancer management. Front. Pharmacol. 2023, 14, 1154034. [Google Scholar] [CrossRef]
- Kusano, M.; Yang, Z.; Okazaki, Y.; Nakabayashi, R.; Fukushima, A.; Saito, K. Using metabolomic approaches to explore chemical diversity in rice. Mol. Plant 2015, 8, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Ju, H.J.; Bang, J.-H.; Chung, J.-W.; Hyun, T.K. Variation in essential oil composition and antimicrobial activity among different genotypes of Perilla frutescens var. crispa. J. Appl. Biol. Chem. 2021, 64, 127–131. [Google Scholar] [CrossRef]
- Ju, H.J.; Kim, K.C.; Kim, H.; Kim, J.-S.; Hyun, T.K. Variability of polyphenolic compounds and biological activities among Perilla frutescens var. crispa genotypes. Horticulturae 2021, 7, 404. [Google Scholar] [CrossRef]
- Kim, D.; Kim, K.Y. Adenophora triphylla var. japonica inhibits candida biofilm formation, increases susceptibility to antifungal agents and reduces infection. Int. J. Mol. Sci. 2021, 22, 12523. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Lee, E.H.; Lee, T.J.; Kim, S.W.; Kim, B.H. Anti-obesity effect and action mechanism of Adenophora triphylla root ethanol extract in C57BL/6 obese mice fed a high-fat diet. Biosci. Biotechnol. Biochem. 2013, 77, 544–550. [Google Scholar] [CrossRef]
- Park, K.; Kim, Y.; Hwangbo, K.; Gil, J.; Chung, H.; Park, S.; Hong, C.P.; Lee, Y. Development of simple sequence repeat markers from Adenophora triphylla var. japonica (Regel) H. Hara using next generation sequencing. Korean J. Med. Crop Sci. 2017, 25, 411–417. [Google Scholar] [CrossRef]
- Park, H.J.; Park, S.H. Hexane fraction of Adenophora triphylla var. japonica root extract induces apoptosis of human lung cancer cells by inactivating Src/STAT3 pathway. Nat. Prod. Res. 2023, 37, 2924–2928. [Google Scholar] [CrossRef]
- Lee, H.R.; Kim, H.M.; Jeong, H.W.; Kim, G.G.; Na, C.I.; Oh, M.M.; Hwang, S.J. Growth Characteristics of Adenophora triphylla var. japonica Hara Seedlings as Affected by Growing Medium. Plants 2019, 8, 466. [Google Scholar] [CrossRef]
- Han, S.; Sebastin, R.; Wang, X.; Lee, K.J.; Cho, G.-T.; Hyun, D.Y.; Chung, J.-W. Identification of Vicia species native to South Korea using molecular and morphological characteristics. Front. Plant Sci. 2021, 12, 608559. [Google Scholar] [CrossRef] [PubMed]
- Le, A.V.; Parks, S.E.; Nguyen, M.H.; Roach, P.D. Improving the vanillin-sulphuric acid method for quantifying total saponins. Technologies 2018, 6, 84. [Google Scholar] [CrossRef]
- Yoon, S.Y.; Kim, K.H.; Hyun, T.K. Antioxidant and anticancer activities of Adenophora triphylla leaf and root extracts. J. Plant Biotechnol. 2023, 50, 137–141. [Google Scholar] [CrossRef]
- Ohga, K.; Muroi, M.; Hayakawa, H.; Yokoyama, J.; Ito, K.; Tebayashi, S.-I.; Arakawa, R.; Fukuda, T. Morphological and anatomical analyses of the serpentine ecotype of Adenophora triphylla var. japonica (Campanulaceae). J. Plant Stud. 2012, 1, 180. [Google Scholar] [CrossRef]
- Moon, B.C.; Kim, W.J.; Han, K.S.; Yang, S.; Kang, Y.; Park, I.; Piao, R. Differentiating authentic Adenophorae radix from its adulterants in commercially-processed samples using multiplexed ITS sequence-based SCAR markers. Appl. Sci. 2017, 7, 660. [Google Scholar] [CrossRef]
- Ohga, K.; Muroi, M.; Hayakawa, H.; Yokoyama, J.; Ito, K.; Tebayashi, S.-I.; Arakawa, R.; Fukuda, T. Coastal adaptation of Adenophora triphylla var. japonica (Campanulaceae). Am. J. Plant Sci. 2013, 4, 596–601. [Google Scholar] [CrossRef]
- Feng, S.; Jiang, Y.; Wang, S.; Jiang, M.; Chen, Z.; Ying, Q.; Wang, H. Molecular identification of Dendrobium Species (Orchidaceae) based on the DNA barcode ITS2 region and Its application for phylogenetic study. Int. J. Mol. Sci. 2015, 16, 21975–21988. [Google Scholar] [CrossRef]
- De Boer, H.J.; Ghorbani, A.; Manzanilla, V.; Raclariu, A.-C.; Kreziou, A.; Ounjai, S.; Osathanunkul, M.; Gravendeel, B. DNA metabarcoding of orchid-derived products reveals widespread illegal orchid trade. Proc. Biol. Sci. 2017, 284, 20171182. [Google Scholar] [CrossRef]
- Hebert, P.D.; Ratnasingham, S.; de Waard, J.R. Barcoding animal life: Cytochrome C oxidase subunit 1 divergences among closely related species. Proc. Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef]
- Li, H.; Xiao, W.; Tong, T.; Li, Y.; Zhang, M.; Lin, X.; Zou, X.; Wu, Q.; Guo, X. The specific DNA barcodes based on chloroplast genes for species identification of orchidaceae plants. Sci. Rep. 2021, 11, 1424. [Google Scholar] [CrossRef]
- Acharya, G.C.; Mohanty, S.; Dasgupta, M.; Sahu, S.; Singh, S.; Koundinya, A.V.V.; Kumari, M.; Naresh, P.; Sahoo, M.R. Molecular phylogeny, DNA barcoding, and ITS2 secondary structure predictions in the medicinally important Eryngium Genotypes of east coast region of India. Genes 2022, 13, 1678. [Google Scholar] [CrossRef] [PubMed]
- Yong, W.; Chunbo, X.; Laga, T.; Xiaoming, Z.; Mingjiu, W. Genetic evaluation and germplasm identification analysis on ITS2, trnL-F, and psbA-trnH of alfalfa varieties germplasm resources. Open Life Sci. 2023, 18, 20220582. [Google Scholar] [CrossRef] [PubMed]
- Mannino, G.; Gentile, C.; Maffei, M.E. Chemical partitioning and DNA fingerprinting of some pistachio (Pistacia vera L.) varieties of different geographical origin. Phytochemistry 2019, 160, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.I.; Zhao, B.T.; Lee, J.H.; Lee, D.-U.; Kim, Y.S.; Min, B.S.; Son, J.K.; Woo, M.H. Quantitative and classification analyses of lupenone and β-Sitosterol by GC-FID in Adenophora triphylla var. japonica Hara and Codonopsis lanceolata. Nat. Prod. Sci. 2014, 20, 243–250. [Google Scholar]
- Kim, H.-J.; Son, D.C.; Kim, H.-J.; Choi, K.; Oh, S.-H.; Kang, S.-H. The chemotaxonomic classification of Korean campanulaceae based on triterpene, sterol, and polyacetylene contents. Biochem. Syst. Ecol. 2017, 74, 11–18. [Google Scholar] [CrossRef]
- Dashbaldan, S.; Pączkowski, C.; Szakiel, A. Variations in triterpenoid deposition in cuticular waxes during development and maturation of selected fruits of Rosaceae family. Int. J. Mol. Sci. 2020, 21, 9762. [Google Scholar] [CrossRef] [PubMed]
- Vrábl, D.; Nezval, J.; Pech, R.; Volná, A.; Mašková, P.; Pleva, J.; Kuzniciusová, N.; Provazová, M.; Štroch, M.; Špunda, V. Light drives and temperature modulates: Variation of phenolic compounds profile in relation to photosynthesis in spring barley. Int. J. Mol. Sci. 2023, 24, 2427. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Y.; Zhao, H.; Fu, X.; Fang, S. Localization and dynamic change of saponins in Cyclocarya paliurus (Batal.) Iljinskaja. PLoS ONE 2019, 14, e0223421. [Google Scholar] [CrossRef]
- Kabashima, K.; Honda, T.; Ginhoux, F.; Egawa, G. The immunological anatomy of the skin. Nat. Rev. Immunol. 2019, 19, 19–30. [Google Scholar] [CrossRef]
- Bogdan, C. Nitric oxide and the immune response. Nat. Immunol. 2001, 2, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-H. Ethyl Acetate Fraction of Adenophora triphylla var. japonica Inhibits Migration of Lewis Lung Carcinoma Cells by Suppressing Macrophage Polarization toward an M2 Phenotype. J. Pharmacopunct. 2019, 22, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.K.; Park, S.K.; Kang, J.Y.; Kim, J.M.; Park, S.H.; Kwon, B.S.; Lee, C.J.; Kang, J.E.; Park, S.B.; Lee, U.; et al. Skin Whitening effect of ethyl acetate fraction of Adenophora triphylla var. japonica Sprout. Korean J. Plant Res. 2017, 30, 352–363. [Google Scholar]
- Chun, J.; Kang, M.; Kim, Y.S. A triterpenoid saponin from Adenophora triphylla var. japonica suppresses the growth of human gastric cancer cells via regulation of apoptosis and autophagy. Tumor Biol. 2014, 35, 12021–12030. [Google Scholar] [CrossRef] [PubMed]
- Pillaiyar, T.; Manickam, M.; Jung, S.H. Downregulation of melanogenesis: Drug discovery and therapeutic options. Drug Discov. Today 2017, 22, 282–298. [Google Scholar] [CrossRef]
- Manzoor, Z.; Koh, Y.-S. Mitogen-activated protein kinases in inflammation. J. Bacteriol. Virol. 2012, 42, 189–195. [Google Scholar] [CrossRef]


| Genotypes | Size (bp)/Accession Number (NABIC) | ||
|---|---|---|---|
| ITS2 | matK | psbA-trnH | |
| NG | 259 bp/NU-1869 | 601 bp/NU-1875 | 312 bp/NU-1882 |
| BT | 258 bp/NU-1870 | 601 bp/NU-1878 | 311 bp/NU-1880 |
| BG | 257 bp/NU-1871 | 601 bp/NU-1877 | 311 bp/NU-1879 |
| L | 257 bp/NU-1872 | 601 bp/NU-1874 | 311 bp/NU-1881 |
| NT | 259 bp/NU-1873 | 601 bp/NU-1876 | 311 bp/NU-1883 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ajayi, O.E.; Yoon, S.Y.; Moon, S.; Kim, K.H.; Kim, J.H.; Chung, J.-W.; Jang, K.-I.; Hyun, T.K. Variability in Phytochemical Contents and Biological Activities among Adenophora triphylla Genotypes. Appl. Sci. 2023, 13, 11184. https://doi.org/10.3390/app132011184
Ajayi OE, Yoon SY, Moon S, Kim KH, Kim JH, Chung J-W, Jang K-I, Hyun TK. Variability in Phytochemical Contents and Biological Activities among Adenophora triphylla Genotypes. Applied Sciences. 2023; 13(20):11184. https://doi.org/10.3390/app132011184
Chicago/Turabian StyleAjayi, Oluwadamilola Elizabeth, Seon Young Yoon, Suyun Moon, Ki Hyun Kim, Jung Hwan Kim, Jong-Wook Chung, Keum-Il Jang, and Tae Kyung Hyun. 2023. "Variability in Phytochemical Contents and Biological Activities among Adenophora triphylla Genotypes" Applied Sciences 13, no. 20: 11184. https://doi.org/10.3390/app132011184
APA StyleAjayi, O. E., Yoon, S. Y., Moon, S., Kim, K. H., Kim, J. H., Chung, J.-W., Jang, K.-I., & Hyun, T. K. (2023). Variability in Phytochemical Contents and Biological Activities among Adenophora triphylla Genotypes. Applied Sciences, 13(20), 11184. https://doi.org/10.3390/app132011184






