Abstract
Timely and relevant information enables clinicians to make informed decisions about patient care outcomes. However, discovering related and understandable information from the vast medical literature is challenging. To address this problem, we aim to enable the development of search engines that meet the needs of medical practitioners by incorporating text difficulty features. We collected a dataset of 209 scientific research abstracts from different medical fields, available in both English and German. To determine the difficulty aspects of readability and technical level of each abstract, 216 medical experts annotated the dataset. We used a pre-trained BERT model, fine-tuned to our dataset, to develop a regression model predicting those difficulty features of abstracts. To highlight the strength of this approach, the model was compared to readability formulas currently in use. Analysis of the dataset revealed that German abstracts are more technically complex and less readable than their English counterparts. Our baseline model showed greater efficacy than current readability formulas in predicting domain-specific readability aspects. Conclusion: Incorporating these text difficulty aspects into the search engine will provide healthcare professionals with reliable and efficient information retrieval tools. Additionally, the dataset can serve as a starting point for future research.
1. Introduction
Search engines play a crucial role in facilitating information retrieval (IR) and supporting users’ information-seeking tasks across various domains. However, the field of healthcare poses unique challenges for medical practitioners when it comes to accessing timely and comprehensible search results as the information they acquire can significantly impact their decision making processes and ultimately influence patient care outcomes. Despite the widespread availability of search engines, medical practitioners often face difficulties in finding the precise and contextually relevant information they need within the constraints of their demanding schedules. These challenges arise from the specialized nature of medical knowledge, the vast amount of scientific literature available, and the requirement for accurate and easily understandable information.
Research conducted by Entin and Klare [1] has revealed the significant influence of factors such as a reader’s level of interest, prior knowledge, and the readability of a text on their comprehension of the material. It is essential to clarify that ease of reading primarily pertains to text understandability, while technicality is more focused on the concepts and domain-specific knowledge within the text [2,3,4]. This understanding underscores the need to address the readability and technicality aspects in the development of search engines tailored specifically to the needs of medical practitioners. To improve IR for healthcare professionals, it is necessary to incorporate these aspects into search engines. By retrieving comprehensible information based on their language proficiency and domain knowledge, these search engines can enhance the efficiency and effectiveness of the information provided, leading to informed decisions and optimal care. Similarly, Ref. [5] proposed to use the readability aspect for accepting or revising health-related documents.
While previous studies [6,7,8] have focused primarily on developing personalized search engines for health information consumers, such as laypeople and patients, there is a clear gap in adapting to the specific requirements of medical practitioners. Unlike laypeople, medical practitioners possess specialized expertise and language proficiency in their respective fields. Therefore, our research project aims to contribute to filling this gap by developing a model capable of extracting and classifying the ease of reading and technicality levels of medical research articles.
It is important to note that ease of reading and technicality are not mutually exclusive aspects [3]. Texts can exhibit varying degrees of both characteristics, leading to different combinations that may arise between these aspects. For instance, a text can be easy to read while still containing high technicality, or it can be difficult to read with low technicality, as shown in the following examples. (All examples have been reviewed and approved by a senior medical practitioner. Examples of easy-to-read content are from [9], while examples of harder-to-read content are from [10]).
- Easy to read and high technicalityAutologous hematopoietic stem cell transplantation has emerged as a promising therapeutic intervention for individuals with refractory multiple myeloma. This treatment approach has shown remarkable advancements in terms of progression-free survival and overall response rates, signifying its potential in improving patient outcomes.
- Hard to read and high technicalityThe pathophysiological mechanisms underlying idiopathic pulmonary fibrosis involve aberrant activation of transforming growth factor-beta signaling pathways, leading to excessive deposition of extracellular matrix components and subsequent progressive scarring of lung tissue.
- Easy to read and low technicalityRegular physical exercise has been widely recognized as a key lifestyle intervention for the prevention of cardiovascular diseases, with numerous studies demonstrating its positive impact on reducing the risk of heart attacks, stroke, and hypertension.
- Hard to read and low technicalityCarcinogenesis is a multifactorial process characterized by the dysregulation of cellular homeostasis, involving intricate interactions between oncogenes and tumor suppressor genes that disrupt normal cell growth control mechanisms, resulting in uncontrolled proliferation and the formation of malignant tumors.
Integrating difficulty aspects into search engines empowers medical practitioners with tailored and relevant search results that are aligned with their expertise and language proficiency. Without these aspects, search engines may fail to effectively address the specific needs of medical professionals, leading to limitations and challenges. For example, search results may include highly technical papers and articles with varying levels of readability, requiring manual sifting and wasting valuable time. The lack of customization based on technicality and ease of reading hinders precision, relevance, and quick access to necessary information. Furthermore, complex language and dense scientific jargon can impede comprehension for practitioners without specialized expertise, hindering decision making [11]. By considering difficulty aspects, search engines not only improve information accessibility and ensure comprehensibility but also optimize decision making and patient care. Similarly, studies [12,13] presented other features that could potentially enhance personalization in medical search engines. This enhancement in IR empowers medical professionals by making it easier for them to identify their target audience, thus facilitating the efficient utilization of their valuable time and expertise [14].
Readability formulas have already been developed for measuring the readability of a given text. However, the most commonly used readability formulas were not developed for technical materials [15]. Moreover, traditional readability formulas are oversimplified to deal with technical materials [16]. Therefore, to accomplish our objective, we leverage the power of pre-trained language models and fine-tuning techniques for predicting the difficulty aspects of a given document.
An intriguing application of our research lies in the integration of these assessed aspects into the IR process. This can be achieved by either filtering out search results that exceed a certain threshold of technicality or ease of reading, ensuring that the retrieved documents align with the user’s preferred level of comprehension. Additionally, these aspects can contribute to the calculation of relevance scores [17], allowing documents that match the desired technicality and readability criteria to be ranked higher in search results. This integration has the potential to enhance the efficiency and precision of IR for medical professionals, aiding them in accessing documents that align with their specific requirements and facilitating informed decision making.
However, we encountered a challenge in finding datasets specifically designed for medical practitioners, as most existing datasets target laypeople. Therefore, we compiled a dataset of medical abstracts from PubMed (PubMed is an extensive online database that grants users access to a diverse range of scientific literature in the field of biomedical and life sciences. It serves as a valuable resource for researchers, medical practitioners, and individuals seeking in-depth scholarly articles, abstracts, and citations pertaining to diverse medical disciplines, https://pubmed.ncbi.nlm.nih.gov/, accessed on 1 June 2023) and sought the expertise of medical doctors and medical students to annotate each article with ease of reading and technicality scores.
Through our research, we aim to demonstrate the ability of language models to capture and classify the ease of reading and technicality levels of medical documents. Furthermore, we investigate the differences in language complexity and technicality between English and German abstracts, finding German abstracts tend to be harder to read and exhibit higher levels of technicality when compared to their English counterparts.
To sum up, our research article makes contributions in the following areas: (a) we address a problem concerning the trade-off between comprehensibility and relevance within the field of IR for medical practitioners; (b) we are presenting a new dataset containing medical research articles annotated with ease of reading and technicality scores; (c) we are developing models capturing these aspects using pre-trained language models and comparing them with known readability formulas.
By addressing the specific needs of medical practitioners and integrating the difficulty aspects into search engines, we strive to enhance the accessibility and relevance of search results, ultimately empowering medical professionals with efficient and reliable IR tools.
2. Literature Review
2.1. Comprehensibility Aspects in Information Retrieval
The consideration of comprehension aspects in IR, particularly in domain-specific contexts, has received significant attention. Researchers have recognized the challenges faced by both domain experts and average users when searching for domain-specific information, such as medical and health-related content, from online resources [18].
A common issue encountered by users in IR systems is the presence of search results that encompass documents with varying levels of readability [16,19,20]. This poses a challenge, particularly for users with limited domain knowledge or lower education levels, as well as those facing physical, psychological, or emotional stress [16]. Consequently, there is a need for IR systems that not only retrieve relevant documents but also prioritize those with higher readability, adapting to the diverse needs of users [21].
To address this challenge, various approaches have been explored. Some studies [22,23] have investigated computational models of readability, aiming to develop efficient methods for assessing the readability of technical materials encountered in domain-specific IR. Traditional readability formulas, although widely used, are often insufficient for handling technical texts. On the other hand, more advanced algorithms, such as textual coherence models, may offer improved accuracy but suffer from computational complexity when applied to large-scale document re-ranking scenarios.
The importance of domain-specific readability computation in IR has been emphasized [24]. Technical terms and the need for efficient computations for large document collections are among the challenges identified. By integrating concept-based readability and domain-specific knowledge into the search process, researchers aim to enhance the accessibility and relevance of search results. These efforts contribute to empowering users, including both domain experts and average users, with efficient and reliable IR tools [16].
2.2. Readability Formulas
Since the early 20th century, researchers in this field have developed a variety of readability formulas aimed at laypeople. Many of these formulas are still widely used today [25]. Among the most commonly used formulas are Simple Measure of Gobbledygook (SMOG), the Dale-Chall Readability formula, the Flesch Reading Ease formula, the Fog Index, and the Fry Readability Graph. More details about readability methods can be found in [26].
These formulas typically analyze syntactic complexity and semantic difficulty. Syntactic complexity is often evaluated by examining sentence length, while semantic difficulty is measured using factors such as syllable count or word frequency lists. Other factors that have been found to influence readability include the presence of prepositional phrases, the use of personal pronouns, and the number of indeterminate clauses. However, it is important to note that readability formulas specifically targeting medical practitioners are currently lacking. Further research is needed to develop readability formulas tailored to the unique needs and expertise of medical professionals.
2.2.1. Simple Measure of Gobbledygook (SMOG)
The SMOG Index was introduced by clinical psychologist G. Harry McLaughlin in 1969 [27]. It is designed to estimate the years of education required to comprehend a piece of written text accurately by counting the words of three or more syllables in three ten-sentence samples. The formula calculates the reading grade level based on a simple mathematical equation that incorporates the count of polysyllabic words within a sample text.
The SMOG formula has been widely used in various fields, including education, healthcare, and IR [28]. Its simplicity and ease of application make it a popular choice for estimating readability levels. However, it is important to note that the SMOG formula may have limitations when applied to specific domains, such as technical or scientific texts, as it does not consider the domain-specific terminology and nuances that might impact comprehension [29].
Nonetheless, SMOG stands out as a well-suited formula for healthcare applications. It consistently aligns results with expected comprehension levels, employs validation criteria, and maintains simplicity in its application [28]. These factors make it a reliable choice for assessing the readability of healthcare-related documents. Researchers frequently use the SMOG formula as a reference point when evaluating alternative readability models or proposing new formulas tailored to specific domains.
2.2.2. Dale-Chall Readability Formula
The Dale-Chall Readability Formula is a widely used readability measure that provides an estimate of the comprehension difficulty of a given text. Developed by Edgar Dale and Jeanne Chall in 1948 [30], this formula takes into account both the length of sentences and the familiarity of words to determine the readability level.
The Dale-Chall readability formula calculates its final score by examining the proportion of words in a given text that do not belong to a predefined list of commonly known words. This list comprises 3000 words that are generally understood by fourth-grade students. The formula calculates the readability score by incorporating the average sentence length and the percentage of unfamiliar words.
In summary, the Dale-Chall formula’s notable advantage lies in its focus on word familiarity, enhancing its ability to assess readability, especially for less experienced readers or those with limited vocabulary. This practical and accessible approach considers both word familiarity and sentence length, offering insights into comprehension difficulty for readers of different proficiency levels. However, it is crucial to acknowledge the formula’s limitations and its suitability for specific contexts.
2.3. Readability for Health Consumers
Health literacy and effective communication of health information are crucial in ensuring that patients can access and understand important medical content [31]. With the increasing reliance on online resources for health-related information, it is essential to assess the readability of online materials, particularly those aimed at health consumers.
Several studies have examined the readability and quality of medical content targeting health consumers [32,33,34]. These studies highlight the challenges associated with readability in health-related content, indicating that a significant portion of medical content is difficult for the average layperson to understand. This issue extends to other languages as well, such as German [35].
The findings from these studies underscore the need for greater attention to readability and clear communication in online health resources. Collaborative efforts among healthcare professionals, researchers, and organizations are crucial for enhancing the readability of health materials, including Wikipedia pages and patient education resources. By improving the readability of these resources, we can enhance health literacy and empower patients to make informed decisions about their health [33].
In conclusion, the related work in this subsection underscores the significance of comprehensibility in domain-specific IR. The exploration of computational models, readability formulas, and approaches tailored for specific domains, such as health and medical IR, reflects a growing recognition of the importance of addressing comprehension aspects. The results of experiments conducted in this domain showcase the potential benefits of integrating readability considerations into IR systems. However, further research is needed to generalize these findings to other domains and explore additional factors influencing word-level relatedness, document cohesion, and sentence-level readability computation.
3. Materials and Methods
Our process started with the creation of a dataset. This required extracting articles from PubMed and then having medical experts annotate them. Next, we analyzed the dataset for its general characteristics, level of difficulty, and language variations. Finally, we utilized these data to refine our pretrained BERT model.
3.1. Data Collection
The dataset creation process started with the extraction of 10,000 articles from PubMed, specifically targeting those with available abstracts in both German and English. The data were then stored in a MongoDB and afterwards extended with information about the readability and technicality of those abstracts. This was completed in an annotation process by 216 medical students and practitioners using an annotation tool developed only for this purpose. In this process, a total number of 209 annotated articles could be gathered. An overview of the data acquisition is shown in Figure 1.
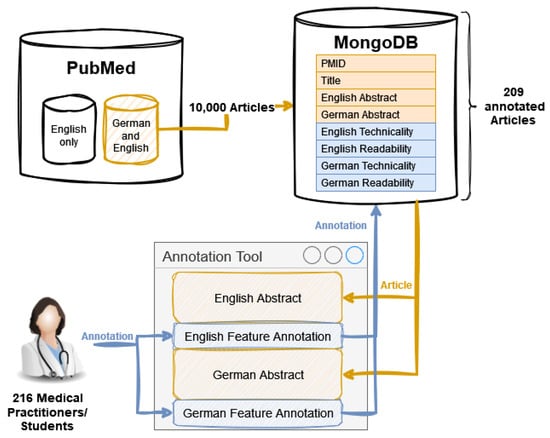
Figure 1.
The dataset creation process includes extracting articles from PubMed and expertly annotating abstracts for technicality and readability assessment.
3.1.1. Documents
We have chosen to focus on research articles’ abstracts as they serve as concise summaries of the main research findings and are widely utilized for initial screening and IR purposes. In light of this, the PubMed database proves to be an ideal resource for our specific use case.
PubMed consists of an extensive collection of scientific literature spanning various medical disciplines, making it a comprehensive repository of valuable medical information. To ensure a manageable dataset, we opted to download a subset of articles from the vast PubMed database. As selection criteria, we chose 10,000 random articles that had abstracts available and were written by the respective authors in both the German and English languages.
3.1.2. Participants
A total of 216 participants were recruited from university hospitals for the annotation process. Those can be divided into four categories, as shown in Table 1: 70 medical students up to the 6th semester (junior students), 59 medical students in the 7th semester or higher (senior students), 59 medical doctors with up to 2 years of experience (junior doctor), and 28 doctors with more than 2 years of experience (senior doctor). All participants either studied at a German university or worked in a German hospital, ensuring they possessed the necessary domain knowledge. Additionally, all participants were asked to assess their German and English language proficiency based on the Common European Framework of Reference (CEFR) (CEFR is an international standard for describing language ability. It describes language ability on a six-point scale, from A1 for beginners, up to C2 for those who have mastered a language), ranging from B1 (Intermediate level) to C2 (Native level). Furthermore, in accordance with institutional regulations and to maintain complete anonymity, no further questions were asked.

Table 1.
Annotation process participants’ distribution among four categories.
3.1.3. Annotation
In the initial phase of the study, we developed a web-based application using Python and MongoDB to facilitate the evaluation process. This application allowed participants to log in anonymously, access clear guidelines, and review the criteria for rating abstracts’ readability and technicality. Participants had no time constraints, providing flexibility in completing the evaluation. Each participant assessed 2–3 different abstracts, rating them for ease of reading and technicality on a scale from 0 to 100 in 5-point increments (0, 5, 10, ⋯, 100). They also identified relevant medical disciplines. To ensure reliability and minimize bias, three independent participants evaluated each abstract, and their scores were averaged to represent text complexity and technicality fairly. Annotations were conducted independently, enhancing the reliability and consistency of ratings.
3.2. Data Analysis
The analysis subsection provides insights into the composition of the dataset and sheds light on the ease of reading and technicality aspects of the abstracts.
3.2.1. Dataset Overview
The annotation process proved to be a resource-intensive task, primarily due to the challenges associated with securing time from busy medical professionals, including both practicing physicians and medical students. With their commitments ranging from extended working hours and on-call duties to direct patient care responsibilities and rigorous exam preparation, allocating time for annotation was constrained. As a result, only 209 abstracts were annotated for technicality and readability aspects. Each annotated abstract included both English and German versions, ensuring comprehensive coverage of the research literature across languages. The dataset encompassed various medical disciplines, creating a representative subset of medical research publications.
3.2.2. Descriptive Statistics
To gain a better understanding of the dataset, we conducted descriptive statistics on the abstracts. The average length of the abstracts was found to be 215 (SD = 90) words for English and 190 (SD = 77) words for German abstracts. It is noteworthy that, while English abstracts have a higher word count, German abstracts tend to be longer in terms of character count. This observation is due to the nature of the German language, where words often contain more characters compared to English [36]. Specifically, the average character count for English abstracts was 1480 (SD = 585), while, for German abstracts, it was 1587 (SD = 616) characters. Additionally, the annotations were accompanied by significant statistical insights. The average annotation time for each document was 176 (SD = 73) seconds, highlighting variability in annotation durations. The annotation process was also conducted at an average rate of 113 (SD = 12) words per minute (WPM), emphasizing diverse annotation speeds, which is consistent with Klatt et al.’s study [37]. Furthermore, the mean intraclass correlation coefficient (ICC) for annotations was found to be 0.81 (SD = 0.08) (according to Koo et al. guideline [38], ICC values below 0.5, between 0.5 and 0.75, between 0.75 and 0.9, and exceeding 0.90 indicate poor, moderate, good, and excellent reliability, respectively), indicating substantial consistency and agreement among the annotations provided by medical professionals.
3.2.3. Ease of Reading Analysis
The ease of reading aspect was assessed by medical professionals, who assigned ease of reading scores to each abstract, where 0 means hard to read and 100 means easy to read. The average ease of reading scores across all abstracts were found to be 64.21 (SD = 21.54) and 61.50 (SD = 21.67) for English and German readability scores, respectively. Figure 2a shows a visual representation of the distribution of ease of reading scores and identifies any potential outliers or patterns.
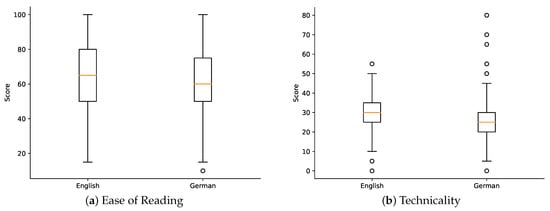
Figure 2.
Aspects of scores’ distributions on English and German abstracts.
3.2.4. Technicality Analysis
The technicality level of the abstracts was evaluated based on the ratings provided by the medical professionals, where 0 means high technicality and 100 low technicality found in the abstract. The average technicality scores for the dataset were 30.55 (SD = 10.02) and 26.72 (SD = 12.81) for English and German readability scores, respectively. Figure 2b shows a visual representation of the distribution of technicality scores and identifies any potential outliers or patterns.
3.2.5. Comparison of German and English Abstracts
For gaining deeper insights into the dataset, we conducted a comparative analysis of the technicality and ease of reading levels between German and English medical abstracts using paired t-tests. The goal was to investigate potential differences in the linguistic characteristics of the two languages and their impact on the ease of reading and technicality of the abstracts.
For the technicality level, our analysis revealed that German medical abstracts tended to exhibit higher levels of technicality compared to their English counterparts. This finding aligns with previous studies [39,40] highlighting the inherent complexity of the German language, particularly in the medical domain. The higher technicality level of German abstracts can be attributed to the frequent usage of specialized medical terminology and the structural intricacies of the German language itself. Figure 3a shows the difference between English and German technicality, where the positive side of the graph shows articles with higher technicality in German, and articles with higher technicality in English on the negative side.
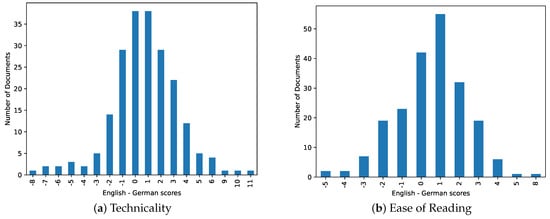
Figure 3.
Difference between English and German scores per article. Scores are between 0 and 20.
In terms of readability, we observed that English medical abstracts were slightly easier to read compared to German abstracts. This disparity can be attributed to several factors. First, the English language generally exhibits a more straightforward and concise writing style, which may enhance readability for a wider audience. Second, English has a larger presence in the global scientific community, leading to greater standardization and familiarity among medical practitioners. Consequently, English abstracts may be tailored to a broader readership, including non-native English speakers. Figure 3b shows the difference between English and German ease of reading, where the positive side of the graph shows articles easier to read in English, and articles are harder to read in German on the negative side.
3.2.6. Limitations
It is important to acknowledge the limitations of the dataset analysis. Out of the 10,000 downloaded abstracts, the sample size of 209 may be inadequate to represent the entire breadth of medical research literature. Moreover, the annotations provided by medical professionals might introduce some level of subjectivity or bias. Nevertheless, we took steps to minimize such limitations by involving multiple annotators per abstract. These limitations must be considered when interpreting the results and generalizing findings from the dataset. Furthermore, we should acknowledge that our analysis focused only on technicality and readability and did not explore other factors that might impact the document’s understandability.
3.3. Model
The model subsection provides insights into the developed models and evaluation matrix.
3.3.1. Our Model
To establish a baseline for our dataset, we employed pretrained BERT models designed for medical text, “PubMedBERT” for English abstracts [41] and “German-MedBERT” for German abstracts [42]. BERT, known for its impressive performance in various Natural Language Processing (NLP) tasks, was a fitting choice for our project.
The fine-tuning process for the pretrained BERT models, specifically “BERT-Readability” for assessing ease of reading and “BERT-Technicality” for evaluating technicality, involved using our dataset, which includes annotated medical abstracts, each assigned scores on a scale from 0 to 100 for ease of reading and technicality. Our main objective was to train these models to predict these scores based on the textual content within the abstracts.
To assess the performance of “BERT-Readability” and “BERT-Technicality,” we employed the root mean square error (RMSE) metric. RMSE measures the average difference between predicted and actual scores, with lower RMSE values indicating better alignment between predictions and actual scores.
By leveraging pretrained BERT models, we established a foundational framework for predicting ease of reading and technicality in medical abstracts. “BERT-Readability” and “BERT-Technicality” serve as baseline models and serve as reference points for future analyses. This enables us to assess the effectiveness of any forthcoming advancements or novel techniques introduced into our work.
3.3.2. Common Readability Formulas
To judge the performance of our models, we specifically selected readability formulas that have been utilized in the domain of medical literature [43,44]. These commonly used readability formulas were tested on the same test set as our models.
Prior to evaluation, we took the necessary steps to ensure compatibility between the outputs of the readability formulas and the ground truth values of our dataset. To achieve this, we employed rescaling/normalization techniques to align the results of each formula with the range and distribution of the dataset’s ground truth scores. This approach allowed us to establish a fair and consistent basis for comparison. Subsequently, we evaluated the performance of each readability formula using the same evaluation metrics employed for our models (“BERT-Readability” and “BERT-Technicality”).
By incorporating a comparison with these readability formulas, we are able to gain a broader perspective on the strengths and limitations of our models. This comparative analysis allows us to assess whether our models outperform or align with the established readability formulas in the specific context of medical documents. The objective is to position the performance of our models within the broader landscape of readability assessment methods utilized in the medical domain.
4. Results
In this study, we can divide our contributions into two parts, which are a dataset and regression models for predicting the ease of reading and technicality of a scientific research abstract.
4.1. Dataset
For this study, we curated a comprehensive dataset of scientific research abstracts written in English and German from various medical disciplines (such as Immunology, Dermatology, Radiology, Emergency medicine, Internal medicine, Neurology, etc.). The dataset comprises 209 abstracts collected from the PubMed database. Each abstract was presented to three medical practitioners (student or doctor). This dataset can be used to improve the readability aspect of any NLP or IR system targeting the medical domain.
4.2. BERT-Readability and BERT-Technicality
We developed BERT-Readability and BERT-Technicality regression models to predict ease of reading and technicality levels in scientific research abstracts on a prediction scale between 0 and 100. These models were trained using the curated dataset, which included annotations for ease of reading and technicality scores for each abstract. We evaluated the performance of BERT-Readability and BERT-Technicality using the RMSE metric. The results demonstrated the models’ effectiveness in predicting ease of reading and technicality scores.
As shown in Table 2, the RMSE for ease of reading is 10.61 for English abstracts and 11.80 for German abstracts. For technicality, the RMSE is 9.42 for English abstracts and 10.07 for German abstracts.

Table 2.
Formulas and our models’ performance on the dataset: RMSE scores comparison.
Furthermore, we conducted a comparative assessment, pitting our models against widely adopted and well-established readability formulas commonly utilized in healthcare literature [45,46], as shown in Table 2. The results consistently demonstrated that our models outperformed these traditional formulas in terms of prediction accuracy. This underscores the critical advantage of employing specialized models tailored explicitly for medical experts.
The existing readability formulas assessed in our comparison rely on a set of general features, such as sentence length, syllable count, and word complexity, which are designed for assessing text readability across various domains. In contrast, our models have been meticulously fine-tuned to account for the specific needs and nuances of medical research abstracts, offering a more precise and effective solution for this specialized context.
Overall, the dataset and models presented in this study offer valuable resources for assessing the ease of reading and technicality of scientific research abstracts in both English and German. These models serve as valuable tools for personalized search engines, whether by enabling the use of filtering-based techniques or contributing to the ranking algorithm used in the retrieval process. This enables medical practitioners to access relevant research findings that align with their language proficiency and expertise.
5. Discussion
The primary objective of this research paper was to tackle the challenges faced by medical practitioners when seeking timely and comprehensible search results within the healthcare domain. We specifically delved into the realms of ease of reading and technicality in medical research articles, with the ultimate aim of enhancing IR for medical professionals, thereby augmenting their decision making capabilities and improving patient care outcomes.
The dataset compiled for this study consists of 209 scientific research abstracts from diverse medical disciplines, available in both English and German. Each abstract underwent annotation by medical practitioners, who assigned ease of reading and technicality scores. The dataset analysis provided valuable insights into the linguistic characteristics of medical abstracts and revealed differences in technicality and ease of reading between English and German abstracts. Notably, our findings revealed that German abstracts tended to be more technically challenging, while English abstracts were slightly easier to read.
Our study introduced two regression models, namely BERT-Readability and BERT- Technicality, which proved highly effective in predicting ease of reading and technicality scores for the abstracts. These models outperformed existing readability formulas commonly employed in the literature, underscoring their significance in predicting domain-specific readability aspects. This highlights the critical importance of employing domain-specific models tailored to scientific research abstracts for precise readability assessment.
The integration of ease of reading and technicality aspects into search engines has practical implications for medical practitioners. Personalized search results, based on language proficiency and expertise, empower medical professionals with efficient and relevant IR tools. This customization enhances the relevance and accessibility of information, optimizing decision making and patient care. Our research paper underscores the significance of tailoring readability assessment to the specific needs of medical practitioners, leading to improved information utilization and overall usability of search engines in the medical domain.
5.1. Future Directions
A promising avenue for future research is to explore more sophisticated models and to utilize advanced transfer learning techniques. These efforts aim to improve the accuracy and applicability of readability assessment in the context of medical research abstracts.
Another compelling area of future study involves investigating the direct influence of readability assessment on the decision making processes and patient care outcomes of medical professionals. Understanding the tangible benefits of improved readability in medical literature can further underscore the importance of our research.
One interesting possibility for future research is the incorporation of text complexity factors into the IR ranking algorithm. Although our study has demonstrated their potential advantages, additional research is required to examine the feasibility of this integration. A future study could focus on implementing text difficulty aspects into IR ranking algorithms to improve the retrieval of relevant medical information for practitioners. The study should address algorithmic refinement, adaptability, and user experience assessment for practical implementation.
5.2. Limitations
One notable limitation of this study concerns the dataset’s sample size. Although the dataset offered useful insights, its limited size may restrict the generalizability of our findings to a wider medical literature context and various medical disciplines.
Our research focused on English and German abstracts, which may not represent the full linguistic diversity of medical literature. Future studies could expand to include a more extensive range of languages to enhance the scope of applicability.
While our regression models demonstrated superior performance, their complexity may pose challenges in real-world implementation. Future research should address ways to streamline these models for practical use.
6. Conclusions
In conclusion, this research paper addresses a crucial gap in the field of healthcare IR and readability assessment. By providing a comprehensive dataset and introducing the integration of ease of reading and technicality aspects into personalized search engines, we have taken significant strides toward enhancing the tools available to medical practitioners. Our work not only offers efficient and reliable IR solutions but also contributes to the broader goal of improving patient care and facilitating informed decision making within the healthcare domain.
The dataset compiled for this study serves as a valuable resource for future research and development in the realm of medical literature analysis. Its provision underscores our commitment to advancing the state of the art in IR and readability assessment.
Author Contributions
Conceptualization, C.L.B., E.M.H., S.F. and N.F.; annotation protocol, C.L.B.; software, S.F., C.L.B. and E.M.H.; validation, S.F. and N.F.; formal analysis, S.F.; supervising annotation process, S.F.; writing—original draft preparation, S.F., C.L.B., E.M.H. and N.F.; writing—review and editing, S.F., C.L.B. and E.M.H.; supervision, N.F.; project administration, N.F.; funding acquisition, S.F. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Ph.D. grants from the DFG Research Training Group 2535 ‘Knowledge- and data-based personalization of medicine at the point of care (WisPerMed)’, University of Duisburg-Essen, Germany. We also acknowledge support by the Open Access Publication Fund of the University of Duisburg-Essen.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Annotation web framework, dataset, annotations, and analysis scripts can be found on the following GitHub repository. https://github.com/samehfrihat/TechnicalityLevelAnnotationTool, accessed on 1 August 2023.
Acknowledgments
We extend our sincere gratitude to Georg Lodde for his invaluable support and insights as a medical practitioner, which greatly enriched the quality of this research project.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| IR | Information Retrieval |
| NLP | Natural Language Processing |
| RMSE | Root Mean Square Error |
| BERT | Bidirectional Encoder Representations from Transformers |
| CEFR | Common European Framework of Reference |
| SD | Standard Deviation |
| ICC | Intraclass Correlation Coefficient |
| WPM | Words Per Minute |
| SMOG | Simple Measure Of Gobbledygook |
References
- Entin, E.B.; Klare, G.R. Relationships of Measures of Interest, Prior Knowledge, and Readability to Comprehension of Expository Passages. Adv. Read./Lang. Res. 1985, 3, 9–38. [Google Scholar]
- Vydiswaran, V.V.; Mei, Q.; Hanauer, D.A.; Zheng, K. Mining consumer health vocabulary from community-generated text. In Proceedings of the AMIA Annual Symposium Proceedings, American Medical Informatics Association, San Diego, CA, USA, 30 October–3 November 2014; Volume 2014, p. 1150. [Google Scholar]
- Chall, J. Readability: An Appraisal of Research and Application; Bureau of Educational Research Monographs: Columbus, OH, USA, 1958. [Google Scholar]
- Hätty, A.; Schlechtweg, D.; Dorna, M.; im Walde, S.S. Predicting degrees of technicality in automatic terminology extraction. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, London, UK, 5–10 July 2020; pp. 2883–2889. [Google Scholar]
- Hedman, A.S. Using the SMOG formula to revise a health-related document. Am. J. Health Educ. 2008, 39, 61–64. [Google Scholar] [CrossRef]
- Liu, Y.; Ji, M.; Lin, S.S.; Zhao, M.; Lyv, Z. Combining readability formulas and machine learning for reader-oriented evaluation of online health resources. IEEE Access 2021, 9, 67610–67619. [Google Scholar] [CrossRef]
- Goeuriot, L.; Jones, G.J.; Kelly, L.; Leveling, J.; Hanbury, A.; Müller, H.; Salanterä, S.; Suominen, H.; Zuccon, G. ShARe/CLEF eHealth Evaluation Lab 2013, Task 3: Information retrieval to address patients’ questions when reading clinical reports. CLEF Online Work. Notes 2013, 4, 191–201. [Google Scholar]
- O’Sullivan, L.; Sukumar, P.; Crowley, R.; McAuliffe, E.; Doran, P. Readability and understandability of clinical research patient information leaflets and consent forms in Ireland and the UK: A retrospective quantitative analysis. BMJ Open 2020, 10, e037994. [Google Scholar] [CrossRef]
- Veltri, L.W.; Milton, D.R.; Delgado, R.; Shah, N.; Patel, K.; Nieto, Y.; Kebriaei, P.; Popat, U.R.; Parmar, S.; Oran, B.; et al. Outcome of autologous hematopoietic stem cell transplantation in refractory multiple myeloma. Cancer 2017, 123, 3568–3575. [Google Scholar] [CrossRef]
- Wynn, T.A.; Ramalingam, T.R. Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat. Med. 2012, 18, 1028–1040. [Google Scholar] [CrossRef]
- Ott, N.; Meurers, D. Information retrieval for education: Making search engines language aware. Themes Sci. Technol. Educ. 2011, 3, 9–30. [Google Scholar]
- Tomažič, T.; Čelofiga, A.K. The Role of Different Behavioral and Psychosocial Factors in the Context of Pharmaceutical Cognitive Enhancers’ Misuse. Healthcare 2022, 10, 972. [Google Scholar] [CrossRef]
- Frihat, S. Context-sensitive, personalized search at the Point of Care. In Proceedings of the 22nd ACM/IEEE Joint Conference on Digital Libraries, Cologne, Germany, 20–24 June 2022; pp. 1–2. [Google Scholar]
- Basch, C.H.; Fera, J.; Garcia, P. Readability of influenza information online: Implications for consumer health. Am. J. Infect. Control 2019, 47, 1298–1301. [Google Scholar] [CrossRef]
- Klare, G.R. The formative years. In Readability: Its Past, Present, and Future; International Reading Association: Newark, DE, USA, 1988; pp. 14–34. [Google Scholar]
- Yan, X.; Song, D.; Li, X. Concept-based document readability in domain specific information retrieval. In Proceedings of the 15th ACM International Conference on Information and Knowledge Management, Arlington, VA, USA, 13–16 August 2006; pp. 540–549. [Google Scholar]
- Ceri, S.; Bozzon, A.; Brambilla, M.; Della Valle, E.; Fraternali, P.; Quarteroni, S.; Ceri, S.; Bozzon, A.; Brambilla, M.; Della Valle, E.; et al. An introduction to information retrieval. Web Inf. Retr. 2013, 3, 3–11. [Google Scholar]
- Selvaraj, P.; Burugari, V.K.; Sumathi, D.; Nayak, R.K.; Tripathy, R. Ontology based recommendation system for domain specific seekers. In Proceedings of the 2019 Third International Conference on I-SMAC (IoT in Social, Mobile, Analytics and Cloud) (I-SMAC), Palladam, India, 12–14 December 2019; pp. 341–345. [Google Scholar]
- Jameel, S.; Qian, X. An unsupervised technical readability ranking model by building a conceptual terrain in LSI. In Proceedings of the 2012 Eighth International Conference on Semantics, Knowledge and Grids, Beijing, China, 22–24 October 2012; pp. 39–46. [Google Scholar]
- Palotti, J.; Goeuriot, L.; Zuccon, G.; Hanbury, A. Ranking health web pages with relevance and understandability. In Proceedings of the 39th International ACM SIGIR Conference on Research and Development in Information Retrieval, Pisa, Italy, 17–21 July 2016; pp. 965–968. [Google Scholar]
- van der Sluis, F.; van den Broek, E.L. Using complexity measures in information retrieval. In Proceedings of the Third Symposium on Information Interaction in Context, New Brunswick, NJ, USA, 18–21 August 2010; pp. 383–388. [Google Scholar]
- Kane, L.; Carthy, J.; Dunnion, J. Readability applied to information retrieval. In Proceedings of the European Conference on Information Retrieval, London, UK, 4–10 April 2006; Springer: Berlin/Heidelberg, Germany, 2006; pp. 523–526. [Google Scholar]
- Taranova, A.; Braschler, M. Textual complexity as an indicator of document relevance. In Proceedings of the Advances in Information Retrieval: 43rd European Conference on IR Research, ECIR 2021, Virtual Event, 28 March–1 April 2021; Springer: Berlin/Heidelberg, Germany, 2021; Volume 43, Part II. pp. 410–417. [Google Scholar]
- Lopes, C.T. Health Information Retrieval—State of the art report. arXiv 2022, arXiv:2205.09083. [Google Scholar]
- Fung, A.C.H.; Lee, M.H.L.; Leung, L.; Chan, I.H.Y.; Kenneth, W. Internet Health Resources on Nocturnal Enuresis—A Readability, Quality and Accuracy Analysis. Eur. J. Pediatr. Surg. 2023. [Google Scholar] [CrossRef] [PubMed]
- DuBay, W.H. The Principles of Readability; Online Submission; Impact Information: Costa Mesa, CA, USA, 2004. [Google Scholar]
- Mc Laughlin, G.H. SMOG grading-a new readability formula. J. Read. 1969, 12, 639–646. [Google Scholar]
- Wang, L.W.; Miller, M.J.; Schmitt, M.R.; Wen, F.K. Assessing readability formula differences with written health information materials: Application, results, and recommendations. Res. Soc. Adm. Pharm. 2013, 9, 503–516. [Google Scholar] [CrossRef]
- Willis, L.; Gosain, A. Readability of patient and family education materials on pediatric surgical association websites. Pediatr. Surg. Int. 2023, 39, 156. [Google Scholar] [CrossRef]
- Dale, E.; Chall, J.S. A formula for predicting readability: Instructions. Educ. Res. Bull. 1948, 5, 37–54. [Google Scholar]
- Basch, C.H.; Mohlman, J.; Hillyer, G.C.; Garcia, P. Public health communication in time of crisis: Readability of on-line COVID-19 information. Disaster Med. Public Health Prep. 2020, 14, 635–637. [Google Scholar] [CrossRef]
- Diviani, N.; van den Putte, B.; Giani, S.; van Weert, J.C. Low health literacy and evaluation of online health information: A systematic review of the literature. J. Med. Internet Res. 2015, 17, e112. [Google Scholar] [CrossRef]
- Modiri, O.; Guha, D.; Alotaibi, N.M.; Ibrahim, G.M.; Lipsman, N.; Fallah, A. Readability and quality of wikipedia pages on neurosurgical topics. Clin. Neurol. Neurosurg. 2018, 166, 66–70. [Google Scholar] [CrossRef]
- Tan, S.S.L.; Goonawardene, N. Internet health information seeking and the patient-physician relationship: A systematic review. J. Med. Internet Res. 2017, 19, e9. [Google Scholar] [CrossRef] [PubMed]
- Zowalla, R.; Pfeifer, D.; Wetter, T. Readability and topics of the German Health Web: Exploratory study and text analysis. PLoS ONE 2023, 18, e0281582. [Google Scholar] [CrossRef] [PubMed]
- Behrens, H. How Difficult are Complex Verbs? Evidence from German, Dutch and English. Linguistics 1998, 36, 679–712. [Google Scholar] [CrossRef]
- Klatt, E.C.; Klatt, C.A. How much is too much reading for medical students? Assigned reading and reading rates at one medical school. Acad. Med. 2011, 86, 1079–1083. [Google Scholar] [CrossRef] [PubMed]
- Koo, T.K.; Li, M.Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef]
- Hockett, C.F. A Course in Modern Linguistics; The Macmillan Company: New York, NY, USA, 1958. [Google Scholar]
- Grimm, A.; Hübner, J. Nonword repetition by bilingual learners of German: The role of language-specific complexity. Biling. Specif. Lang. Impair. Bi-SLI 2017, 201, 288. [Google Scholar]
- Gu, Y.; Tinn, R.; Cheng, H.; Lucas, M.; Usuyama, N.; Liu, X.; Naumann, T.; Gao, J.; Poon, H. Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing. arXiv 2020, arXiv:2007.15779. [Google Scholar] [CrossRef]
- Deepset-AI. State-of-the-Art German BERT Model Trained from Scratch. Available online: https://www.deepset.ai/german-bert (accessed on 1 August 2023).
- Worrall, A.P.; Connolly, M.J.; O’Neill, A.; O’Doherty, M.; Thornton, K.P.; McNally, C.; McConkey, S.J.; De Barra, E. Readability of online COVID-19 health information: A comparison between four English speaking countries. BMC Public Health 2020, 20, 100231. [Google Scholar] [CrossRef]
- Fajardo, M.A.; Weir, K.R.; Bonner, C.; Gnjidic, D.; Jansen, J. Availability and readability of patient education materials for deprescribing: An environmental scan. Br. J. Clin. Pharmacol. 2019, 85, 1396–1406. [Google Scholar] [CrossRef]
- Powell, L.; Krivanek, T.; Deshpande, S.; Landis, G. Assessing Readability of FDA-Required Labeling for Breast Implants. Aesthetic Surg. J. Open Forum 2023, 5, ojad027-009. [Google Scholar] [CrossRef]
- Szmuda, T.; Özdemir, C.; Ali, S.; Singh, A.; Syed, M.T.; Słoniewski, P. Readability of online patient education material for the novel coronavirus disease (COVID-19): A cross-sectional health literacy study. Public Health 2020, 185, 21–25. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).