The Inhibition Activities of the Fruit Extract of Plinia cauliflora against Melanoma Cells and the Single-Stranded DNA-Binding Protein (SSB) from Klebsiella pneumoniae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Expression and Isolation of the Recombinant Protein
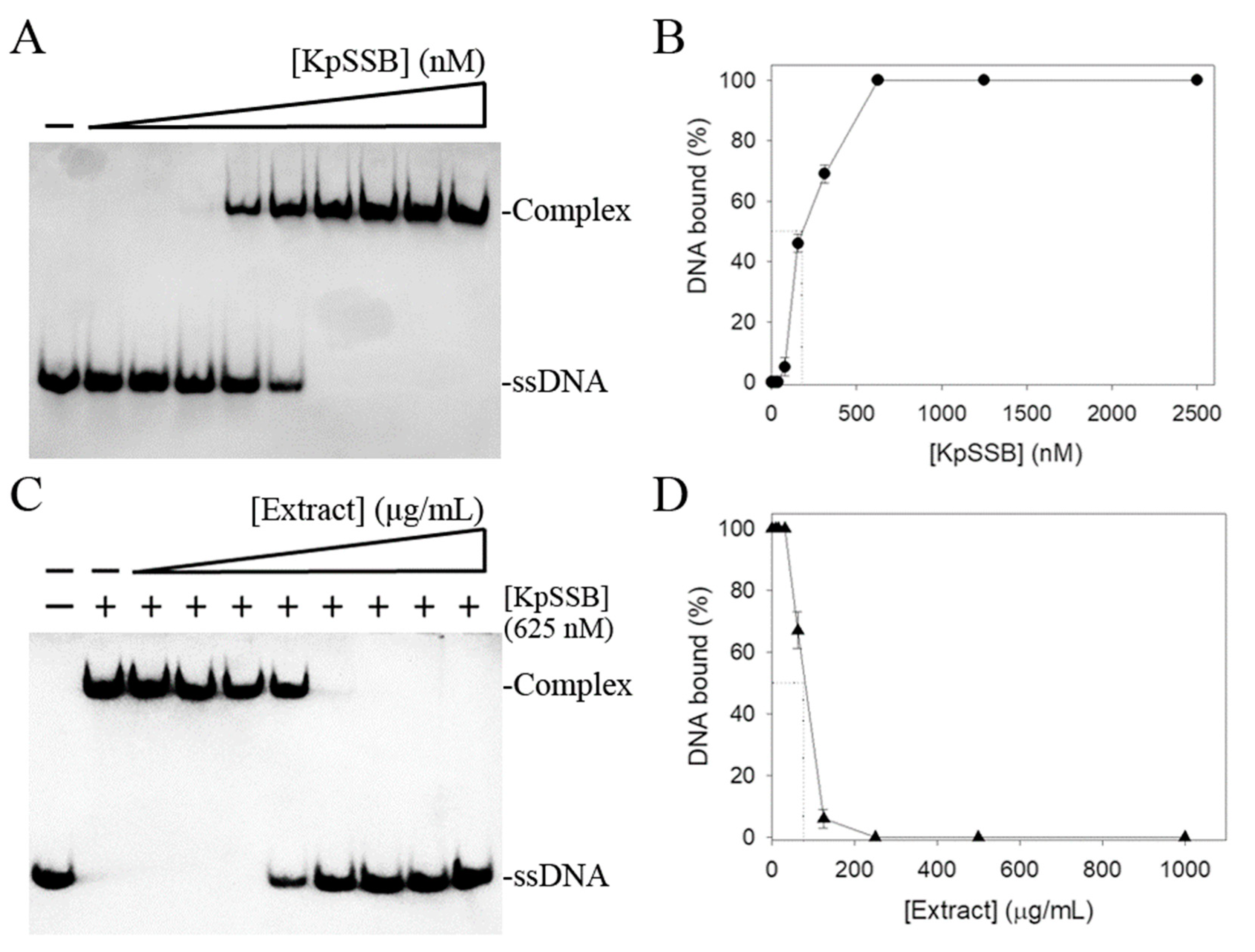
2.3. Electrophoretic Mobility Shift Analysis (EMSA)
2.4. Inhibition Assay
2.5. Plant Material and Fruit Harvest
2.6. Extract Preparation
2.7. GC–MS Analysis
2.8. Agar Well Diffusion Assay
2.9. Total Phenolic Content and Flavonoid Content
2.10. Antioxidant Activity Analysis
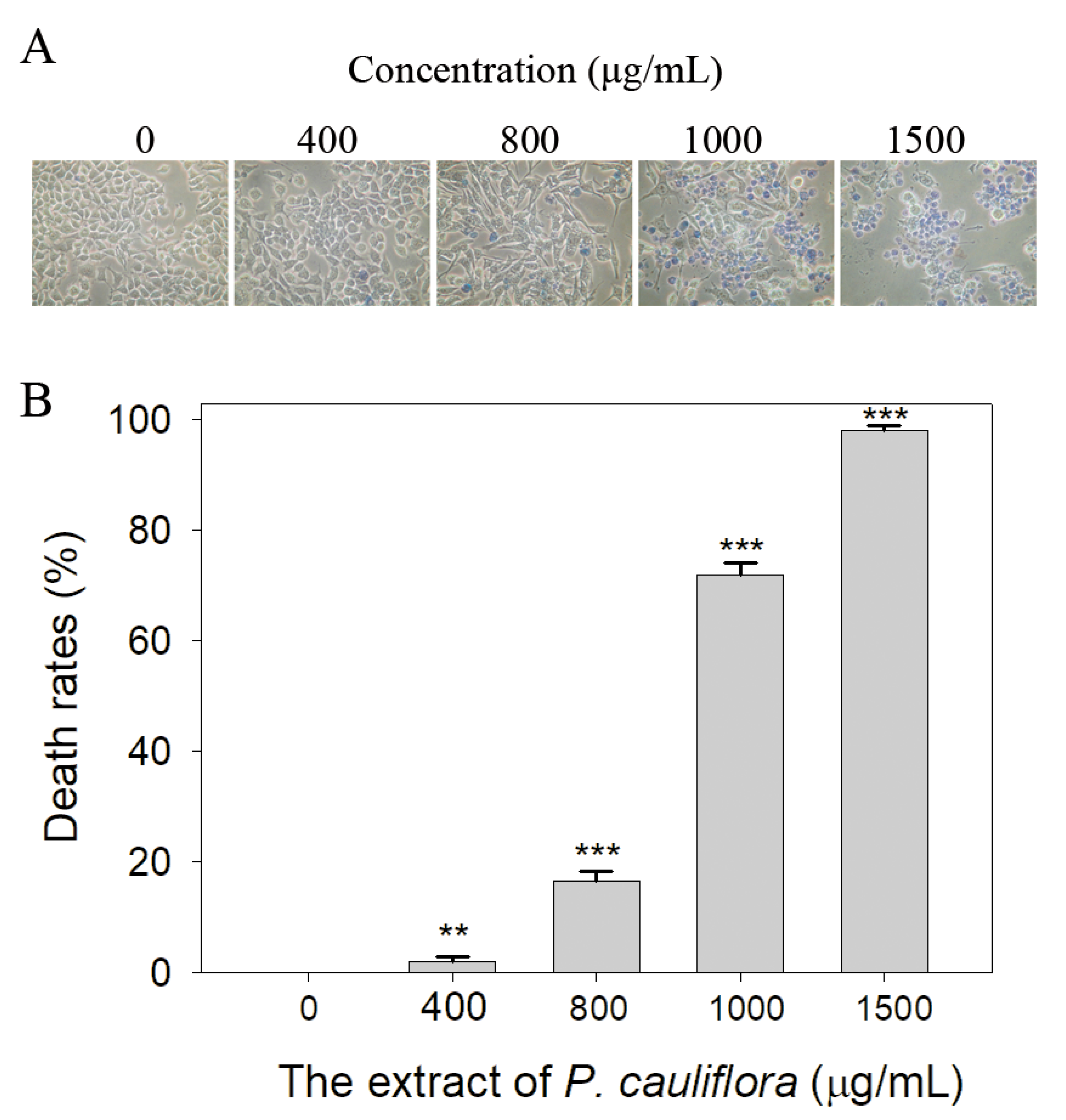
2.11. Trypan Blue Cytotoxicity Assay
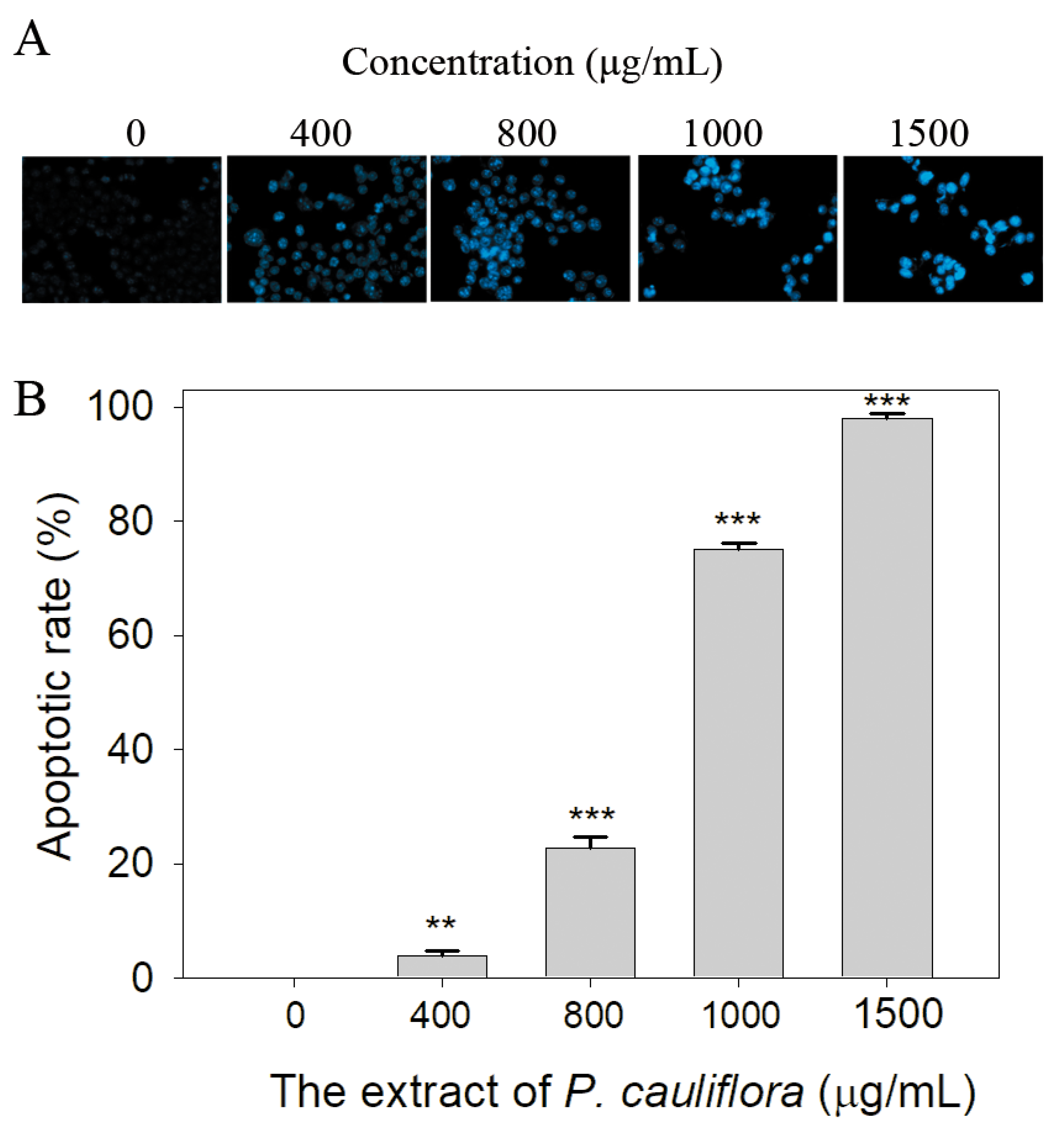
2.12. Chromatin Condensation Assay
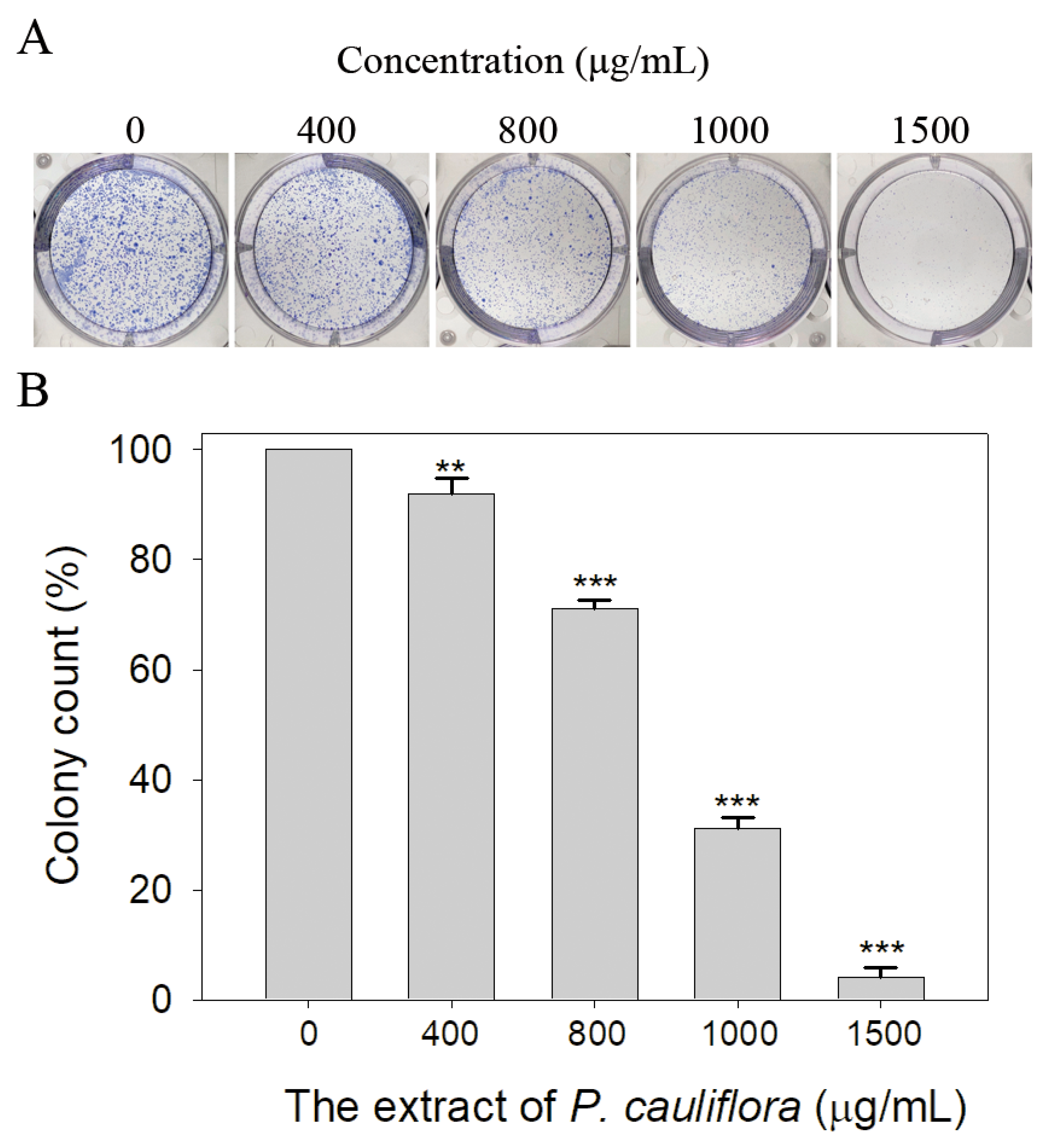
2.13. Clonogenic Formation Assay
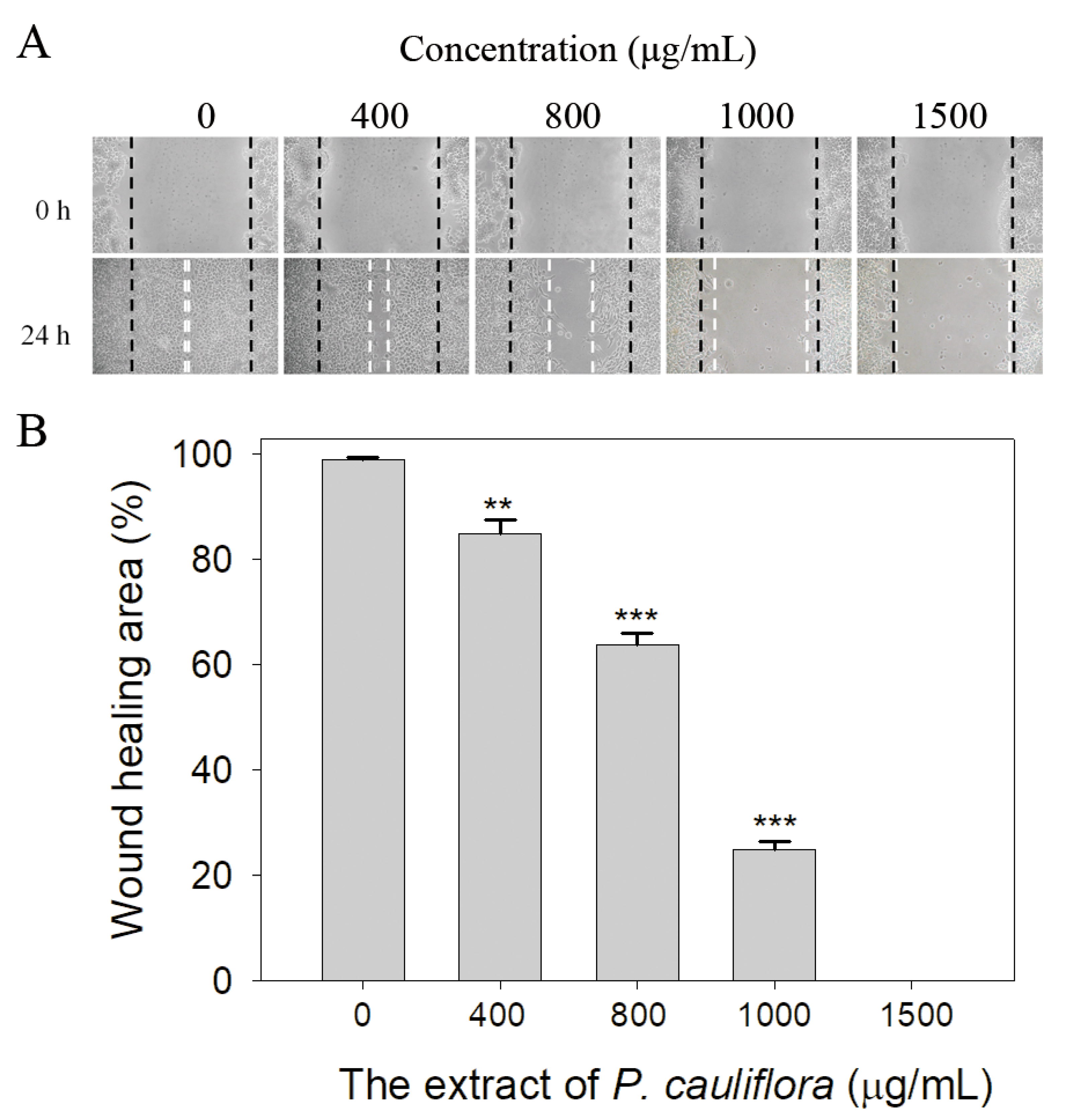
2.14. Wound-Healing Assay
2.15. MOE-Dock Analysis
2.16. Biorad Protein (Bradford) Assay
2.17. Statistical Analysis
3. Results
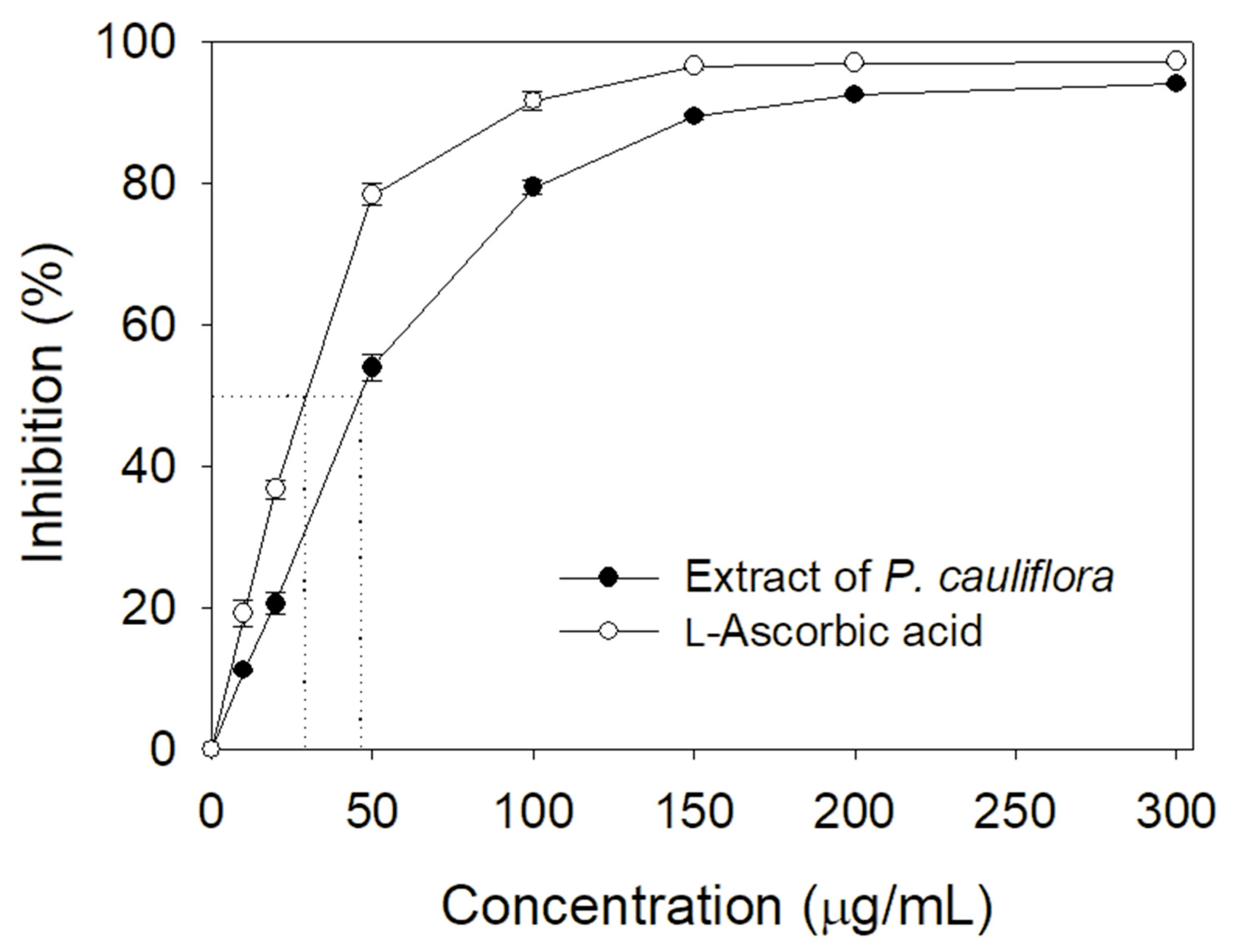
3.1. Inhibition of KpSSB by the Extract of Plinia cauliflora
3.2. Inhibition of KpSSB by Rutin
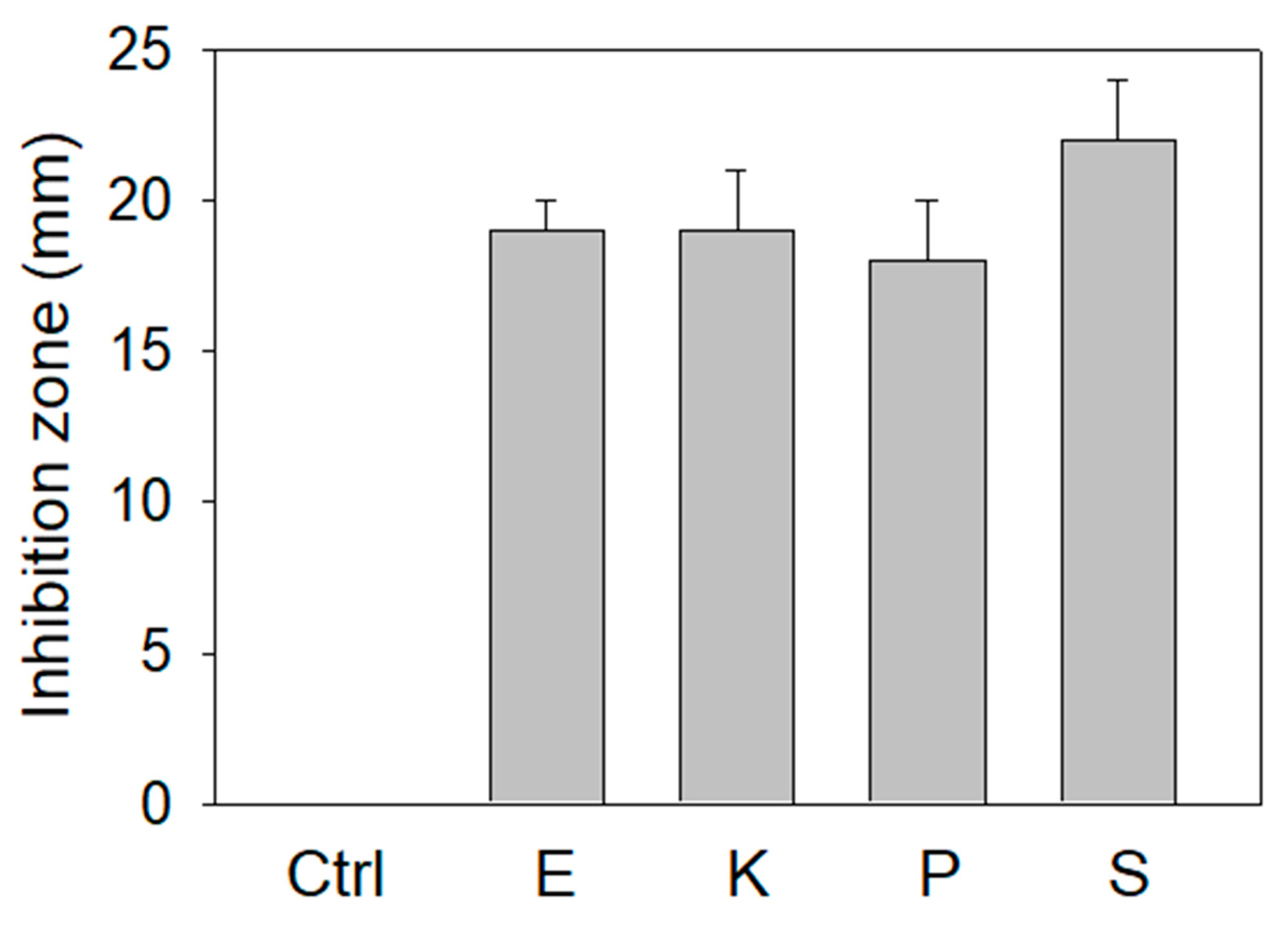
3.3. Antibacterial Activity of the Extract of P. cauliflora
3.4. TPC, TFC, and Antioxidant Ability of P. cauliflora
3.5. GC–MS Analysis
3.6. Cytotoxic Effect of the Extract of P. cauliflora
3.7. The Extract of P. cauliflora Induced Apoptosis of B16F10 Cells
3.8. The Extract of P. cauliflora Inhibited the Migration of B16F10 Cells
3.9. The Extract of P. cauliflora Inhibited the Proliferation of B16F10 Cells
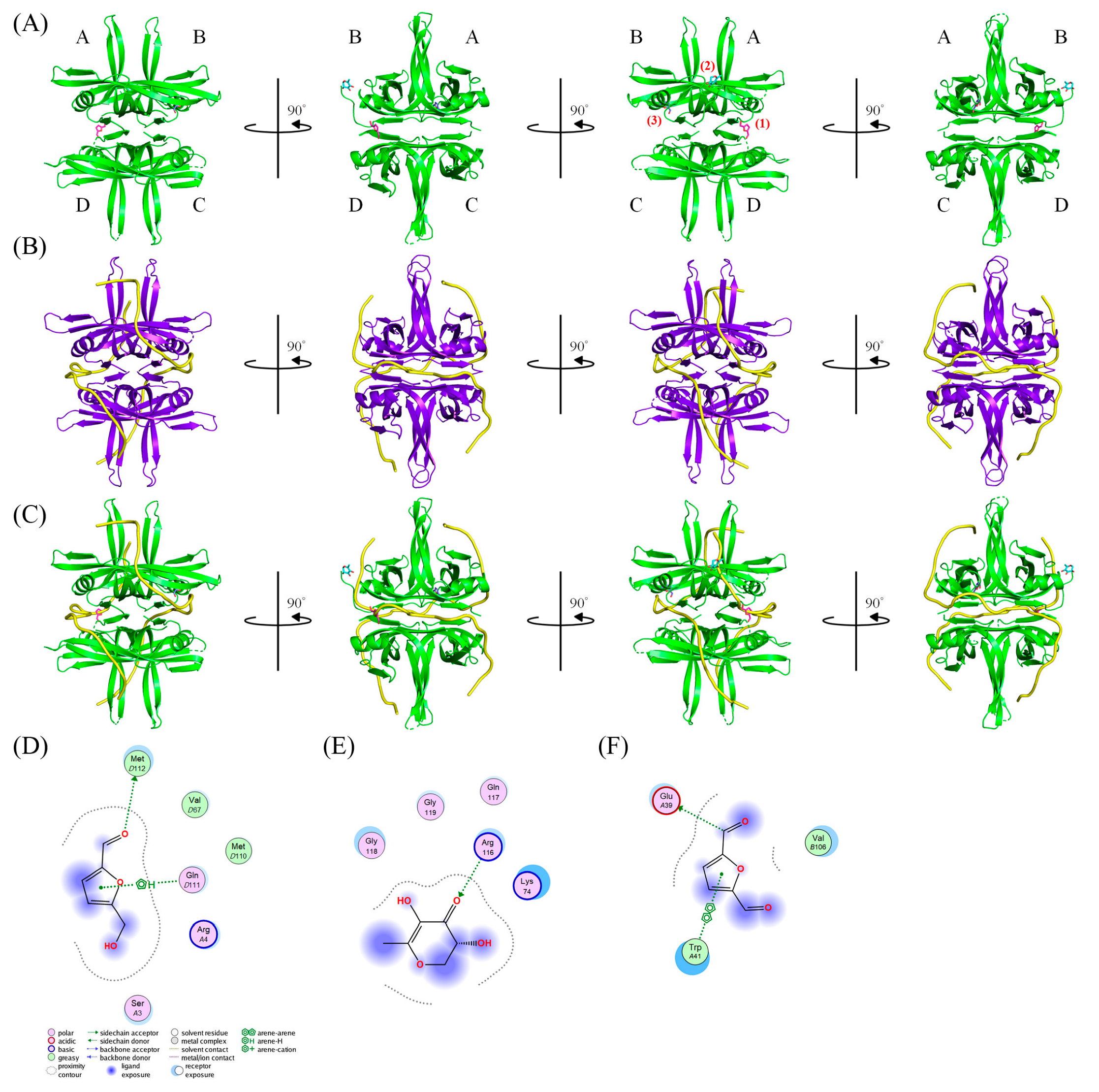
3.10. Molecular Docking
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roszkowski, S. Application of Polyphenols and Flavonoids in Oncological Therapy. Molecules 2023, 28, 4080. [Google Scholar] [CrossRef] [PubMed]
- Lobiuc, A.; Pavăl, N.E.; Mangalagiu, I.I.; Gheorghiță, R.; Teliban, G.C.; Amăriucăi-Mantu, D.; Stoleru, V. Future Antimicrobials: Natural and Functionalized Phenolics. Molecules 2023, 28, 1114. [Google Scholar] [CrossRef] [PubMed]
- Jakobušić Brala, C.; Karković Marković, A.; Kugić, A.; Torić, J.; Barbarić, M. Combination Chemotherapy with Selected Polyphenols in Preclinical and Clinical Studies-An Update Overview. Molecules 2023, 28, 3746. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- WHO. Traditional medicine growing needs and potential. WHO Policy Perspect. Med. 2002, 2, 1–6. [Google Scholar]
- Rufino-Palomares, E.E.; Pérez-Jiménez, A.; García-Salguero, L.; Mokhtari, K.; Reyes-Zurita, F.J.; Peragón-Sánchez, J.; Lupiáñez, J.A. Nutraceutical Role of Polyphenols and Triterpenes Present in the Extracts of Fruits and Leaves of Olea europaea as Antioxidants, Anti-Infectives and Anticancer Agents on Healthy Growth. Molecules 2022, 27, 2341. [Google Scholar] [CrossRef]
- Khan, A.W.; Farooq, M.; Haseeb, M.; Choi, S. Role of Plant-Derived Active Constituents in Cancer Treatment and Their Mechanisms of Action. Cells 2022, 11, 1326. [Google Scholar] [CrossRef] [PubMed]
- Gligorov, J.; Richard, S. Breast cancer: Weekly paclitaxel—Still preferred first-line taxane for mBC. Nat. Rev. Clin. Oncol. 2015, 12, 508–509. [Google Scholar] [CrossRef]
- da Veiga Correia, V.T.; da Silva, P.R.; Ribeiro, C.M.S.; Ramos, A.; Mazzinghy, A.; Silva, V.D.M.; Júnior, A.H.O.; Nunes, B.V.; Vieira, A.L.S.; Ribeiro, L.V.; et al. An Integrative Review on the Main Flavonoids Found in Some Species of the Myrtaceae Family: Phytochemical Characterization, Health Benefits and Development of Products. Plants 2022, 11, 2796. [Google Scholar] [CrossRef]
- Gasparotto Junior, A.; de Souza, P.; Lívero, F. Plinia cauliflora (Mart.) Kausel: A comprehensive ethnopharmacological review of a genuinely Brazilian species. J. Ethnopharmacol. 2019, 245, 112169. [Google Scholar] [CrossRef]
- Chang, S.K.; Alasalvar, C.; Shahidi, F. Superfruits: Phytochemicals, antioxidant efficacies, and health effects—A comprehensive review. Crit. Rev. Food Sci. Nutr. 2019, 59, 1580–1604. [Google Scholar] [CrossRef]
- Inada, K.O.P.; Leite, I.B.; Martins, A.B.N.; Fialho, E.; Tomás-Barberán, F.A.; Perrone, D.; Monteiro, M. Jaboticaba berry: A comprehensive review on its polyphenol composition, health effects, metabolism, and the development of food products. Food Res. Int. 2021, 147, 110518. [Google Scholar] [CrossRef]
- Wu, S.B.; Dastmalchi, K.; Long, C.; Kennelly, E.J. Metabolite profiling of jaboticaba (Myrciaria cauliflora) and other dark-colored fruit juices. J. Agric. Food Chem. 2012, 60, 7513–7525. [Google Scholar] [CrossRef]
- Reguengo, L.M.; Nascimento, R.P.D.; Machado, A.; Marostica Junior, M.R. Signaling pathways and the potential anticarcinogenic effect of native Brazilian fruits on breast cancer. Food Res. Int. 2022, 155, 111117. [Google Scholar] [CrossRef]
- Machado, A.; Alves, M.D.R.; Nascimento, R.P.D.; Reguengo, L.M.; Marostica Junior, M.R. Antiproliferative effects and main molecular mechanisms of Brazilian native fruits and their by-products on lung cancer. Food Res. Int. 2022, 162, 111953. [Google Scholar] [CrossRef]
- Filho, A.V.; Avila, L.B.; Lacorte, D.H.; Martiny, T.R.; Rosseto, V.; Moraes, C.C.; Dotto, G.L.; Carreno, N.L.V.; da Rosa, G.S. Brazilian Agroindustrial Wastes as a Potential Resource of Bioative Compounds and Their Antimicrobial and Antioxidant Activities. Molecules 2022, 27, 6876. [Google Scholar] [CrossRef]
- Avila, L.B.; Barreto, E.R.C.; Souza, P.K.; Silva, B.Z.; Martiny, T.R.; Moraes, C.C.; Morais, M.M.; Raghavan, V.; Rosa, G.S.D. Carrageenan-Based Films Incorporated with Jaboticaba Peel Extract: An Innovative Material for Active Food Packaging. Molecules 2020, 25, 5563. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.R.; Laine, P.S. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990, 54, 342–380. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.R. The Biochemical Mechanism of Fork Regression in Prokaryotes and Eukaryotes-A Single Molecule Comparison. Int. J. Mol. Sci. 2022, 23, 8613. [Google Scholar] [CrossRef] [PubMed]
- Antony, E.; Lohman, T.M. Dynamics of E. coli single stranded DNA binding (SSB) protein-DNA complexes. Semin. Cell Dev. Biol. 2019, 86, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Shereda, R.D.; Kozlov, A.G.; Lohman, T.M.; Cox, M.M.; Keck, J.L. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 289–318. [Google Scholar] [CrossRef] [PubMed]
- Heller, R.C.; Marians, K.J. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol. 2006, 7, 932–943. [Google Scholar] [CrossRef] [PubMed]
- Marians, K.J. PriA-directed replication fork restart in Escherichia coli. Trends Biochem. Sci. 2000, 25, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Lohman, T.M.; Ferrari, M.E. Escherichia coli single-stranded DNA-binding protein: Multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994, 63, 527–570. [Google Scholar] [CrossRef]
- Bianco, P.R. The mechanism of action of the SSB interactome reveals it is the first OB-fold family of genome guardians in prokaryotes. Protein Sci. 2021, 30, 1757–1775. [Google Scholar] [CrossRef]
- Voter, A.F.; Killoran, M.P.; Ananiev, G.E.; Wildman, S.A.; Hoffmann, F.M.; Keck, J.L. A High-Throughput Screening Strategy to Identify Inhibitors of SSB Protein-Protein Interactions in an Academic Screening Facility. SLAS Discov. 2017, 23, 94–101. [Google Scholar] [CrossRef]
- Glanzer, J.G.; Endres, J.L.; Byrne, B.M.; Liu, S.; Bayles, K.W.; Oakley, G.G. Identification of inhibitors for single-stranded DNA-binding proteins in eubacteria. J. Antimicrob. Chemother. 2016, 71, 3432–3440. [Google Scholar] [CrossRef]
- Marceau, A.H.; Bernstein, D.A.; Walsh, B.W.; Shapiro, W.; Simmons, L.A.; Keck, J.L. Protein interactions in genome maintenance as novel antibacterial targets. PLoS ONE 2013, 8, e58765. [Google Scholar] [CrossRef]
- Iftode, C.; Daniely, Y.; Borowiec, J.A. Replication protein A (RPA): The eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999, 34, 141–180. [Google Scholar] [CrossRef]
- Byrne, B.M.; Oakley, G.G. Replication protein A, the laxative that keeps DNA regular: The importance of RPA phosphorylation in maintaining genome stability. Semin. Cell Dev. Biol. 2019, 86, 112–120. [Google Scholar] [CrossRef]
- Tommasi, R.; Brown, D.G.; Walkup, G.K.; Manchester, J.I.; Miller, A.A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 2015, 14, 529–542. [Google Scholar] [CrossRef]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 2023, 21, 280–295. [Google Scholar] [CrossRef] [PubMed]
- Du Toit, A. The cost of resistance. Nat. Rev. Microbiol. 2022, 20, 317. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Yang, X.; Chan, E.W.; Zhang, R.; Chen, S. Klebsiella species: Taxonomy, hypervirulence and multidrug resistance. eBioMedicine 2022, 79, 103998. [Google Scholar] [CrossRef]
- Arato, V.; Raso, M.M.; Gasperini, G.; Berlanda Scorza, F.; Micoli, F. Prophylaxis and Treatment against Klebsiella pneumoniae: Current Insights on This Emerging Anti-Microbial Resistant Global Threat. Int. J. Mol. Sci. 2021, 22, 4042. [Google Scholar] [CrossRef] [PubMed]
- Franzolin, M.R.; Courrol, D.D.S.; Silva, F.R.O.; Courrol, L.C. Antimicrobial Activity of Silver and Gold Nanoparticles Prepared by Photoreduction Process with Leaves and Fruit Extracts of Plinia cauliflora and Punica granatum. Molecules 2022, 27, 6860. [Google Scholar] [CrossRef]
- Machado, G.H.A.; Marques, T.R.; de Carvalho, T.C.L.; Duarte, A.C.; de Oliveira, F.C.; Gonçalves, M.C.; Piccoli, R.H.; Corrêa, A.D. Antibacterial activity and in vivo wound healing potential of phenolic extracts from jaboticaba skin. Chem. Biol. Drug Des. 2018, 92, 1333–1343. [Google Scholar] [CrossRef]
- Mott, M.L.; Berger, J.M. DNA replication initiation: Mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 2007, 5, 343–354. [Google Scholar] [CrossRef]
- Nižnanský, Ľ.; Osinová, D.; Kuruc, R.; Hengerics Szabó, A.; Szórádová, A.; Masár, M.; Nižnanská, Ž. Natural Taxanes: From Plant Composition to Human Pharmacology and Toxicity. Int. J. Mol. Sci. 2022, 23, 15619. [Google Scholar] [CrossRef]
- Merecz-Sadowska, A.; Sitarek, P.; Kowalczyk, T.; Zajdel, K.; Kucharska, E.; Zajdel, R. The Modulation of Melanogenesis in B16 Cells Upon Treatment with Plant Extracts and Isolated Plant Compounds. Molecules 2022, 27, 4360. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventos, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Methods Enzymol. 1999, 299, 152–178. [Google Scholar]
- Chang, C.C.; Yang, M.H.; Wen, H.M.; Chern, J.C. Estimation of total flavonoid content in propolis by two complementary colorimetric methods. J. Food Drug Anal. 2002, 10, 178–182. [Google Scholar]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 2001, 21, A-3B. [Google Scholar] [CrossRef]
- Larsson, R.; Nygren, P. A rapid fluorometric method for semiautomated determination of cytotoxicity and cellular proliferation of human tumor cell lines in microculture. Anticancer Res. 1989, 9, 1111–1119. [Google Scholar]
- Chen, M.H.; Yang, W.L.; Lin, K.T.; Liu, C.H.; Liu, Y.W.; Huang, K.W.; Chang, P.M.; Lai, J.M.; Hsu, C.N.; Chao, K.M.; et al. Gene expression-based chemical genomics identifies potential therapeutic drugs in hepatocellular carcinoma. PLoS ONE 2011, 6, e27186. [Google Scholar] [CrossRef]
- Liang, C.C.; Park, A.Y.; Guan, J.L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef]
- Reynertson, K.A.; Yang, H.; Jiang, B.; Basile, M.J.; Kennelly, E.J. Quantitative analysis of antiradical phenolic constituents from fourteen edible Myrtaceae fruits. Food Chem. 2008, 109, 883–890. [Google Scholar] [CrossRef]
- Dahiya, D.; Nigam, P.S. Biotherapy Using Probiotics as Therapeutic Agents to Restore the Gut Microbiota to Relieve Gastrointestinal Tract Inflammation, IBD, IBS and Prevent Induction of Cancer. Int. J. Mol. Sci. 2023, 24, 5748. [Google Scholar] [CrossRef]
- Choi, J.R.; Kozalak, G.; di Bari, I.; Babar, Q.; Niknam, Z.; Rasmi, Y.; Yong, K.W. In Vitro Human Cancer Models for Biomedical Applications. Cancers 2022, 14, 2284. [Google Scholar] [CrossRef] [PubMed]
- Dzialo, M.; Mierziak, J.; Korzun, U.; Preisner, M.; Szopa, J.; Kulma, A. The Potential of Plant Phenolics in Prevention and Therapy of Skin Disorders. Int. J. Mol. Sci. 2016, 17, 160. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Halma, M.T.J.; Wuite, G.J.L. Unravelling How Single-Stranded DNA Binding Protein Coordinates DNA Metabolism Using Single-Molecule Approaches. Int. J. Mol. Sci. 2023, 24, 2806. [Google Scholar] [CrossRef] [PubMed]
- Sugino, A.; Peebles, C.L.; Kreuzer, K.N.; Cozzarelli, N.R. Mechanism of action of nalidixic acid: Purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. USA 1977, 74, 4767–4771. [Google Scholar] [CrossRef]
- Gellert, M.; O’Dea, M.H.; Itoh, T.; Tomizawa, J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl. Acad. Sci. USA 1976, 73, 4474–4478. [Google Scholar] [CrossRef]
- Pandey, P.; Khan, F.; Qari, H.A.; Oves, M. Rutin (Bioflavonoid) as Cell Signaling Pathway Modulator: Prospects in Treatment and Chemoprevention. Pharmaceuticals 2021, 14, 1069. [Google Scholar] [CrossRef]
- Richard, D.J.; Bolderson, E.; Cubeddu, L.; Wadsworth, R.I.; Savage, K.; Sharma, G.G.; Nicolette, M.L.; Tsvetanov, S.; McIlwraith, M.J.; Pandita, R.K.; et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature 2008, 453, 677–681. [Google Scholar] [CrossRef]
- Raghunathan, S.; Kozlov, A.G.; Lohman, T.M.; Waksman, G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000, 7, 648–652. [Google Scholar] [CrossRef]
- Dickey, T.H.; Altschuler, S.E.; Wuttke, D.S. Single-stranded DNA-binding proteins: Multiple domains for multiple functions. Structure 2013, 21, 1074–1084. [Google Scholar] [CrossRef]
- Su, X.C.; Wang, Y.; Yagi, H.; Shishmarev, D.; Mason, C.E.; Smith, P.J.; Vandevenne, M.; Dixon, N.E.; Otting, G. Bound or free: Interaction of the C-terminal domain of Escherichia coli single-stranded DNA-binding protein (SSB) with the tetrameric core of SSB. Biochemistry 2014, 53, 1925–1934. [Google Scholar] [CrossRef]
- Pinc, M.M.; Dalmagro, M.; da Cruz Alves Pereira, E.; Donadel, G.; Thomaz, R.T.; da Silva, C.; Macruz, P.D.; Jacomassi, E.; Gasparotto Junior, A.; Hoscheid, J.; et al. Extraction Methods, Chemical Characterization, and In Vitro Biological Activities of Plinia cauliflora (Mart.) Kausel Peels. Pharmaceuticals 2023, 16, 1173. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.H.; Tyan, Y.C.; Chen, Z.S.; Lin, C.G.; Yang, M.H.; Yuan, S.S.; Tsai, W.C. Evaluation of the antioxidant activity and antiproliferative effect of the jaboticaba (Myrciaria cauliflora) seed extracts in oral carcinoma cells. Biomed. Res. Int. 2014, 2014, 185946. [Google Scholar] [CrossRef]
- Mendonça de Assis, P.; Cypriano Dutra, R.; Amarante, C.B.D.; Afonso Miranda Chaves, M.D.G.; Moreira, C.P.S.; Brandão, M.A.F.; Raposo, N.R.B. Plinia cauliflora (Mart.) Kausel: Toxicological assays, biological activities, and elemental analysis of organic compounds. Nat. Prod. Res. 2021, 35, 1727–1731. [Google Scholar] [CrossRef] [PubMed]
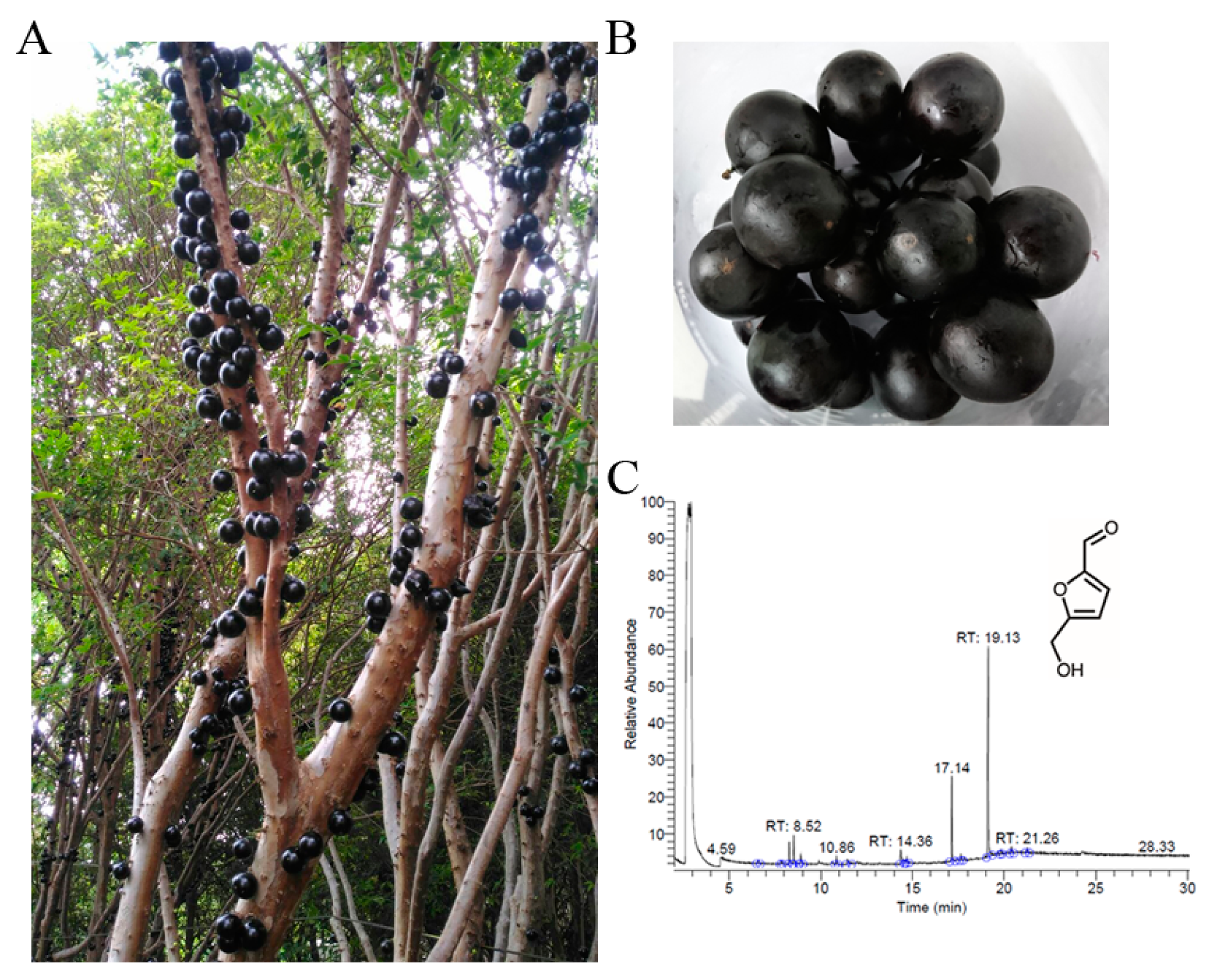
| Peak No. | RT (min) | Name of Compounds | MF | CS | MW | Area (%) |
|---|---|---|---|---|---|---|
| 1 | 19.13 | 5-Hydroxymethylfurfural | C6H6O3 | 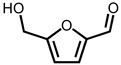 | 126 | 53.73% |
| 2 | 17.14 | 2,3-Dihydro-3,5-dihydroxy-6-methyl-4h-pyran-4-one | C6H8O4 | 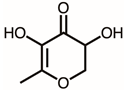 | 144 | 14.43% |
| 3 | 14.36 | 2,5-Diformylfuran | C6H4O3 | 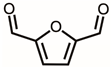 | 124 | 6.06% |
| 4 | 8.52 | Furfural | C5H4O2 | 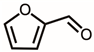 | 96 | 5.73% |
| 5 | 8.28 | Acetic acid | C2H4O2 | 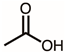 | 60 | 4.80% |
| 6 | 11.46 | Citraconic anhydride | C5H4O3 | 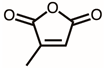 | 112 | 2.98% |
| 7 | 8.92 | Formic acid | CH2O2 | 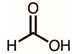 | 46 | 2.24% |
| 8 | 20.39 | Ethyl 4-hydroxy-3-methylbut-2-enoate | C7H12O3 | 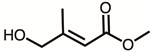 | 144 | 2.04% |
| 9 | 10.86 | Furfuryl alcohol | C5H6O2 | 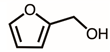 | 98 | 1.53% |
| 10 | 14.68 | Furyl hydroxymethyl ketone | C6H6O3 | 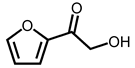 | 126 | 1.49% |
| 11 | 17.65 | 3-Acetyl-3-hydroxyoxolane-2-one | C6H8O4 | 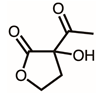 | 144 | 1.27% |
| 12 | 21.26 | 2,3-Dihydro-5-hydroxy-2-(hydroxymethyl)-4H-pyran-4-one | C6H8O4 | 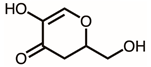 | 144 | 1.20% |
| 13 | 19.83 | 2(3H)-Furanone, dihydro-4-hydroxy- | C4H6O3 | 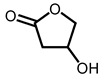 | 102 | 0.87% |
| 14 | 6.59 | Hydroxyacetone | C3H6O2 | 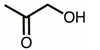 | 74 | 0.86% |
| 15 | 7.85 | 1-Hydroxybut-3-en-2-one | C4H6O2 | 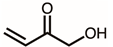 | 86 | 0.77% |
| S Score | Receptor Residue | Interaction | Distance (Å) | E (kcal/mol) | |
|---|---|---|---|---|---|
| (1) 5-Hydroxymethylfurfural | −4.3646 | (D) M112 | H-donor | 2.97 | −1.5 |
| (D) Q111 | pi–H | 4.56 | −0.5 | ||
| (D) Q111 | pi–H | 4.18 | −0.5 | ||
| (2) 2,3-Dihydro-3,5-dihydroxy-6-methyl-4h-pyran-4-one | −3.1840 | (B) R116 | H-acceptor | 2.94 | −4.0 |
| (3) 2,5-Diformylfuran | −3.5429 | (A) E39 | H-donor | 3.34 | −0.6 |
| (A) W41 | pi–pi | 3.59 | −0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, E.-S.; Huang, C.-Y. The Inhibition Activities of the Fruit Extract of Plinia cauliflora against Melanoma Cells and the Single-Stranded DNA-Binding Protein (SSB) from Klebsiella pneumoniae. Appl. Sci. 2023, 13, 11061. https://doi.org/10.3390/app131911061
Lin E-S, Huang C-Y. The Inhibition Activities of the Fruit Extract of Plinia cauliflora against Melanoma Cells and the Single-Stranded DNA-Binding Protein (SSB) from Klebsiella pneumoniae. Applied Sciences. 2023; 13(19):11061. https://doi.org/10.3390/app131911061
Chicago/Turabian StyleLin, En-Shyh, and Cheng-Yang Huang. 2023. "The Inhibition Activities of the Fruit Extract of Plinia cauliflora against Melanoma Cells and the Single-Stranded DNA-Binding Protein (SSB) from Klebsiella pneumoniae" Applied Sciences 13, no. 19: 11061. https://doi.org/10.3390/app131911061
APA StyleLin, E.-S., & Huang, C.-Y. (2023). The Inhibition Activities of the Fruit Extract of Plinia cauliflora against Melanoma Cells and the Single-Stranded DNA-Binding Protein (SSB) from Klebsiella pneumoniae. Applied Sciences, 13(19), 11061. https://doi.org/10.3390/app131911061






