Abstract
The incidence of melanoma cases continues to rise, underscoring the critical need for early detection and treatment. Recent studies highlight the significance of deep learning in melanoma detection, leading to improved accuracy. The field of computer-assisted detection is extensively explored along all lines, especially in the medical industry, as the benefit in this field is to save hu-man lives. In this domain, this direction must be maximally exploited and introduced into routine controls to improve patient prognosis, disease prevention, reduce treatment costs, improve population management, and improve patient empowerment. All these new aspects were taken into consideration to implement an EHR system with an automated melanoma detection system. The first step, as presented in this paper, is to build a system based on the fusion of decisions from multiple neural networks, such as DarkNet-53, DenseNet-201, GoogLeNet, Inception-V3, InceptionResNet-V2, ResNet-50, ResNet-101, and compare this classifier with four other applications: Google Teachable Machine, Microsoft Azure Machine Learning, Google Vertex AI, and SalesForce Einstein Vision based on the F1 score for further integration into an EHR platform. We trained all models on two databases, ISIC 2020 and DermIS, to also test their adaptability to a wide range of images. Comparisons with state-of-the-art research and existing applications confirm the promising performance of the proposed system.
1. Introduction
Melanoma is a form of tumor that emerges when melanocytes experience a malignant alteration. This sort of cancer initiates in the skin, yet as the tumor advances, it can extend across the body as cancerous metastases [1]. One of the key factors that add to the formation of melanoma is exposure to ultraviolet (UV) radiation, whether from natural sunlight or artificial sources. This radiation can induce skin burns that result in structural changes, potentially leading to scars that could eventually evolve into melanomas. Furthermore, UV radiation can impact the skin’s healing process at a genetic level, causing modifications to the DNA. This, in time, can trigger mutations in normal cells, causing them to become cancerous. Genetic predisposition also plays a vital part, with a history of cancer in the family indicating susceptibility to UV radiation, especially in individuals with characteristics such as fair skin, blue eyes, or a higher number of skin features, such as moles, freckles, or patches. Another contributing factor is the presence of benign growths with an elevated concentration of melanocytes and melanin, presenting as dark areas or moles [2,3]. Nevertheless, the most prevalent cause of melanoma is, as previously mentioned, exposure to UV radiation, specifically from sunlight or artificial tanning devices that emit such radiation.
Prolonged or brief yet intense exposure to this form of radiation can lead to mutations within melanocytes, which contributes further to the formation of melanoma. In approximately 25% of instances, melanocytes arise from the transformation of moles, exhibiting characteristics, such as irregular shapes, alterations in color, significant sizes, compromised skin, or itching [4,5]. In more advanced cases, there might even be instances of bleeding or inflammation. Melanomas can manifest in areas of the skin devoid of visible indications, on recently formed moles, or other skin marks. While certain moles can surpass a size of 5 mm, certain melanomas can be smaller than 5 mm, and their size does not necessarily indicate malignancy. Interestingly, these melanomas can arise even in regions that have not been exposed to UV radiation [6]. Melanoma ranks among the leading five causes of death among men and the top seven among women. It develops either from pre-existing lesions that undergo mutations or can originate in healthy skin [7]. According to [8], estimates for the year 2023 in the United States indicate around 97,610 new cases of melanoma, with 58,120 occurring in males and 39,490 in females. Additionally, there are projections of approximately 7990 deaths attributed to melanoma, comprising 5420 in males and 2570 in females [9].
Melanoma undergoes two distinct growth phases: a vertical phase and a radial phase. During the vertical growth phase, cancer cells penetrate deeply into the epidermis and, with time, progress through vertical expansion. At this stage, the cells infiltrate the dermis and give rise to metastasis, spreading throughout the body. Lesions resulting from melanoma are categorized based on their depth: thin (up to one millimeter thick), moderate (between 1–4 mm thick), and thick (exceeding 4 mm in thickness) [9,10]. In the study outlined in [11], it was reported that in 2020, skin lesions in the form of melanoma accounted for 1.3% of all cancer-related deaths in European Union member states. Additionally, this type of cancer constituted 4% of all newly identified cancer cases. These statistics consequently positioned melanoma as the 15th most prevalent cause of cancer-related fatalities within European Union member states [11].
On a global scale, approximately 57,000 deaths were recorded in 2020, reflecting a mortality rate of 0.4 per 100,000 for women and 0.7 per 100,000 for men [12]. Presently, the evaluation of such tumors involves a visual examination conducted by a specialized medical professional utilizing a dermatoscope. The diagnostic process takes into account factors such as asymmetry, irregular edges, color variations, size, and thickness. Additionally, blood tests may be conducted to determine the disease’s stage. Biopsy results can also provide insights into potential resection margins [13]. However, this type of examination is time-consuming and susceptible to human errors.
The survival rate for this form of cancer is heavily dependent on its stage. In stage one, where cancer is immediately detected, the survival rate stands at 90%. In stage two, this rate drops to approximately 45%, and in stage three, it further declines to about 27% [14]. Consequently, delayed detection significantly increases the risk of fatality. The primary solution in such cases is early detection and analysis of skin damage. Currently, this necessitates a visit to a medical professional and involves a battery of tests for accurate diagnosis. These tests often consume a considerable amount of time and may be invasive, causing inconvenience and anxiety for the patient. Thus, the development of a computer-assisted automatic system for detecting skin lesions with high accuracy becomes imperative. In recent years, researchers have dedicated considerable effort to examining this topic from both clinical and technical perspectives, seeking an optimal approach to skin lesion detection. From a technical standpoint, various methods have been explored, with Deep Learning (DL) techniques emerging as the most effective. In the following sections, we will present the present state of research by delving into some studies conducted between 2020 and 2023 that utilized DL techniques to create systems capable of achieving high-performance automatic detection of skin diseases.
Within the scope of the OPTIM Project (https://optim.upb.ro, accessed on 2 November 2022), our central aim was to create an Electronic Health Record (EHR) information system seamlessly integrating advanced artificial intelligence (AI)-powered classification solutions for medical images. Moreover, the project sought to introduce novel applications and functionalities tailored to the intricate workflow demands of medical institutions. This initiative was born out of the necessity to establish an information system that houses all the essential modules essential for patient diagnosis and streamlined communication. Given the modern emphasis on efficiency, consolidating interactions within a single platform proves to be advantageous, especially considering the challenges and stress that ailing patients experience while navigating multiple avenues for diagnostic purposes. In essence, this research paper unveils a systematic, criterion-based approach to decision fusion from multiple networks for melanoma detection. This work distinguishes itself through the utilization of F1-score-based weighting, adaptability to dual databases, and a comprehensive empirical assessment, thereby solidifying its superiority over existing applications in the field. In this study, we introduce a multi-network system rooted in decision fusion for melanoma detection, achieved by combining various high-performance Convolutional Neural Networks (CNNs). Subsequently, we juxtapose this system against several web-based tools designed for constructing machine learning models, assessing their compatibility for integration into alternative platforms.
Our primary endeavor centers on the establishment of an EHR system intertwined with an automated melanoma detection mechanism. This involves the creation of a bespoke decision fusion classifier that seamlessly incorporates an array of neural networks, including DarkNet-53, DenseNet-201, GoogLeNet, Inception-V3, InceptionResNet-V2, ResNet-50, and ResNet-101. The core of our research focuses on implementing an EHR system fortified with an automated melanoma detection system. This innovation harmonizes cutting-edge advancements in machine learning, neural networks, and medical diagnostics to address a critical healthcare challenge. The process entails intricately merging a diverse range of neural networks within a tailored decision fusion classifier. The significance of our research emanates from the urgency to enhance melanoma detection’s efficiency and accuracy within the framework of an EHR system. Timely and accurate melanoma detection can profoundly impact patient outcomes, thereby enhancing healthcare interventions. By automating the detection process, we empower medical professionals with a potent tool for swift diagnosis and informed decision-making, ultimately elevating patient care.
Furthermore, our research stands out for its comprehensive approach. It extends beyond crafting the custom decision fusion classifier to encompass a comprehensive comparison with four prominent applications—Google Teachable Machine, Microsoft Azure Machine Learning, Google Vertex AI, and Salesforce Einstein Vision. This comparative analysis serves as a benchmark, affirming the effectiveness and superiority of our approach in automating melanoma detection. In summary, our research introduction shines a light on pivotal components: our ambition to integrate automated melanoma detection within an EHR system, the development of a tailored decision fusion classifier utilizing diverse neural networks, and the profound implications of enhancing medical diagnostics. This is validated through a comprehensive comparison that underscores the efficacy of our system when measured against established applications.
All the abbreviations in this study can be found in Abbreviation.
2. Related Work
In reference to [15], a CNN classifier was detailed and optimized using metaheuristic techniques for pre-trained network models. The optimization of biases and weights within the CNN models was executed through the implementation of a whale optimization algorithm. Experimental results attained from the application of these algorithms to the Dermquest and DermIS databases showcased superior performance metrics compared to other Deep Learning (DL) techniques. Notably, the achieved metrics included a Sensitivity of 0.95, Specificity of 0.92, and Accuracy of 0.91. Similarly, in the year 2020, Ref. [16] proposed a system integrating an AlexNet CNN designed to detect four distinct classes: Urticaria, Acne, Eczema herpeticum, and Keratosis. The system’s training on the DermNet database, followed by testing on 69 images, demonstrated a remarkable accuracy ranging from 98.6% to 99.04% [16].
Another approach surfaced in the work by [17], which introduced an automatic system that employed hyperspectral data and GoogLeNet CNN. This method entailed utilizing a hyperspectral imager to gather position and wavelength information, and GoogLeNet was employed for the classification of skin lesions into two classes. To accommodate the network’s three input channels, a “Mini Network” layer was added to reduce channel dimensions from 84 to 3. Subsequent evaluation of the system after data augmentation yielded a specificity of 81.2%, accuracy of 77.2%, and sensitivity of 72.3% [17].
For instance, in [16], a CNN comprising three hidden layers with 64, 32, and 16 channels for each layer was proposed. Various optimizers, including Nadam, RMSprop, Adam, and SGD Optimizers, were tested using a learning rate of 0.001. The Adam optimizer emerged as the most effective for skin lesion detection. This system successfully differentiated four distinct classes: melanoma, dermatofibroma, squamous cell carcinoma, and nevus pigments. It achieved an accuracy of 99% and an F1 score close to unity when applied to the ISIC database [18].
In another study, Ref. [19] employed a ResNet-50 CNN trained on 1072 acral benign nevi images from a Korean hospital. The system aimed to detect acral lentiginous melanoma. A comparative analysis was performed across three survey types: dermoscopic images analyzed solely by 60 physicians, dermoscopic images combined with clinical information, and all previous information supplemented by the CNN’s predictions. The conclusion drawn indicated that detection performance increased progressively across the stages, culminating in an accuracy of 86.9% [19].
Conducting a comprehensive overview in [20], researchers analyzed 5112 studies on melanoma detection using DL techniques. Out of these, 55 papers were deemed reputable and selected for in-depth analysis. The study offered insights into recent trends, solutions, opportunities, and challenges concerning melanoma detection via DL techniques. The authors also proposed a taxonomy to summarize various detection solutions [20].
Furthermore, a study by [21] delved into CNN models for melanoma detection. The study employed TensorFlow and Keras to build the model, involving varying layers of different network architectures. The system underwent data augmentation, normalization, and transfer learning using CNNs such as InceptionResNet-V2, VGG16, MobileNet, Inception-V3, and ResNet-50. Results demonstrated that InceptionResNet achieved the highest performance, boasting an accuracy, recall, precision, and F1 score of 91%. The system successfully differentiated seven categories of skin lesions [21].
The evaluation of an approved system in the European market for certain types of skin lesions was conducted by [22], revealing favorable diagnostic support from the CNN. However, performance limitations were noted for specific lesion types, such as subungual or mucosal lesions. The study used six dermoscopic image sets, including 100 benign lesions and 30 melanoma images each. High specificities and sensitivities were achieved for nodular and superficial melanomas, while subungual and mucosa melanoma detection presented lower performance [22]. Transitioning to 2021, Ref. [23] introduced a system incorporating a MobileNet pre-trained CNN for feature extraction. Support Vector Machine (SVM) and linear discriminant analysis (LDA) were utilized for classifying skin lesions into melanoma and non-melanoma classes. By applying cropping and boundary localization to PH2 database images, the proposed system achieved an exceptional specificity of 97.91% and an accuracy of 95% [23].
In a similar vein, Ref. [24] proposed two systems involving a CNN model and a combination of CNN and Neural Network NN. The combined model exhibited higher accuracy (92.34%) compared to the standalone CNN model (73.69%). Performance assessments were conducted on 1200 images from the ISIC database [24]. Additionally, Ref. [25] assessed the performance of their CNN on the HAM10000 database. The system, incorporating 45 layers and utilizing InceptionResNet-V2, achieved an impressive accuracy of 99.69%, with low false negatives. The system accurately classified images into nevi, non-nevi, and melanoma categories [25].
In another study, Ref. [26] focused on pre-processing approaches to enhance a CNN-based skin lesion detection system. Region of interest (ROI) extraction was accomplished using a proposed algorithm, leading to improved CNN model training and evaluation efficiency. InceptionResNet-V2’s performance notably improved by 2.18% when trained on ROI images. ISIC-2019 served as the database for this study [26]. In a study by [27], the author employed the EfficientNet-B6 CNN to capture fine-grained features, yielding competitive results with an AUC-ROC score of 0.917 when applied to the ISIC 2020 database [27].
As research extended into 2022, Ref. [28] employed Deep Learning to accurately extract skin lesions. A process involving Super-Resolution Generative Adversarial Networks (ESRGAN) improved image quality, followed by the extraction of Regions of Interest (ROI). Employing a CNN and a modified version of ResNet-50, the proposed method displayed high potential with metrics such as F-score, recall, accuracy, and precision, reaching 0.86, 0.86, 0.86, and 0.84, respectively [28].
Alternatively, Ref. [29] introduced a novel approach considering a third dimension, namely the depth of the skin lesion. This dimension, captured from light-field images, was combined with two-dimensional (2D) information using Morlet scattering transform and CNN. The combined three-dimensional (3D)–two-dimensional approach yielded the highest accuracy of 94%, while 2D-only and 3D-only dimensions achieved accuracies of 84% and 74%, respectively [29].
A novel Fuzzy GrabCut-stacked convolutional neural network GC-SCNN-based model was proposed by [30] for skin lesion detection. Utilizing PH2 and HAM10000 databases, fuzzy logic enabled boundary detection, and stacked CNNs (GC-SCNN) facilitated feature extraction. Segmentation-enhanced SVMs were then used for segmentation and classification, achieving high metrics, including 100% specificity, sensitivity, and 99.75% accuracy [30].
Continuing into [31], the ISIC 2018 database was leveraged to train and test the proposed method. Pre-processing, subsampling, and down-sampling steps were conducted to balance data, and VGG16 and ResNet-50 CNNs were used to detect melanoma. The system demonstrated a dice coefficient of 97.4%, accuracy of 92.3%, recall of 90%, Jaccard index of 99.8%, and precision of 93.3% [31]. By 2023, Ref. [32] proposed a CNN strategy for early melanoma detection involving five convolution layers, five pooling layers, a fully connected layer, an input layer, and an output layer. Employing 10,000 and 1000 images for training and testing, respectively, the system achieved an accuracy of 91% [33].
Lastly, in [33], two decision fusion methodologies based on CNN models were introduced, involving the extraction of Regions of Interest (ROI) using the Geometric Active Contour (GAC) algorithm. Extracted features were then employed for classification through Random Forest (RF) and NN. The second method included the fusion of features from various CNN models, yielding an accuracy of 96.1%, an ACC of 94.41%, a precision of 88.69%, a sensitivity of 88.9%, and a specificity of 99.44% [33].
In [34], a model is presented that utilizes level-set segmentation for effectively segmenting images with non-uniform properties. This model, featured in the current research, boasts exceptional attributes, including high accuracy, speed, rapid convergence, resilience to varying starting contours, and resistance to noise interference. The proposed approach delivers results characterized by a true detection rate of 94%, a false negative rate of 3.62%, and an error rate of 3.3%. The testing outcomes substantiate the assertion that the recommended method for lesion segmentation is robust and remains unaffected by factors such as hair, blood vessels, changes in brightness, and variations in contrast.
In [35], the authors explore the application of various machine-learning techniques to construct a high-performance ensemble classifier for six distinct skin lesions. Specifically, the researchers utilize Adaboost, voted ensemble, random forest, boosted Gaussian Mixture Model (GMM), voted Convolutional Neural Network (CNN), and boosted Support Vector Machine (SVM). The authors recognize an issue of imbalanced classification across all classes, prompting them to adopt ensemble transfer learning and transfer learning techniques. This strategic choice effectively mitigates the challenge posed by class imbalance.
The study encompasses a total of 19 classifiers, which include 5 ensemble transfer learning models, 6 ensemble models, 4 Machine Learning (ML) and Deep Learning (DL) models, and four transfer learning models. Performance metrics are thoroughly assessed to gauge the effectiveness of each classifier. In the paper’s conclusions, the authors note that the Ensemble voted, GMM, and random forest classifiers exhibit comparatively lower performance than the Adaboost and boosted SVM classifiers. Notably, the ensemble CNN achieves an impressive accuracy rate of 98.67% [35]. In conclusion, the field of melanoma detection through CNNs and DL techniques has seen significant exploration over the years, with various methodologies offering promising results. However, the challenge remains open due to the need for systems trained on sufficient images that ensure high accuracy and minimal chance of misclassification.
These studies explore various methodologies, architectures, and databases for the purpose of identifying skin lesions, particularly melanoma. Each study tackles the challenge of accurate detection from different angles, often achieving promising results. However, despite the multitude of efforts, the researchers point out that achieving high accuracy and minimizing false detections remains an ongoing challenge due to factors such as dataset size and complexity. In contrast, our paper takes a distinct approach by focusing on enhancing the performance of melanoma detection through a multi-network system utilizing decision fusion. We consider several high-performance CNN architectures, including DarkNet-53, DenseNet-201, GoogLeNet, Inception-V3, InceptionResNet-V2, ResNet-50, ResNet-101, and Xception. The selection of these networks is guided by specific criteria outlined in the methodology section, indicating a methodical approach to network selection.
The key innovation lies in the establishment of decision fusion weights based on F1-scores from each CNN combined with the utilization of specific criteria to select the CNN demonstrating the highest performance based on state-of-the-art analysis. By leveraging these scores, we optimize the performance of our proposed system. We test the system’s effectiveness on two distinct databases, ISIC 2020 and DermIS, highlighting its adaptability to varying datasets. Our approach involves training, validation, and testing on 700, 300, and 200 images, respectively, showcasing rigorous empirical evaluation. Moreover, we conduct a comprehensive comparison of our system’s performance against four other existing applications that utilize CNN combinations, such as Google Teachable Machine, Microsoft Azure Machine Learning (ML), Google Vertex AI, and Salesforce Einstein Vision. This comparison underscores our proposed system’s superior performance.
The structure of this study is as follows: Section 3 provides insights into the utilized databases and outlines the various CNN architectures applied for skin lesion detection. The methodology employed for validating system performance is also detailed. Section 4 delves into the experimental results. Section 5 engages in a discussion of the proposed methodologies and includes a comparison with existing systems. Finally, Section 6 presents the conclusion of this research.
3. Materials and Methods
To begin, we provide an overview of the databases employed in this study. Subsequently, we delve into the methodologies employed for skin lesion detection. From a hardware perspective, all the software architecture we proposed was executed on a computing device equipped with 16 GB RAM and a 4.5 GHz Core i7 Intel processor. The architecture for our study flow is presented in Figure 1.
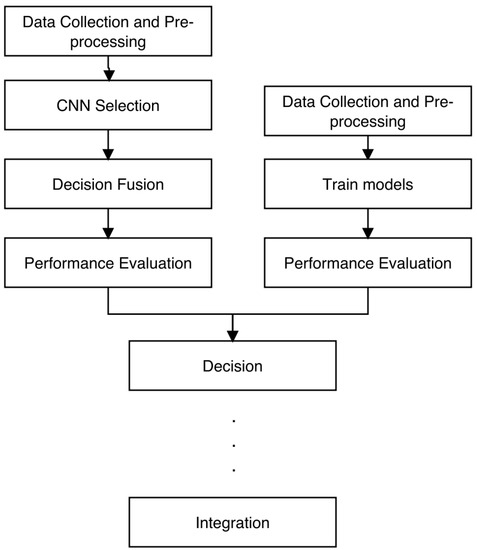
Figure 1.
Architecture for the proposed method for the study.
3.1. Databases and Image Pre-Processing
Images from two different dermoscopic image databases, namely, ISIC 2020 and DermIS, were used to test and train the proposed architectures.
3.1.1. ISIC Database
As stated in reference [36], the complete ISIC database comprises a total of 93,083 images, with 71,372 designated for training purposes and 21,711 earmarked for testing, constituting the most extensive publicly available collection of dermoscopic skin lesion images [35]. In our previous study conducted in 2022 [37], we demonstrated that ISIC is the preferred database among researchers for developing high-performance melanoma detection systems. This is due to the meticulous expert labeling of images in the field and the annual challenge that offers monetary rewards.
One of ISIC’s primary objectives is to foster the discovery of optimal computational solutions for skin lesion detection, achieved by offering training and testing databases. The repository primarily encompasses several smaller databases, such as MSK, HAM10000, and BCN2000 [37,38]. As previously mentioned, this current paper employs training and testing data drawn from the ISIC 2020 database. This repository contains 33,126 images of skin lesions designated for system training and 10,982 images intended for system testing. These images have been sourced from various institutions situated in diverse global regions and involve over 2000 patients [36,39,40].
For our study, we selected a subset of 1000 images from the ISIC 2020 database, out of which 700 images were utilized for training the proposed architectures, while the remaining 200 images were used for testing purposes. Examples of images from the ISIC 2020 Database are presented in Figure 2 and Figure 3.
Figure 2.
Non-Melanoma images extracted from the ISIC 2020 database.
Figure 3.
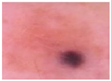
Melanoma images extracted from the ISIC 2020 database.
3.1.2. DermIS Database
The DermIS database stands as the most comprehensive repository of dermatological information available on the internet. It encompasses a wide array of images depicting nearly all categories of skin conditions and diagnoses, supplemented with differential diagnoses, case reports, and additional pertinent data such as medical journals. This collaborative project is conducted in conjunction with the Department of Socio-Clinical Medicine at the University of Heidelberg and the Department of Dermatology at the University of Erlangen. Within this database, there are 500 images portraying melanoma cases and another 500 images depicting healthy skin conditions [41,42].
Much like our utilization of the ISIC database, this paper similarly leverages the entire collection of 1000 images within the DermIS database. Out of these images, 700 were allocated for training purposes, while the remaining 200 were dedicated to testing the proposed algorithms. Examples of images drawn from the DermIS Database are showcased in Figure 4 and Figure 5.
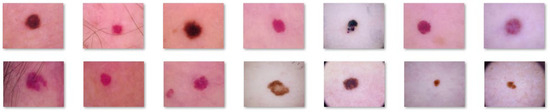
Figure 4.
Non-melanoma images extracted from the DermIS database.
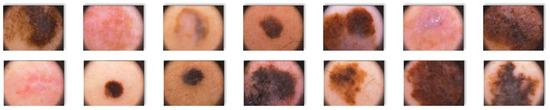
Figure 5.
Melanoma images extracted from the ISIC database.
As commonly understood, the development of a highly effective medical diagnosis system for image classification necessitates the utilization of high-quality input images. Given the absence of suitable databases, image pre-processing becomes an imperative step. Enhancing image quality is pivotal, as it facilitates subsequent processing stages. Initially, this entails adjustments to brightness and contrast, often followed by converting color images to grayscale. The selection of specific image processing techniques hinges on the nature of the classification task at hand. These techniques encompass noise reduction, image resizing, color correction, segmentation, and feature extraction [43,44,45]. In this study, we employ pre-processing techniques that involve adjusting image dimensions through resizing and eliminating hair artifacts. The resizing is tailored to the specific requirements of each CNN utilized. For instance, in the case of the GoogLeNet architecture, images were resized to dimensions of 256 × 256 × 3 pixels, and the aspect ratio was 1:1 (this is the case with all the images we pre-processed). Concerning skin lesion images, certain elements can be regarded as image artifacts, as they impede the computer-aided diagnosis process. Notably, some images suffer from blurriness due to either human movements (by the operator or patient) or the presence of hair or water droplets. Additionally, annotations, such as measuring scales, might be present in certain images, potentially misleading the system.
Our research is dedicated to propelling the field of dermatology and the precise classification and diagnosis of skin conditions through meticulous analysis of skin lesions. In pursuit of this goal, we have deliberately chosen to incorporate the ISIC 2020 and DermIS datasets into our study. The selection of the ISIC 2020 and DermIS datasets for our research is substantiated by a range of factors. Firstly, these datasets hold considerable prominence and are widely employed in the realm of dermatology and the analysis of skin images. They have undergone exhaustive curation and validation by domain experts, instilling confidence in the quality and dependability of the data. Furthermore, the ISIC 2020 dataset centers specifically on images of skin lesions, presenting an extensive compilation of diverse cases encompassing melanoma, nevi, and various other prevalent skin conditions. This dataset showcases a diverse array of images, facilitating the training and assessment of our models across a broad spectrum of skin ailments, thereby amplifying the applicability of our findings. In tandem, the DermIS dataset supplements the ISIC 2020 dataset by furnishing an extensive trove of dermatological images, spanning both clinical and dermoscopic views. This dataset bestows invaluable insights into the visual attributes and patterns of diverse skin disorders, allowing us to delve into and dissect multifaceted aspects of dermatological ailments. Our intent in utilizing these datasets is to harness the collective wisdom and dexterity embedded within the data to engineer robust and accurate algorithms for analyzing skin images. Both the ISIC 2020 and DermIS datasets have undergone rigorous curation procedures, validating the accuracy and authenticity of the data. The involvement of esteemed institutions and experts in dermatology in the compilation of these datasets bolsters their credibility.
Both datasets showcase a diverse array of skin lesions spanning different types, severities, and anatomical locations. This diversity guarantees the precision of our analysis while concurrently ensuring its adaptability to a wide spectrum of skin conditions, thus enhancing the reliability of our conclusions. These datasets have gained substantial recognition as benchmarks within the dermatology community. By incorporating the ISIC 2020 and DermIS datasets, we ensure that our research is benchmarked against established studies, enabling meaningful assessments of the advancements we propose. Moreover, both datasets are openly accessible to the research community, certifying the reproducibility of our experiments and extending the opportunity for fellow researchers to authenticate our findings. This transparent approach bolsters the integrity of our study and cultivates an environment of collaboration.
Fortunately, over time, numerous researchers have addressed the issue of skin lesion image pre-processing, providing effective solutions. In [46], the authors introduced the DullRazor application, which has gained popularity as a software dedicated to hair removal in skin lesion images. This software employs morphological closing operations followed by the replacement of extracted pixels through bilinear interpolation. The replaced pixels are then smoothed using a median filter. Other techniques encompass various morphological operations along with operators such as bottom-hat [47], Sobel [48], Tophat [49], Laplacian [50], Radon transform (E-shaver) [51], and multiscale skeleton-morphological operators (“Virtual Shaver” method) [52]. In this study, we applied the DullRazor algorithm to eliminate hair artifacts from the selected images sourced from the ISIC 2020 and DermIS databases. This software was chosen due to its superior performance in hair removal, as acknowledged in the current state of the art. The outcomes of employing the DullRazor algorithm on the chosen images are depicted in Figure 6 and Figure 7.
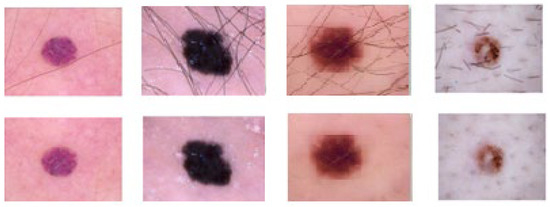
Figure 6.
Results obtained after applying the DullRazor algorithm to the DermIS skin lesion images database.
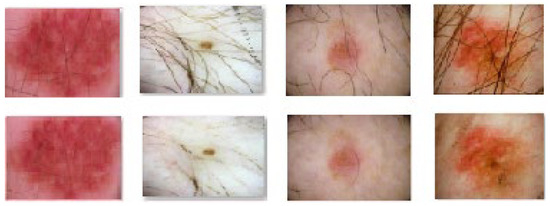
Figure 7.
Results obtained after applying the DullRazor algorithm to the ISIC 2020 skin lesion images database.
The results achieved are notably favorable, although it is worth mentioning that in the case of the ISIC 2020 database, for certain images, we found it necessary to apply the DullRazor algorithm twice. This decision stemmed from the initial outcomes not meeting our satisfaction. The approach involved initially applying the algorithm to the original image. Subsequently, the resulting image underwent the same algorithm again to yield improved outcomes. This two-step process was undertaken because images in the ISIC 2020 database tend to exhibit more noise, annotations, and even thicker hairs intersecting the lesion area. During the initial run, annotations were sometimes eliminated, whereas during the second run, hair removal was the focus. As demonstrated in Figure 6, due to the thickness of certain hairs, complete removal was not always achieved.
In this research, we will utilize a dataset comprising 1200 images for the purposes of training, validation, and testing. However, this quantity might not provide an ample number of examples for the CNNs to learn effectively. These images have undergone comprehensive labeling, enabling us to possess precise knowledge about each image, including both the predicted and actual labels. Non-random data selection implies that the testing dataset has been intentionally designed to mirror real-world scenarios. This ensures that the model’s performance evaluation is based on data that truly reflect the situations it will encounter in practical use and that we have a robust understanding of the true outcomes for each instance in our testing set. This high-quality ground truth information enables a meaningful assessment of the model’s predictions. Considering these factors, it is possible that the current testing setup, which benefits from non-random data and thorough labeling, may provide reliable insights without the immediate need for larger sample sizes or other validation methods.
3.2. Skin Lesion Detection Using Various CNNs
We employed MATLAB R2023b to implement the various CNN methodologies described. The process of selecting software tools for a project entails a thorough assessment of several factors, which includes analyzing the particular task’s requirements, the availability of relevant functions, and the ease of putting the solution into practice. In our situation, our choice to employ MATLAB and construct the decision fusion classifier within this environment stemmed from a blend of practicality, our existing expertise, and the distinctive features that MATLAB provides. Our team possesses a strong background in MATLAB, which has been developed over years of experience. This familiarity with MATLAB’s programming language, built-in functions, and toolboxes allowed us to expedite the development process and focus on the core aspects of our research.
MATLAB boasts an array of specialized toolboxes catered to diverse scientific and engineering domains. These toolboxes offer a comprehensive selection of functions and algorithms purpose-built for tasks such as data analysis, signal processing, and machine learning. Given the intricate decision fusion techniques involved in our project, the Signal Processing Toolbox and Statistics and Machine Learning Toolbox within MATLAB were invaluable in efficiently implementing these methods. The visual capabilities of MATLAB empower us to visualize and meticulously examine the outcomes of the decision fusion process. The capability to create insightful plots, graphs, and visual presentations enhances our comprehension of the classifier’s performance and aids in conveying our findings to colleagues and stakeholders. MATLAB’s programming environment affords a considerable level of flexibility when it comes to realizing intricate algorithms. This was especially advantageous for our decision fusion classifier, which necessitated complex data manipulation and fusion strategies. The comprehensive array of functions within MATLAB enabled us to fine-tune the implementation to our precise needs. MATLAB’s interactive nature and immediate feedback loop expedite swift prototyping and experimentation. This proved pivotal during the iterative progression of our decision fusion classifier, allowing us to promptly evaluate various approaches, refine parameters, and evaluate performance. Furthermore, MATLAB’s optimization capabilities, which encompass vectorized operations, streamlined the execution of our decision fusion classifier. This efficiency is of utmost significance, particularly when grappling with extensive datasets or algorithms demanding significant computational resources.
The CNNs used were initially pre-trained on the ImageNet database. Therefore, the outcomes we achieved were based on the application of transfer learning techniques. These pre-trained CNNs already possess the ability to differentiate between various fundamental objects. To adapt these CNNs for skin lesion detection, we followed a sequence of steps. Initially, we fine-tuned the pre-trained CNNs to enable the classification of skin lesions into two categories: melanoma and non-melanoma. Subsequently, we initiated the data loading process, specifying the image locations. As elaborated in the preceding section, we partitioned the data into training and testing datasets, each accounting for approximately 70% and 20% of the data, respectively. Following this, we loaded the pre-trained networks. For network retraining, we substituted the final two layers with a new fully connected layer and a classification layer. To accelerate the learning process, we adjusted the weight learn rate factor by increasing its value. To facilitate the training procedure, image resizing was performed. All necessary training options were then configured. Validation images were subjected to classification, and we additionally generated a confusion matrix for each executed architecture [53,54].
3.2.1. DarkNet-53
Darknet is an open-source neural network framework specifically designed for tasks related to object detection and recognition in the field of computer vision. Joseph Redmon is the developer behind Darknet, which is renowned for its effectiveness and accuracy in real-time object detection. This framework is coded in C and CUDA languages, rendering it compatible with both CPU and GPU processors. It has widespread applications, spanning surveillance systems, autonomous vehicles, robotics, and more. One of its notable features is the inclusion of pre-trained models, such as YOLO (You Only Look Once), which is used for object detection tasks [37,38].
In a study conducted by [50], a hybrid approach was introduced that combined Residual Network, YOLOv2, and Darknet-19. This network configuration encompasses a total of 53 convolutional layers, employing consecutive 1 × 1 and 3 × 3 convolutional layers. The outcomes presented by [55] indicate that this architecture outperforms others, including ResNet-152, Darknet-19, and ResNet-101. Darknet is predominantly utilized for YOLO workflows and addressing object detection challenges [56]. The architectural depiction of DarkNet-53 is illustrated in Figure 8.
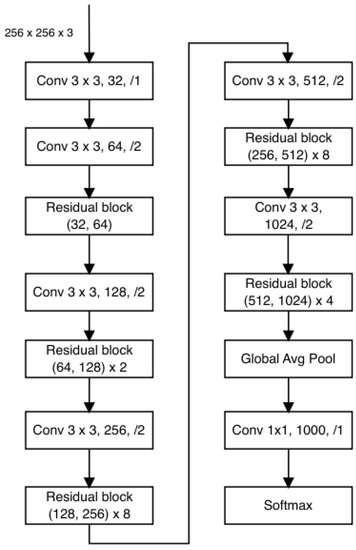
Figure 8.
DarkNet-53 architecture.
3.2.2. DenseNet 201
DenseNet is a commonly employed architecture in approaches related to Skin Lesion detection due to its remarkable accuracy and efficiency [37]. The design of DenseNet centers around dense blocks, serving as fundamental components. An illustration of this structure can be observed in Figure 8. Within each dense block, four densely connected layers are incorporated, characterized by a growth rate of 4. A skip-connection scheme is employed, in which the current layer is connected to all the preceding layers. This configuration efficiently tackles challenges associated with the vanishing gradient problem [57], Ref. [58] by aggregating input from previous layers and utilizing it iteratively with fewer convolution kernels. The architectural layout of DenseNet-201 is visualized in Figure 9.
Figure 9.
DenseNet-201 architecture.
3.2.3. GoogLeNet
GoogLeNet, also recognized as Inception v1, is a convolutional neural network (CNN) framework crafted by Google researchers. It was developed for the 2014 ImageNet Large-Scale Visual Recognition Challenge (ILSVRC), where it showcased notable performance enhancements compared to prior models. The distinctive feature of GoogLeNet lies in its “Inception module”, which integrates nine such modules side by side. This design choice enables the network to capture information across diverse spatial scales by utilizing multiple filters with varying receptive field sizes. A significant innovation occurs at the culmination of the final Inception module, where global average pooling is employed. This technique adeptly extracts both local and global features from images. Additionally, the Inception module incorporates 1 × 1 convolutions, which serve to reduce the dimensionality of feature maps and enhance computational efficiency.
GoogLeNet is composed of numerous stacked Inception modules, complemented by supplementary layers for classification purposes. In total, the architecture comprises 22 layers and replaces fully connected layers at the network’s end with global average pooling. This strategy curtails the number of parameters within the model and mitigates the risk of overfitting. Despite incorporating 100 independent building blocks, the network retains computational efficiency and adaptability for diverse label sets [59]. The architectural configuration of GoogLeNet is depicted in Figure 10.
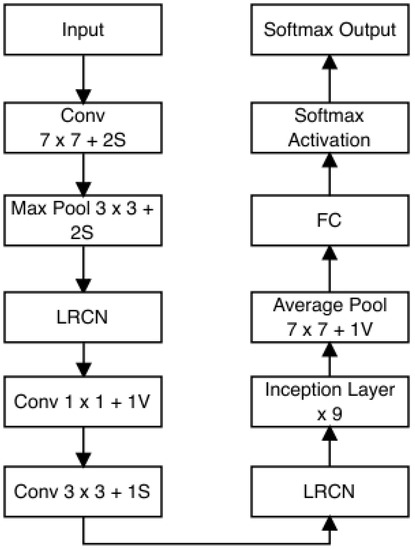
Figure 10.
GoogLeNet architecture.
The Inception architecture is a complex neural network featuring recurrent modules (illustrated in Figure 11) accompanied by subsequent max-pooling layers, which effectively halve the grid resolution. For the purpose of optimizing training efficiency, Inception modules are exclusively employed in the higher layers, while lower layers incorporate standard convolution layers. This approach permits the augmentation of units at each stage without compounding computational intricacies.
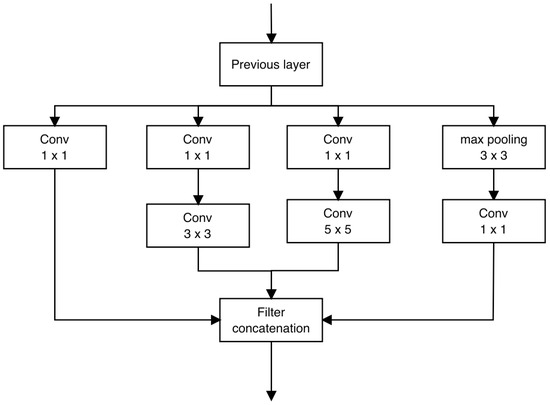
Figure 11.
Inception module.
3.2.4. Inception V3
Inception V3 was developed with the aim of reducing the number of parameters and connections while preserving the network’s efficiency. In this iteration, the 7 × 7 convolutional layer has been decomposed into three consecutive 3 × 3 convolutions. Following each convolution, ReLU activation blocks and Batch Normalization are employed. As for the inception blocks within the network, three 35 × 35 inception modules are integrated, each equipped with 288 filters. Additionally, five other grids with dimensions of 17 × 17 and a total of 768 filters, along with two more grids of size 8 × 8 × 1280, are utilized [37,60,61]. The architectural arrangement of Inception V3 is depicted in Figure 12.
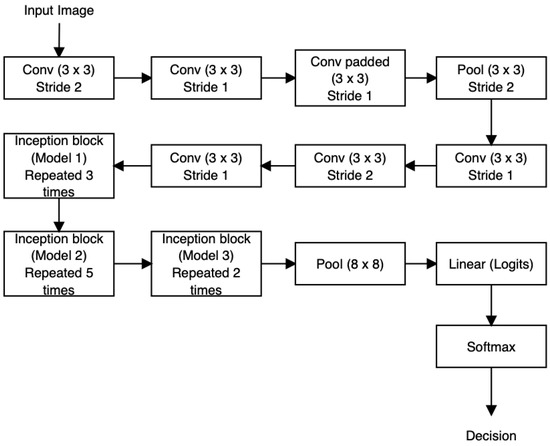
Figure 12.
Inception V3 architecture.
3.2.5. ResNet-50 and ResNet-101
ResNet stands as a widely adopted convolutional neural network architecture that employs residual blocks to construct a network capable of progressively comprehending intricate features as data pass through its layers. This architecture has garnered remarkable success in diverse tasks such as image recognition, object detection, and image segmentation. Originating from the endeavors of Microsoft Research researchers in 2015, ResNet addresses the challenge of training profoundly deep neural networks while evading the performance degradation that often accompanies depth.
The foundational version of ResNet, ResNet-34, encompasses 34 layers and integrates shortcut connections, as depicted in Figure 13a. These connections imbue the network with residual properties, resulting in fewer filters and a streamlined complexity. Further iterations include ResNet-50 and ResNet-101, housing 50 and 101 layers, respectively, and adopting residual connections [48]. In ResNet-50, a three-layer bottleneck block supplants each two-layer block found in ResNet-34, yielding a network of 50 layers. Similarly, the integration of more three-layer blocks produces ResNet-101. It is commonly recognized that both ResNet-101 and ResNet-50 exhibit superior accuracy compared to ResNet-34. Figure 13b visually illustrates the architecture of ResNet [62].
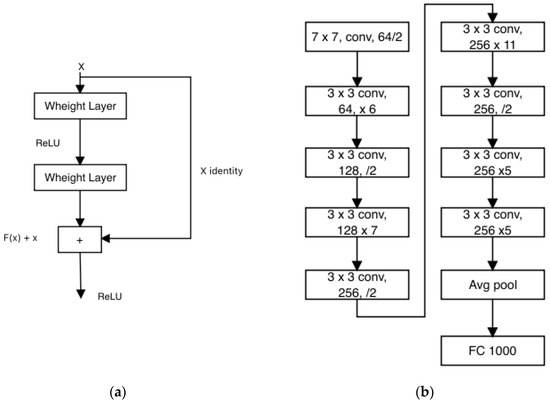
Figure 13.
(a) Residual Learning: Building Block; (b) ResNet Architecture.
3.2.6. Xception
Xception, a convolutional neural network architecture unveiled by François Chollet in 2016, stands as a variation of the Inception architecture, dubbed “Extreme Inception.” Designed to enhance the efficiency and effectiveness of conventional Inception models, Xception introduces the concept of depthwise separable convolutions. In conventional convolutional layers, each filter carries out both spatial convolution and channel mixing. However, Xception adopts depthwise separable convolutions, which disentangle these operations into distinct layers.
Depthwise convolutions focus on performing spatial convolution separately for each input channel, while pointwise convolutions execute 1 × 1 convolutions to intermingle the channels. This segmentation of processes mitigates computational intricacies and fosters more streamlined learning. By incorporating depthwise separable convolutions, Xception attains a commendable equilibrium between model intricacy and accuracy. Its adeptness has been demonstrated across a spectrum of image classification tasks, rendering it a staple in numerous computer vision applications [49]. The architecture of Xception is visually outlined in Figure 14 [63].
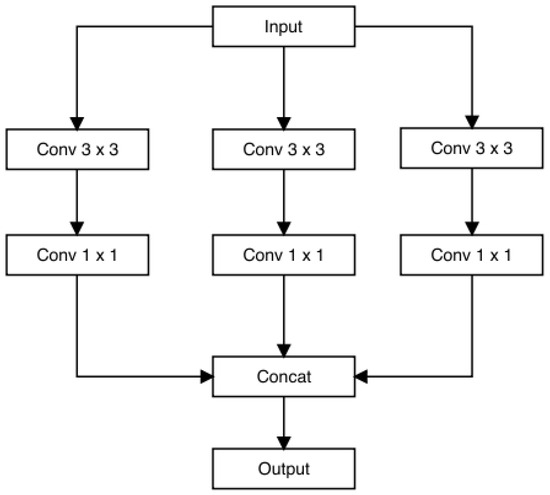
Figure 14.
Xception Architecture.
3.2.7. Inception-ResNet-V2
Inception-ResNet-V2 stands as a sophisticated CNN architecture that amalgamates the attributes of both Inception and ResNet models. Its development aimed at enhancing the efficacy of image recognition endeavors, such as object classification and detection. The essence of Inception-ResNet-V2 revolves around the synergy of inception modules and residual connections as seen in Figure 15, which collectively facilitate efficient and precise extraction of features from images. This architecture has gained widespread traction in the realm of computer vision applications, consistently attaining cutting-edge results in various image recognition challenges. The structure of Inception-ResNet-V2 encompasses an array of layers and modules that collaborate to extract features from input images. The journey begins with a sequence of convolutional layers dedicated to initial feature extraction. Subsequently, the architecture integrates the Inception module, characterized by parallel convolutional branches equipped with varying filter sizes. This arrangement empowers the network to apprehend features across diverse scales.
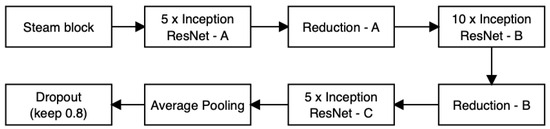
Figure 15.
Inception-ResNet-V2.
Moreover, Inception-ResNet-V2 integrates the ingenious concept of residual connections from the ResNet paradigm. These connections facilitate the learning of residual mappings, which proves instrumental in training deeper networks and mitigating the vanishing gradient predicament. The architectural repertoire extends to encompass other vital components, such as batch normalization, pooling layers, and fully connected layers tailored for classification purposes. Collectively, Inception-ResNet-V2 manifests as an intricate and profound network, capitalizing on the strengths of both Inception and ResNet models to attain remarkable performance in the domain of image recognition tasks [64].
3.3. Methodology for Skin Lesion Detection Using Web-Based Tools for Machine Learning Models
Our future objective is to research the possibility of integrating a melanoma classification system into an EHR medical information system to optimize the diagnosis and treatment process. As mentioned earlier, we tried four web-based tools for creating machine learning models. This type of application has some advantages:
- Ease of Use: Web-based tools are designed to be user-friendly, making them accessible to individuals without extensive programming or technical expertise. Google Teachable Machine, for example, provides a user-friendly interface that allows users to create and train machine learning models using their own data without writing complex code;
- Accessibility: Web-based tools are accessible from any device with an internet connection, making them widely available to users worldwide. This accessibility enables researchers, developers, and enthusiasts to access and utilize the tools without the need for specialized hardware or software installations;
- Rapid Prototyping: Web-based tools often provide a streamlined process for quickly prototyping and testing machine learning models. These tools offer pre-trained models, pre-processing capabilities, and simplified workflows that facilitate the development and iteration of models for various applications;
- Visualization and Interactivity: Web-based tools typically offer visualizations and interactive features that enhance the understanding and exploration of machine learning models. These tools often provide visual feedback, real-time predictions, and interactive controls, enabling users to interact with their models and gain insights into their behavior;
- Collaboration and Sharing: Web-based tools foster collaboration and knowledge sharing among users. They often provide functionalities for sharing models, databases, and project files, allowing researchers and developers to collaborate on projects, exchange ideas, and learn from each other’s work;
- Cloud Infrastructure: Many web-based tools leverage cloud computing infrastructure, enabling users to offload the computational requirements of training and inference to powerful servers. This eliminates the need for local high-performance hardware and allows users to leverage scalable cloud resources for faster and more efficient model development;
- Community Support: Web-based tools often have active communities of users and developers who share resources, provide guidance, and offer support. This community aspect fosters learning, collaboration, and the exchange of best practices;
- Integration with Other Tools and Services: Web-based tools, such as Google Teachable Machine, often integrate with other services and platforms, allowing users to incorporate their models into broader applications or workflows seamlessly. These integrations may include exporting models for deployment, connecting to APIs, or integrating with popular frameworks and libraries.
These advantages make web-based tools valuable for individuals and organizations seeking to explore and utilize machine learning without extensive technical barriers. They enable rapid prototyping, collaboration, and accessibility, opening doors for innovation and experimentation in various domains. On the other hand, software-wise, because we used MATLAB to create our classifier, this environment offers several advantages as well:
- Comprehensive Toolset: MATLAB provides a comprehensive set of tools, functions, and libraries specifically designed for image processing and analysis. This includes built-in functions for image pre-processing, feature extraction, and machine learning algorithms, making it convenient for developing classifiers for medical images;
- Robust Image Processing Capabilities: MATLAB’s image processing toolbox offers a wide range of functions for handling medical images, such as noise removal, image enhancement, segmentation, and feature extraction. These capabilities enable researchers and developers to pre-process and extract relevant features from medical images effectively, improving the performance of classifiers;
- Integration with Other MATLAB Toolboxes: MATLAB offers various toolboxes, such as the Statistics and Machine Learning Toolbox, which provide additional algorithms and functionality for machine learning tasks. This integration allows for seamless use of different algorithms and techniques to enhance classifier performance;
- Customization and Flexibility: MATLAB provides a flexible environment for developing and customizing classifiers. Users can easily modify and adapt algorithms, fine-tune parameters, and incorporate domain-specific knowledge into their classifiers. This flexibility is particularly valuable in medical imaging, where specific requirements and constraints often exist;
- Extensive Documentation and Community Support: MATLAB has a vast community of users and developers, which provides access to comprehensive documentation, tutorials, and forums. This support network facilitates learning, troubleshooting, and sharing knowledge, accelerating the development process and addressing challenges encountered in classifier development for medical images;
- Visualization and Analysis Tools: MATLAB offers robust visualization tools that enable users to visualize and analyze medical images and classifier outputs. These tools assist in understanding the behavior of the classifier, validating results, and gaining insights from the classification process;
- Deployment Options: MATLAB provides various options for deploying classifiers developed within the MATLAB environment. These include generating standalone executables, creating web applications, or integrating classifiers into other software frameworks or platforms. This flexibility allows for the seamless integration of MATLAB classifiers into existing medical imaging workflows or systems;
- Research and Academic Support: MATLAB is widely used in the research and academic communities, and many medical imaging studies and papers employ MATLAB for developing classifiers. This popularity leads to a wealth of research resources, benchmark databases, and code examples that can serve as references or starting points for developing classifiers in medical imaging. The workflow used to evaluate the creation of models for the Google Teachable Machine, Azure Machine Learning, Google Vertex AI, and SalesForce Einstein is in Figure 16.
 Figure 16. Training Dataset.
Figure 16. Training Dataset.
3.3.1. Google Teachable Machine
Teachable Machine is a web-based tool for creating machine learning models. The models built with TensorFlow (TensorFlow.js library—open source for machine learning and artificial intelligence) are used to train and run the models in a web browser. The created models use transfer learning techniques. There is a pre-trained CNN where the classes that the end-user works with become the last layer of the CNN. The model used for image classification is MobileNet, and in the end, these models can be incorporated into any application that uses JavaScript as the programming language. The advantage of this tool is that it allows modification of the training parameters of the network for optimal results (learning rate, batch size, number of epochs). Teachable Machine uses three types of models for different types of classifications. It uses MobileNet for image classification, Speech Commands for audio classification, and PoseNet for real-time body position classification from a video capture [65].
3.3.2. Microsoft Azure Machine Learning
Azure Machine Learning provides various pre-trained CNN models for image classification, including popular architectures such as ResNet, VGG, and DenseNet. The choice of model depends on the specific requirements of the task and the database used.
A huge benefit of using Azure services (functions and APIs such as Computer Vision) is the elimination of the need for companies to use individual servers, therefore reducing implementation costs. Another benefit is that Microsoft-developed services that use image processing on a very large scale can be used through cognitive services.
The algorithm starts with file upload and triggers an event that sends a notification to trigger Azure functions that call the Azure Computer Vision API to analyze the newly uploaded image. Subsequently, Azure functions store the Azure Computer Vision API response, which includes the classification results, along with the metadata of the image. Finally, the results can be reflected in a web interface or a mobile interface [65]. The main components of the Azure Machine Learning system are [65,66]:
- Computer Vision API: Retrieves information about the uploaded image;
- Azure Functions: Processes the image;
- Azure Event Grid: Triggers the Azure functions;
- Azure Blob Storage: Stores all uploaded images;
- Azure Cosmos DB: Stores metadata about each image and the classification results.
3.3.3. Google Vertex AI
Vertex AI utilizes various neural network architectures, including but not limited to CNNs, RNNs, transformer models, and DNNs. The specific neural network architecture used by Google Vertex AI can vary depending on the task or application being addressed. Google Vertex AI supports various CNN models for image classification tasks [67].
Some commonly used CNN models in Google Vertex AI include:
- EfficientNet: EfficientNet is a family of CNN models that have achieved state-of-the-art performance in image classification tasks while maintaining model size and computational resources efficiency;
- ResNet: as already mentioned earlier in this paper, ResNet is a popular CNN architecture known for its deep structure. It uses residual connections to address the vanishing gradient problem and enables the training of very deep networks;
- Inception: also described earlier, Inception models are characterized by the use of Inception modules, which allow efficient feature extraction at multiple scales;
- MobileNet: MobileNet is a lightweight CNN architecture designed for mobile and embedded devices. It achieves a good balance between model size and accuracy by using depth-wise separable convolutions [67].
3.3.4. Salesforce Einstein Vision
Einstein Vision and Language Model Builder is an application for automated image analysis that performs image classification based on training a model on a database divided into classes. The application is developed using the Apex, Java, Scala, and Node programming languages. The Einstein Vision application uses a deep learning model based on the API programming language, which allows access to pre-trained classifiers or training custom classifiers to solve various practical cases in the field of computer vision. The Einstein Vision package allows not only image classification but also object detection in an image, optical character recognition (OCR), sentiment prediction from written text, and intent categorization of unstructured texts to understand the user’s objective or desired outcome. All these modules are used to enhance message understanding, conversion, workflow optimization, and automation in the business environment.
Einstein Vision and Language Model Builder is a user interface layered over a REST API (API—Application Programming Interface—a set of definitions and protocols for creating and integrating software applications—specifying the required information content for the consumer (call) and producer (response), REST—Representational State Transfer—a set of architectural constraints) [68,69,70,71,72,73,74,75]. When a client makes a request via the REST API, it transfers the representation of the state of the resource to the requester (endpoint). This information is delivered in various formats (JSON—JavaScript Object Notation, XML, Python, PHP, or text) for Einstein, enabling quick dataset uploading, deep learning model training, and model performance testing through an easy-to-use graphical interface. Salesforce Einstein utilizes various neural network architectures such as:
- Convolutional Neural Networks: CNNs are often used for image recognition tasks. They consist of multiple layers, including convolutional layers, pooling layers, and fully connected layers. CNNs are designed to learn and automatically extract features from images;
- Recurrent Neural Networks: RNNs are suitable for processing sequential data, such as natural language processing. They have recurrent connections that allow information persistence across time steps;
- Long Short-Term Memory (LSTM) networks are a type of RNN that can efficiently capture long-term dependencies in sequential data [75];
- These neural networks are used in different components of Salesforce Einstein, such as natural language processing, image recognition, and predictive modeling, to enable advanced AI functionalities. The specific architectures of neural networks used by Salesforce Einstein can vary depending on the task and application and can be customized as needed. The specific details of the neural network architectures and training process used by Salesforce Einstein are not publicly available.
3.4. Methodology for Performance Evaluation
In this paper, we considered the following performance indicators: accuracy (ACC), F1 Score, Sensitivity/Recall, and Specificity, described in Table 1 with their formulas.

Table 1.
Expressions for the performance indicators.
- Accuracy—a commonly used metric to assess the overall performance of a melanoma detection system. It measures the proportion of correctly classified melanoma and non-melanoma cases. However, it is important to consider other metrics as well, as accuracy alone may not provide a comprehensive evaluation;
- F1 Score—a metric that combines precision and recall. It considers both false positive (FP) and false negative (FN) and provides a balanced measure of a model’s performance. Higher F1 scores indicate better performance in terms of both precision and recall;
- Sensitivity/Recall—Sensitivity, also known as recall or true positive rate, measures the proportion of actual melanoma cases correctly identified by the system. It focuses on minimizing false negatives (FN) and is particularly important in medical applications to avoid missing melanoma cases;
- Specificity—Specificity measures the proportion of non-melanoma cases correctly identified as such by the system. It focuses on minimizing false positives (FP) and is essential to reducing unnecessary biopsies or false alarms [76,77,78,79,80,81,82,83].
In the context of this study, our primary emphasis will be on the assessment of the F1 score as opposed to accuracy. This preference stems from the heightened utility of the F1 score, particularly in scenarios where the costs associated with FN and FP significantly differ. As previously mentioned, the F1 score serves as an amalgamation of both recall and precision, whereby achieving a higher F1 score inherently implies improved precision and recall. Notably, the F1 score does not take into account the count of true negatives that were accurately identified. It is particularly recommended to employ the F1 score in situations where the concern about predicting false negatives is of substantial significance. This approach aligns with our focus on optimizing the performance of our melanoma detection system while considering the potential consequences of missing cases of melanoma.
3.5. Methodology for Skin Lesion Detection Using the Decision Fusion Classifier System
In our previous work [6], we introduced a decision fusion framework employing diverse classifiers encompassing Neural Networks (NN), feature-based techniques, and CNN. However, the selection process for these classifiers was not addressed. The fusion weights for this system were established solely on the basis of accuracy [6]. Building upon the insights gleaned from the literature review in Chapter Two, wherein numerous authors demonstrated remarkable outcomes using CNNs, our present endeavor introduces a novel decision fusion classifier. This classifier is designed to amalgamate multiple CNN architectures, thereby striving to attain the utmost performance levels for melanoma detection while also ensuring adaptability to varying databases. The selection of the eight CNN architectures was guided by a comprehensive set of criteria grounded in various considerations:
- Performance—the neural network has a proven track record of achieving high accuracy and performance on melanoma detection tasks and on the specific databases we used;
- Availability of pre-trained models on large-scale databases, such as ImageNet. Pre-trained models can significantly speed up the training process and provide a good starting point for transfer learning;
- Complexity—the neural network is not out of reach complex. The complexity of the CNN architectures should be taken into consideration. Complex models may require more computational resources and longer training times, which can be challenging in practical applications. Hence, it is important to strike a balance between complexity and performance;
- Generalization—the network should have a strong ability to generalize to unseen data and can be applied to a variety of different images. Robustness and generalization are crucial for real-world deployment;
- Diversity of architectural designs—in the selection of the neural networks, we took into consideration diverse design principles. Each architecture has different features and characteristics, such as the use of residual connections, inception modules, or dense connections. Including diverse architectures can help capture a wider range of melanoma characteristics and improve the chances of detecting different types of melanomas. It is beneficial to choose CNNs that exhibit diverse architectural designs and utilize different techniques. This increases the chances of capturing complementary information and avoiding bias or limitations that might be present in a single architecture;
- Previous success—the neural network has demonstrated exceptional performance in melanoma detection or similar medical imaging tasks [76,77,78,79].
Certainly, these factors are profoundly influenced by the databases utilized for CNN training and the pre-processing methodologies implemented. To ascertain the optimal combination of CNN architectures for our specific application, an iterative approach involving experimentation and refinement is indispensable. In this context, our initial step involves the selection of eight CNNs based on the specified criteria, a process informed by a comprehensive examination of the current state of the field and the demonstrated performance on the extensive ImageNet image database [83,84] and pre-processing the images as we mentioned earlier by eliminating the hair and resizing them to fit the input of the CNNs. The chosen CNNs encompass DarkNet-53, DenseNet-201, GoogLeNet, Inception-V3, InceptionResNet-V2, ResNet-50, ResNet-101, and Xception. With the overarching goal of maximizing system performance, we embarked on the exploration of an optimal approach for allocating decision fusion weights to each individual CNN. The resultant architecture of our proposed system is elucidated in Figure 17.
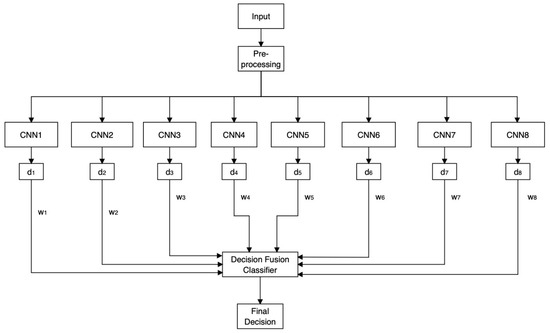
Figure 17.
Proposed system architecture.
Where the determination between melanoma and non-melanoma, executed by the decision fusion classifier, is effectuated through the utilization of the following Equations (1) and (2):
where DM is the decision in case of melanoma, where the false positive is considered as being a non-melanoma predicted as melanoma, and DNM is the decision in case of non-melanoma, where the false negative is considered as being a melanoma predicted as a non-melanoma, i represents the image, and di is the partial decision made by each CNN and wi the weights calculated from the average mean of the F1 score obtained for ISIC and DermIS database as seen in Expressions (3) and (4).
The ultimate verdict is determined by comparing the values of the Expressions (1) and (2), with the higher value dictating the outcome. For example, if DM > DNM, then the given result is melanoma. In the case of DM < DNM, then the result is non-melanoma. In essence, through (1), we consider the objective melanoma, and through (2), we consider the objective non-melanoma. The choice between using the raw scores before the sigmoid activation function versus the binary decisions (0 or 1) for decision fusion can impact the overall performance and information content of the system.
We took into consideration the advantages and disadvantages before taking the decision to use binary decisions. The advantage of raw scores is that they preserve the continuous nature of the model’s output, providing more nuanced information about the model’s confidence in its predictions. It also maps raw scores to probabilities between 0 and 1, allowing for a gradual transition from very confident negative predictions (close to 0) to very confident positive predictions (close to 1). The disadvantage is that raw scores require additional interpretation to determine decision thresholds, potentially adding complexity to the decision-making process. In addition, choosing appropriate thresholds for raw scores might be subjective and could vary depending on the application and dataset.
The advantage of binary decisions is that they simplify the decision-making process as they provide a clear and immediate answer of whether a lesion is classified as positive (1) or negative (0). They are also straightforward to understand, making them more accessible for medical professionals and users. The downside is that binary decisions discard the nuanced information carried by the raw scores, potentially leading to a loss of subtleties in model confidence. We chose the binary decision in the proposed classifier because it yields some benefits that fit better in our end goal to integrate an automated melanoma detection module into an EHR, such as:
- Simplicity and Interpretability: Binary decisions provide a clear and straightforward answer, making it easier for both medical professionals and general users to understand the outcome of the classification. This simplicity is particularly valuable in medical applications where quick and easy interpretation of results is crucial;
- Clinical Decision Support: In a clinical setting, where decisions need to be made rapidly, binary outcomes can serve as valuable decision-support tools. For example, a clear positive (1) or negative (0) result can guide medical professionals in determining the next steps for patient care;
- Standardization: Binary decisions enable standardized reporting and communication of results. A consistent binary output format simplifies data sharing, communication between different medical institutions, and comparisons across different studies or systems;
- Regulatory and Legal Considerations: In medical applications, regulatory agencies often require clear, easily interpretable outcomes. Binary decisions can align with regulatory standards and facilitate compliance;
- Reduced Ambiguity: Binary outcomes reduce ambiguity in scenarios where nuanced classifications might lead to confusion or differing interpretations. By providing a direct answer, binary decisions can mitigate potential misunderstandings;
- Threshold Adaptation: Binary decisions allow for straightforward threshold adaptation. If specific sensitivity or specificity requirements need to be met, adjusting the threshold for positive predictions can be easily performed to achieve the desired balance;
- Efficient Workflow Integration: In automated systems, binary outputs can integrate seamlessly into an HER and other medical record systems, enabling quick and efficient data management and retrieval.
4. Experimental Results
4.1. Experimental Results for Skin Lesion Detection Using Various CNNs
To implement the CNN methodologies, we employed MATLAB. All CNN models were pre-trained on the ImageNet database, leading to results that rely on the principles of transfer learning. As previously indicated, each of the two databases (ISIC 2020 and DermIS) were treated separately. For training, we allocated 700 images, alongside 300 for validation and 200 for testing purposes. The training process encompassed several key aspects, including validation accuracy, iterations per epoch, maximum iterations, and the time taken for training, which was documented for all eight CNNs forming the comprehensive decision fusion system. The specifics of this training progress are summarized in Table 2. Our training configuration included 6 epochs, a learning rate of 0.0001, and a maximum of 276 iterations, with 46 iterations per epoch applied uniformly across all the CNNs.

Table 2.
Training and validation progress.
Looking at Table 2, the results are quite revealing. For the ISIC 2020 database, DarkNet-53 achieved the highest accuracy, while for the DermIS database, GoogLeNet came out on top. In terms of training duration, Inception-ResNet-V2 on the ISIC 2020 database required the longest training time (49 min and 0 s), whereas GoogLeNet on the DermIS database boasted the shortest training time (3 min and 45 s). Notably, many of the CNNs exhibited improved training times when applied to the DermIS database compared to their performance on the ISIC 2020 database. This observation might relate to the database’s quality and characteristics. Numerous factors can impact the duration of the training, but our initial focus when addressing this aspect is the determination of an appropriate learning rate for each CNN. It is established that reducing the learning rate prolongs the CNN’s learning process while excessively elevating it can cause weight divergence, ultimately diminishing training quality. As a prospective endeavor, we intend to devise an efficient method for identifying suitable learning rates for individual CNNs. This is important because assessing various constant learning rates for each CNN is both time-intensive and prone to errors. To compute the F1 score, we employed MATLAB to generate a confusion matrix for each CNN. The F1 score was then calculated using the formula provided in Table 1.
Based on Figure 18, we can observe that for the DermIS database, DarkNet-53 achieved the highest F1 score of 0.8899, closely followed by GoogLeNet with a score of 0.8898 and Inception-ResNet-V2 with a score of 0.859. On the other hand, for the ISIC 2020 database, DarkNet-53 once again stood out with the highest F1 score of 0.818. Following DarkNet-53, ResNet-50 and DenseNet-201 attained F1 scores of 0.804 and 0.77, respectively. Once more, we can note that a majority of CNNs yielded superior outcomes when employed on the DermIS database compared to the ISIC database. This discrepancy further underscores the distinction in image quality between these two databases. Enhanced image clarity, characterized by minimal noise, corresponds to heightened performance levels. Consequently, as part of our future endeavors, we plan to delve into more comprehensive and advanced image-processing techniques to augment the performance of the diverse CNNs. Additionally, we provided some predictions made by the CNNs during the testing phase for both the ISIC and DermIS databases in Table 3 and Table 4. We selected three images (Im1—Image 1, Im2—Image 2, Im3—Image 3) from each database with the real label (NM—Non-Melanoma and M—Melanoma) and tracked the result predicted by the architectures.
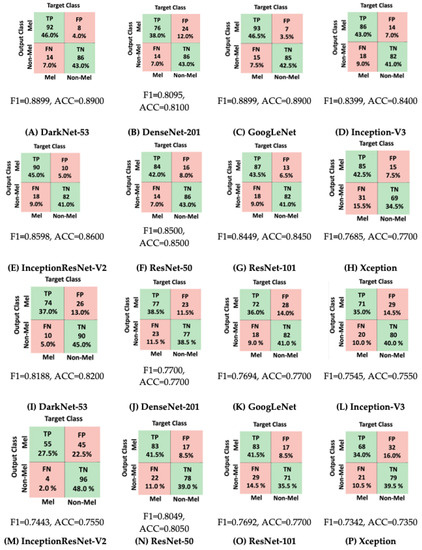
Figure 18.
Confusion matrices for considered CNNs. (A–H): DermIS database, (I–P): ISIC database.

Table 3.
Prediction of the used CNNs in the testing phase on the DermIS database.

Table 4.
Prediction of the used CNNs in the testing phase on the ISIC database.
4.2. Experimental Results for Skin Lesion Detection Using the Automated Applications
For model creation, we utilized the built-in interface provided by each application, creating the dataset and training the model. The outcomes of the testing phase are depicted in Figure 19. As illustrated in Figure 19, for the ISIC 2020 database, the most remarkable accuracy and F1 score were achieved using Microsoft Azure Machine Learning. On the other hand, for the DermIS database, the highest accuracy was observed with Google Vertex AI. Delving into the comparison between FP and FN, Google Teachable Machine yielded the highest number of false positives for the DermIS database, while for the ISIC 2020 database, Google Vertex AI recorded the highest count of false positives.
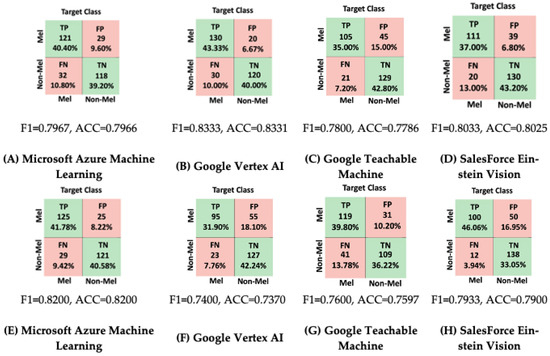
Figure 19.
Confusion matrices for considered automated applications. (A–D): DermIS database, (E–H): ISIC database.
The primary aim of evaluating the performance of different machine learning models and platforms, such as Google Teachable Machine, Google Vertex AI, Salesforce Einstein Vision, and Microsoft Azure Machine Learning, against a custom classifier that combines these CNNs’ decisions, is to determine the most appropriate option for integrating into an EHR system equipped with an automated melanoma detection module. This assessment is a pivotal stage in EHR system development and aids in well-informed decision-making during the planning and solution analysis phases. The main goal is to appraise the accuracy and dependability of each model and platform in accurately identifying cases of melanoma. By considering the combined decisions of various classifiers, decision fusion plays a role in reducing false positives and false negatives. Different CNNs might excel in distinct aspects of melanoma detection due to variations in their designs and training data. Decision fusion capitalizes on classifier diversity to address uncertainties and fluctuations in detection, which is crucial in real-world situations where data can be noisy or exhibit unexpected variations. It is vital to test the models on a diverse dataset encompassing various types of melanoma images to gauge their resilience and ability to apply to different scenarios. Robust models can handle changes in lighting, perspectives, and skin tones. The comparative analysis seeks to assess how clinically applicable each solution is within the healthcare context, as precise and reliable outcomes are crucial for informed clinical decisions.
Evaluating the speed of inference and resource efficiency for each model holds significance, particularly in a real-time healthcare environment. Models that deliver accurate results swiftly are preferred for integration into an EHR system. Real-time performance is essential for seamless incorporation into the fast-paced realm of healthcare delivery. The evaluation also involves scrutinizing the integration potential of each solution into the existing EHR system infrastructure. Factors such as compatibility, API, and deployment choices must be taken into account. A solution that seamlessly aligns with the system’s architecture saves time and resources during implementation.
Furthermore, scalability is assessed for each solution, gauging their ability to accommodate larger datasets and future expansion. Solutions that can scale in accordance with the evolving needs of a growing healthcare practice are sought after. The capacity of the chosen solution to scale alongside increased data and usage is imperative for long-term viability. Cost analysis is crucial to ascertain the expenses associated with adopting each solution and to ensure compatibility with the project’s budget. The cost-effectiveness of each solution is evaluated, encompassing development, deployment, and maintenance expenses. Optimal value delivery within the project’s financial constraints is a pivotal consideration.
Finally, the user interface and user experience of the chosen solution must align with the design and user requisites of the EHR system. By systematically evaluating these aspects, we can determine which of the evaluating models or platforms is the most reliable and effective solution to integrate into the EHR system with an automated melanoma detection module. This analysis will help inform the decision-making process during the planning and solution analysis phases of the EHR system development project. In the present paper, we assessed the way in which we would approach the melanoma detection module by developing a custom decision fusion melanoma classifier and comparing it with some of the well-known web-based models online.
After evaluating the results, Google Teachable Machine emerges as a highly versatile option. The model exhibits commendable accuracy and offers seamless integration into architectures utilizing Java as the programming language. The process of uploading images, training the model, and subsequent cloud-based utilization is provided without any charge. An additional advantage lies in the flexibility to adjust network training parameters, such as learning rate, batch size, and number of epochs, for optimal outcomes. This renders the application suitable for integration into an EHR system. For a comprehensive overview, we present a comparison of accuracy and F1 scores among the four web-based applications in Table 5.

Table 5.
Experimental results for the decision fusion classifier vs. the automated applications.
4.3. Experimental Results for Skin Lesion Detection Using the Decision Fusion Classifier
To be able to create a more performant system, the weights presented in Table 6 were assigned for each implied CNN, where M—Melanoma, NM—Non-Melanoma.

Table 6.
Decision Fusion classifier weights.
The objective of appropriately configuring the weights is to enhance the effectiveness of the decision fusion-oriented system by influencing the ultimate decision made by the classifiers with higher weights. This trend is evident in Table 6, where, among the initial five CNNs, the weight wNM surpasses wM, while for the remaining three CNNs, wM takes precedence over wNM. This indicates that the first five CNNs exhibit a partial inclination toward predicting a Non-Melanoma outcome, whereas the other three display a preference for predicting Melanoma. The confusion matrix on both ISIC 2020 and DermIS databases of the proposed system was generated and presented in Figure 20. Based on these confusion matrices, the F1 score was calculated for each database.
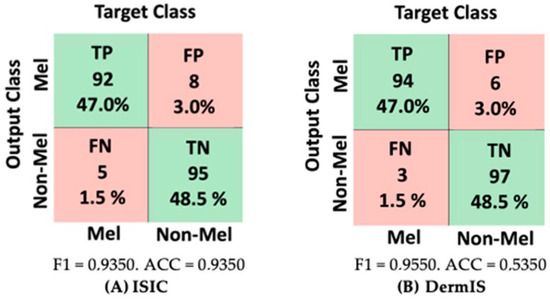
Figure 20.
Confusion Matrix for Decision Fusion Classifier.
The performance of our proposed decision fusion classifier is depicted in Figure 20. Notably, this classifier demonstrates remarkable performance in terms of accuracy and F1 score for both the DermIS and ISIC 2020 databases. The outcomes from the four applications are also illustrated in Figure 20 for comparison. Evidently, our proposed system yields superior results compared to those obtained from the four applications. Additionally, it is apparent that, apart from Microsoft Azure Machine Learning, better outcomes were achieved on the ISIC 2020 database. Our proposed method attains an F1-score of 0.955 for the DermIS database and 0.9350 for ISIC 2020. As previously stated within the paper, our perspective is that a system should ideally produce a higher count of FP than FN. This preference stems from the notion that it is more advantageous for both the system and the patient to recommend further investigations than to incorrectly label a melanoma as a benign lesion, which could potentially elevate the risk of patient mortality. Figure 20 illustrates that our proposed system indeed generated a greater number of False Positives in both the ISIC and DermIS datasets.
5. Discussion
In this paper, we introduced a multi-network system that employs decision fusion for melanoma detection, utilizing eight high-performance CNNs: DarkNet-53, DenseNet-201, GoogLeNet, Inception-V3, InceptionResNet-V2, ResNet-50, ResNet-101, and Xception. The selection of these CNNs was guided by a thorough analysis of the state-of-the-art. The fusion system’s weights were determined based on the F1 scores achieved by each individual CNN. We evaluated the performance of each architecture on two distinct databases, considering both databases when assigning the fusion weights to enhance adaptability.
As mentioned in our earlier work [6], the DullRazor algorithm could not completely remove hair from all ISIC 2020 images, partially due to the presence of annotations and thick hair in some images. In Equations (5) and (6), the variables represented by terms such as “dDarkNet” and “dXception” are decisions made by the neural network architectures. The formula essentially represents a weighted combination of these measurements. Each architecture’s measurement is multiplied by a specific weight (such as 0.8543 for DarkNet and 0.7513 for Xception) and then added together. This combination is represented by “DM” and “DNM” for two different scenarios.
DM = dDarkNet·0.8543 + … + dXceptio·0.7513
DNM = dDarkNet·0.8599 + … + dXception·0.7494
It is notable that results obtained from the DermIS database tend to be superior to those from the ISIC 2020 database, both in the decision fusion classifier and the web-based applications. This could be attributed to the fact that ISIC 2020 comprises images collected from various sources with different equipment, conditions, and populations. To improve results, a pre-processing step involving hair removal and resizing was necessary for ISIC 2020 images. All results obtained are presented in Table 7, alongside recent work by other researchers who evaluated their models based on accuracy and F1 score. Our proposed classifier demonstrates superior performance compared to most evaluated classifiers. Although the web-based applications yielded less accurate results, they hold promise in terms of rapid prototyping without the need for pre-processing steps.

Table 7.
Experimental results for the decision fusion classifier vs. other classifiers vs. the automated applications.
Both MATLAB-made classifiers proposed in this paper and web-based tools have their own advantages and use cases. MATLAB offers extensive capabilities, flexibility, and advanced image processing functionalities suitable for complex medical image analysis. On the other hand, web-based tools provide user-friendly interfaces, rapid prototyping, accessibility, collaboration features, and integration with cloud infrastructure. The choice between the two depends on the specific requirements, expertise, and preferences of the user or development team.
Regarding the easiness of integration vs. performance, web-based tools are often designed with simplicity and ease of use in mind. They typically provide user-friendly interfaces and streamlined workflows that make it easy to train and deploy machine learning models. Integration into another platform can be straightforward, as these tools often offer export options or APIs to facilitate model integration. Integrating MATLAB-made classifiers into another platform may require additional steps, as MATLAB is standalone software. Depending on the platform and desired integration method, it may involve exporting the model, converting it to a compatible format, leveraging MATLAB Compiler to create standalone executables, or integrating MATLAB scripts with other programming languages.
Performance comparison between MATLAB-made classifiers and web-based tools, such as Google Teachable Machine, depends on various factors, including the complexity of the problem, database size, and the specific algorithms and techniques employed. Both MATLAB and web-based tools can achieve good performance, but it ultimately depends on the implementation and customization of the classifiers. MATLAB provides extensive capabilities for customization, algorithm selection, and fine-tuning, allowing users to optimize classifiers for specific tasks. This flexibility may result in better performance for complex or domain-specific applications where advanced image processing or tailored algorithms are required.
Web-based tools, such as Google Teachable Machine, are designed for ease of use and rapid prototyping. They often leverage pre-trained models or simplified workflows, which may provide good performance for simpler tasks or generic image classification. However, they may have limitations in terms of customization and adaptability to specific requirements. Ultimately, the choice between MATLAB-made classifiers and web-based tools for integration into another platform depends on the specific requirements, available expertise, and the desired trade-off between ease of integration and performance optimization. If the integration process needs to be simple and streamlined, with a focus on ease of use, web-based tools, such as Google Teachable Machine, may be a more suitable choice. On the other hand, if customization, advanced algorithms, and fine-tuning are critical for achieving optimal performance, MATLAB, with its extensive toolset and flexibility, may be the preferred option.
The novelty and distinctions of this work lie in the fact that this paper focuses on enhancing the performance of a multi-network system through decision fusion for melanoma detection to leverage the strengths of diverse CNN architectures, potentially leading to improved accuracy. The selection of CNN architectures is not arbitrary but guided by specific criteria outlined in the methodology section, and furthermore, instead of treating all networks equally, the paper introduces a weighted decision fusion system. The weights assigned to each network are based on the F1 scores achieved by each CNN. This weighting approach considers the networks’ individual strengths in melanoma detection. This indicates a deliberate and informed approach to choosing networks that are best suited for the task at hand.
On the other hand, the system’s performance optimization takes into consideration two distinct databases, ISIC 2020 and DermIS, that employ real-world data consisting of 1200 images in total for training, validation, and testing. This practical evaluation underscores the system’s applicability to genuine scenarios. The assignment of weights considers both databases, which contributes to the adaptability of the system to varying datasets. This dual-database approach acknowledges the need for robustness across different data sources.
Also, to establish the system’s effectiveness, a comprehensive comparison is conducted against four existing applications: Google Teachable Machine, Microsoft Azure Machine Learning, Google Vertex AI, and Salesforce Einstein Vision. The comparison showcases the superior performance of the proposed system in melanoma image classification. The quantitative comparison substantiates the advancements achieved by the novel approach, positioning it as a valuable contribution to the field of melanoma detection, taking into consideration that the end goal here is to develop an EHR system that has an automated melanoma detection module in order to introduce a way to make melanoma investigations introduced into routine controls in order to improve patient prognosis, disease prevention, reduce treatment costs, improve population management and improve patient empowerment so that patients would take better control of their health and would educate the population that it is better to prevent than to treat. In order to do this, researchers need to give people the means to do so effectively.
6. Conclusions
In conclusion, the incidence of melanoma cases is on the rise, necessitating the need for early detection to prevent further growth. Deep learning has shown promising results in melanoma detection, with increased accuracy observed over time. Our endeavor to develop an EHR system with an automated melanoma detection system has led us to meticulously assess the computational complexity of our approach, which involves the integration of a custom decision fusion classifier utilizing multiple neural networks. This endeavor necessitates a thorough justification, considering the intricate nature of the problem, the significance of accuracy, and the complexities involved in implementation and integration. Addressing the challenge of automating melanoma detection within an EHR system demands a solution that is both sophisticated and accurate. Our custom decision fusion classifier, which combines neural networks such as DarkNet-53, DenseNet-201, GoogLeNet, Inception-V3, InceptionResNet-V2, ResNet-50, and ResNet-101, emerges as a pioneering resolution. This multifaceted approach harmonizes effectively with the intricacies of melanoma detection, enabling us to exploit the strengths of diverse neural networks to enhance accuracy. Our system, built upon a fusion of high-performance CNN architectures including DarkNet-53, DenseNet-201, GoogLeNet, Inception-V3, InceptionResNet-V2, ResNet-50, ResNet-101, and Xception, demonstrates a clear superiority over existing applications in melanoma detection. The decision fusion procedure, optimized based on the F1 scores of each network, exhibits a level of accuracy, precision, and sensitivity that sets a new standard in this critical domain. The experimental evaluation of our proposed multi-network decision fusion system for melanoma detection yields significant implications for both medical research and clinical practice. The outcomes of our rigorous evaluation underscore the remarkable performance and distinct advantages of our approach.
The empirical evaluation, conducted on diverse datasets, including ISIC 2020 and DermIS, reaffirms our system’s adaptability across different image sources, enhancing its potential for real-world deployment. Our careful consideration of two distinct databases in weight assignment contributes to robust and versatile performance. As indicated in Table 7, our suggested decision fusion classifier displayed the most impressive performance when considering the DermIS database. It achieved a value of 0.9550 for both accuracy and F1 score. Among the other four applications we assessed, Google Vertex AI yielded the next best outcome with an accuracy of merely 0.8333 and an F1 score of 0.8331. This underscores the demonstrated superiority of our proposed system. In the case of ISIC, our system exhibited an accuracy and F1 score of 0.9350.
In light of the decision fusion classifier’s complexity, we conducted an extensive comparison with established applications, namely Google Teachable Machine, Microsoft Azure Machine Learning, Google Vertex AI, and Salesforce Einstein Vision. This comprehensive evaluation substantiated that our approach yields substantial accuracy gains, thereby validating its justification. While the integration complexities presented by the decision fusion classifier are acknowledged, the commendable performance it delivers serves as a counterbalance, accentuating its significance. The implications of these results are substantial. Our proposed multi-network decision fusion system not only advances the state of the art in melanoma detection but also holds immense potential for clinical implementation. By offering superior accuracy and a refined decision-making process, our system stands as a powerful tool for medical practitioners, aiding in early detection and improving patient outcomes.
Given the medical context of melanoma detection, precision is of paramount importance. The combination of neural networks within our custom decision fusion classifier contributes synergistically to elevating accuracy. This heightened accuracy holds direct implications for dependable melanoma detection within the EHR system, potentially assisting medical professionals in prompt diagnosis and treatment decisions. Acknowledging the integration challenges associated with the decision fusion classifier, we are cognizant that incorporating this advanced system may be more intricate compared to alternative options. However, this complexity is offset by the exceptional accuracy it offers. Furthermore, the efficiency and scalability of the decision fusion classifier, despite its complexity, render it aptly suited for the EHR system’s context. The decision to integrate the customized decision fusion classifier bears enduring value. Leveraging multiple neural networks enhances adaptability across a broad spectrum of melanoma cases, ensuring resilience against variations. While the initial integration hurdles are acknowledged, the sustained benefits in terms of accuracy, early detection, and dependable patient care overshadow these challenges.
In conclusion, our work not only introduces a novel methodology but also delivers on the promise of enhanced accuracy and performance. The computational complexity of our tailor-made decision fusion classifier is substantiated by its distinctive and innovative methodology, heightened accuracy in melanoma detection, a well-documented comparative assessment, and the enduring value it brings to the EHR system. Despite the integration challenges, the advantages it confers to medical diagnostics validate this as a prudent and justified choice. The implications of our experimental evaluation point towards a future where melanoma detection can be augmented by our innovative approach, ultimately contributing to more effective and reliable healthcare interventions. The next steps for improving the solution would be to find a way to integrate our classifier obtained using MATLAB to have better accuracy and flexibility to adapt algorithms for an unlimited number of tasks. This will require an integration between MATLAB and the SalesForce Platform.
Author Contributions
H.E.-k. contributed to CNN implementation and obtained the experimental results for the selected CNNs and the implementation of the proposed decision fusion classifier. A.-M.Ș. contributed to the research of applications for automated image analysis that perform image classification and the possibility of integration with an EHR system, and edited the paper and the content. D.P. conceived the paper and validated it. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created.
Acknowledgments
The results presented in this article were obtained with the support of the Ministry of Investments and European Projects through the Human Capital Sectoral Operational Program 2014-2020, Contract no. 62461/03.06.2022, SMIS code 153735.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ACC | Accuracy |
| AI | Artificial Intelligence |
| API | Application Programming Interface |
| CNN | Convolutional Neural Network |
| CPU | Central Processing Unit |
| DB | Database |
| DNA | Deoxyribonucleic Acid |
| DNN | Deep Neural Network |
| EHR | Electronic Health Record |
| FN | False Negative |
| FP | False Positive |
| GAC | Geometric Active Contour |
| GC-SCNN | GrabCut-Stacked Convolutional Neural Network |
| GMM | Gaussian Mixture Model |
| Im | image |
| ILSVRC | ImageNet Large-Scale Visual Recognition Challenge |
| ISIC | International Skin Imaging Collaboration |
| JSON | JavaScript Object Notation |
| LSTM | Long Short-Term Memory |
| M | Melanoma |
| ML | Machine Learning |
| NN | Neural Networks |
| NM | Non-Melanoma |
| OCR | Optical Character Recognition |
| ROI | Region Of Interest |
| REST | Representational State Transfer |
| RF | Random Forest |
| RNN | Recurrent Neural Network |
| SVM | Support Vector Machine |
| TN | True Negative |
| TP | True Positive |
| UV | Ultraviolet Radiation |
| VGG | Visual Geometry Group |
| XML | Extensible Markup Language |
| YOLO | You Only Look Once |
References
- Ott, P.A. Intralesional Cancer Immunotherapies. Hematol. Oncol. Clin. N. Am. 2019, 33, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sheikh, M.S. Melanoma: Molecular Pathogenesis and Therapeutic Management. Mol. Cell. Pharmacol. 2014, 6, 228. [Google Scholar]
- Strashilov, S.; Yordanov, A. Aetiology and Pathogenesis of Cutaneous Melanoma: Current Concepts and Advances. Int. J. Mol. Sci. 2021, 22, 6395. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Melanoma Treatment for Health Professionals (PDQ). Available online: www.cancer.gov/types/skin/hp/melanoma-treatment-pdq (accessed on 2 February 2020).
- Stewart, B.W.; Wild, C.P. World Cancer Report 2014; World Health Organization, International Agency for Research on Cancer, IARC Publications: Geneva, Switzerland, 2014. [Google Scholar]
- El-Khatib, H.; Popescu, D.; Ichim, L. Deep Learning–Based Methods for Automatic Diagnosis of Skin Lesions. Sensors 2020, 20, 1753. [Google Scholar] [CrossRef] [PubMed]
- Ghazawi, F.M.; Darwich, R.; Le, M.; Rahme, E.; Zubarev, A.; Moreau, L.; Burnier, J.V.; Sasseville, D.; Burnier, M.N.; Litvinov, I.V. Uveal melanoma incidence trends in Canada: A national comprehensive population-based study. Br. J. Ophthalmol. 2019, 103, 1872–1876. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Ward, W.H.; Lambreton, F.; Goel, N.; Yu, J.Q.; Farma, J.M. Clinical Presentation and Staging of Melanoma. In Cutaneous Melanoma: Etiology and Therapy [Internet]; Ward, W.H., Farma, J.M., Eds.; Codon Publications: Brisbane, Australia, 2018. [Google Scholar]
- Patil, R.; Bellary, S. Machine learning approach in melanoma cancer stage detection. J. King Saud Univ.—Comput. Inf. Sci. 2022, 34, 3285–3293. [Google Scholar] [CrossRef]
- Koczkodaj, P.; Sulkowska, U.; Didkowska, J.; Rutkowski, P.; Mańczuk, M. Melanoma Mortality Trends in 28 European Countries: A Retrospective Analysis for the Years 1960–2020. Cancers 2023, 15, 1514. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Hayek, S.A.; Munoz, A.; Dove, J.T.; Hunsinger, M.; Arora, T.; Wild, J.; Shabahang, M.; Blansfield, J. Hospital-Based Study of Compliance with NCCN Guidelines and Predictive Factors of Sentinel Lymph Node Biopsy in the Setting of Thin Melanoma Using the National Cancer Database. Am. Surg. 2018, 84, 672–679. [Google Scholar] [CrossRef]
- Lideikaitė, A.; Mozūraitienė, J.; Letautienė, S. Analysis of prognostic factors for melanoma patients. Acta Med. Litu. 2017, 24, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Gao, H.J.; Zhang, J.; Badami, B. Optimization of the Convolutional Neural Networks for Automatic Detection of Skin Cancer. Open Med. 2019, 15, 27–37. [Google Scholar] [CrossRef]
- Shanthi, T.; Sabeenian, R.; Anand, R. Automatic diagnosis of skin diseases using convolution neural network. Microprocess. Microsyst. 2020, 76, 103074. [Google Scholar] [CrossRef]
- Hirano, G.; Nemoto, M.; Kimura, Y.; Kiyohara, Y.; Koga, H.; Yamazaki, N.; Christensen, G.; Ingvar, C.; Nielsen, K.; Nakamura, A.; et al. Automatic diagnosis of melanoma using hyperspectral data and GoogLeNet. Skin Res. Technol. 2020, 26, 891–897. [Google Scholar] [CrossRef]
- Fu’adah, Y.N.; Pratiwi, N.C.; Pramudito, M.A.; Ibrahim, N. Convolutional Neural Network (CNN) for Automatic Skin Cancer Classification System. IOP Conf. Ser. Mater. Sci. Eng. 2020, 982, 012005. [Google Scholar] [CrossRef]
- Lee, S.; Chu, Y.; Yoo, S.; Choi, S.; Choe, S.; Koh, S.; Chung, K.; Xing, L.; Oh, B.; Yang, S. Augmented decision-making for acral lentiginous melanoma detection using deep convolutional neural networks. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 1842–1850. [Google Scholar] [CrossRef]
- Naeem, A.; Farooq, M.S.; Khelifi, A.; Abid, A. Malignant Melanoma Classification Using Deep Learning: Datasets, Performance Measurements, Challenges and Opportunities. IEEE Access 2020, 8, 110575–110597. [Google Scholar] [CrossRef]
- Nahata, H.; Singh, S.P. Deep Learning Solutions for Skin Cancer Detection and Diagnosis. In Machine Learning with Health Care Perspective; Jain, V., Chatterjee, J., Eds.; Learning and Analytics in Intelligent Systems; Springer: Cham, Switzerland, 2020; Volume 13. [Google Scholar] [CrossRef]
- Winkler, J.K.; Sies, K.; Fink, C.; Toberer, F.; Enk, A.; Deinlein, T.; Hofmann-Wellenhof, R.; Thomas, L.; Lallas, A.; Blum, A.; et al. Melanoma recognition by a deep learning convolutional neural network—Performance in different melanoma subtypes and localisations. Eur. J. Cancer 2020, 127, 21–29. Available online: https://www.sciencedirect.com/science/article/pii/S0959804919308640 (accessed on 9 July 2023). [CrossRef]
- Gajera, H.K.; Zaveri, M.A.; Nayak, D.R. Improving the Performance of Melanoma Detection in Dermoscopy Images Using Deep CNN Features. In Artificial Intelligence in Medicine; Tucker, A., Henriques Abreu, P., Cardoso, J., Pereira Rodrigues, P., Riaño, D., Eds.; AIME 2021. Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2021; Volume 12721. [Google Scholar] [CrossRef]
- Ningrum, D.N.; Kung, W.M.; Tzeng, I.S.; Yuan, S.P.; Wu, C.C.; Huang, C.Y.; Muhtar, M.S.; Nguyen, P.A.; Li, J.Y.; Wang, Y.C. A Deep Learning Model to Predict Knee Osteoarthritis Based on Nonimage Longitudinal Medical Record. J. Multidiscip. Healthc. 2021, 14, 2477–2485. [Google Scholar] [CrossRef]
- Özbay, E.; Özbay, F.A. A cnn framework for classification of melanoma and benign lesions on dermatoscopic skin images. Int. J. Adv. Netw. Appl. 2021, 13, 4874–4883. [Google Scholar] [CrossRef]
- Zanddizari, H.; Nguyen, N.; Zeinali, B.; Chang, J.M. A new preprocessing approach to improve the performance of CNN-based skin lesion classification. Med. Biol. Eng. Comput. 2021, 59, 1123–1131. [Google Scholar] [CrossRef]
- Zhang, R. Melanoma Detection Using Convolutional Neural Network. In Proceedings of the 2021 IEEE International Conference on Consumer Electronics and Computer Engineering (ICCECE), Guangzhou, China, 15–17 January 2021; pp. 75–78. [Google Scholar] [CrossRef]
- Alwakid, G.; Gouda, W.; Humayun, M.; Sama, N.U. Melanoma Detection Using Deep Learning-Based Classifications. Healthcare 2022, 10, 2481. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.M.; Thomaz, L.A.; Tavora, L.M.; Assuncao, P.A.; Fonseca-Pinto, R.M.; Paiva, R.P.; de Faria, S.M. Melanoma classification using light-Fields with morlet scattering transform and CNN: Surface depth as a valuable tool to increase detection rate. Med. Image Anal. 2022, 75, 102254. Available online: https://www.sciencedirect.com/science/article/pii/S1361841521002991 (accessed on 9 July 2023). [CrossRef] [PubMed]
- Bhimavarapu, U.; Battineni, G. Skin Lesion Analysis for Melanoma Detection Using the Novel Deep Learning Model Fuzzy GC-SCNN. Healthcare 2022, 10, 962. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, S.; Qayyum, H.; Hassan, F.; Ullah, A.; Nawaz, A.; Ur Rahman, A. Melanoma Detection Using a Deep Learning Approach. Int. J. Innov. Sci. Technol. 2022, 4, 222–232. [Google Scholar] [CrossRef]
- Waheed, S.R.; Saadi, S.M.; Rahim, M.S.M.; Suaib, N.M.; Najjar, F.H.; Adnan, M.M.; Salim, A.A. Melanoma Skin Cancer Classification based on CNN Deep Learning Algorithms. Malays. J. Fundam. Appl. Sci. 2023, 19, 299–305. [Google Scholar] [CrossRef]
- Olayah, F.; Senan, E.M.; Ahmed, I.A.; Awaji, B. AI Techniques of Dermoscopy Image Analysis for the Early Detection of Skin Lesions Based on Combined CNN Features. Diagnostics 2023, 13, 1314. [Google Scholar] [CrossRef]
- Ragab, M.; Choudhry, H.; Al-Rabia, M.W.; Binyamin, S.S.; Aldarmahi, A.A.; Mansour, R.F. Early and accurate detection of melanoma skin cancer using hybrid level set approach. Front. Physiol. 2022, 13, 965630. [Google Scholar] [CrossRef]
- Alshawi, S.A.; Musawi, G.F.K.A. Skin Cancer Image Detection and Classification by CNN based Ensemble Learning. Int. J. Adv. Comput. Sci. Appl. 2023, 14, 2023. [Google Scholar] [CrossRef]
- Cassidy, B.; Kendrick, C.; Brodzicki, A.; Jaworek-Korjakowska, J.; Yap, M.H. Analysis of the ISIC image datasets: Usage, benchmarks and recommendations. Med. Image Anal. 2022, 75, 102305. [Google Scholar] [CrossRef]
- Popescu, D.; El-Khatib, M.; El-Khatib, H.; Ichim, L. New Trends in Melanoma Detection Using Neural Networks: A Systematic Review. Sensors 2022, 22, 496. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.G.; Lima, G.R.; Salomão, A.S.; Krohling, B.; Biral, I.P.; de Angelo, G.G.; Alves, F.C.R., Jr.; Esgario, J.G.; Simora, A.C.; Castro, P.B.; et al. PAD-UFES-20: A skin lesion dataset composed of patient data and clinical images collected from smartphones. Data Brief 2020, 32, 106221. [Google Scholar] [CrossRef] [PubMed]
- Rotemberg, V.; Kurtansky, N.; Betz-Stablein, B.; Caffery, L.; Chousakos, E.; Codella, N.; Combalia, M.; Dusza, S.; Guitera, P.; Gutman, D.; et al. A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci. Data 2021, 8, 34. [Google Scholar] [CrossRef]
- International Skin Imaging Collaboration: 2020. Available online: https://www.isic-archive.com (accessed on 9 July 2023).
- Diepgen, T.L.; Yihune, G. Dermatology Information System—Dermis. 2012. Available online: https://www.dermis.net/dermisroot/en/home/index.htm (accessed on 9 July 2023).
- Moura, N.; Veras, R.; Aires, K.; Machado, V.; Silva, R.; Araújo, F.; Claro, M. ABCD rule and pre-trained CNNs for melanoma diagnosis. Multimed. Tools Appl. 2019, 78, 6869–6888. [Google Scholar] [CrossRef]
- Patel, I.; Patel, S.; Patel, A. Analysis of Various Image Preprocessing Techniques for Denoising of Flower Images. Int. J. Comput. Sci. Eng. 2018, 6, 1111–1117. [Google Scholar] [CrossRef]
- Chithra, P.L.; Bhavani, P. A Study on Various Image Processing Techniques. Int. J. Emerg. Technol. Innov. Eng. 2019, 5, 316–322. Available online: https://ssrn.com/abstract=3388008 (accessed on 9 July 2023).
- Güneş, A.; Kalkan, H.; Durmuş, E. Optimizing the color-to-grayscale conversion for image classification. Signal Image Video Process. 2016, 10, 853–860. [Google Scholar] [CrossRef]
- Lee, T.; Ng, V.; Gallagher, R.; Coldman, A.; McLean, D. Dullrazor®: A software approach to hair removal from images. Comput. Biol. Med. 1997, 27, 533–543. [Google Scholar] [CrossRef]
- Maglogiannis, I.; Doukas, C.N. Overview of Advanced Computer Vision Systems for Skin Lesions Characterization. IEEE Trans. Inf. Technol. Biomed. 2009, 13, 721–733. [Google Scholar] [CrossRef]
- Rondón, C.V.N.; Carvajal, D.A.C.; Delgado, B.M.; Casadiego, S.A.C.; Ibarra, D.G. Body hair noise suppression in skin lesions by differential operator and maximum variance threshold between classes. In Proceedings of the 2021 IEEE 2nd International Congress of Biomedical Engineering and Bioengineering (CI-IB&BI), Bogota, Colombia, 13–15 October 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Bibiloni, P.; González-Hidalgo, M.; Massanet, S. Skin Hair Removal in Dermoscopic Images Using Soft Color Morphology. In Proceedings of the Artificial Intelligence in Medicine: 16th Conference on Artificial Intelligence in Medicine, AIME 2017, Vienna, Austria, 21–24 June 2017; Proceedings 16. Springer: Cham, Switzerland, 2017; pp. 322–326. [Google Scholar] [CrossRef]
- Maglogiannis, I.; Delibasis, K. Hair removal on dermoscopy images. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 2960–2963. [Google Scholar] [CrossRef]
- Kiani, K.; Sharafat, A.R. E-shaver: An improved DullRazor® for digitally removing dark and light-colored hairs in dermoscopic images. Comput. Biol. Med. 2011, 41, 139–145. [Google Scholar] [CrossRef]
- Koehoorn, J.; Sobiecki, A.C.; Boda, D.; Diaconeasa, A.; Doshi, S.; Paisey, S.; Jalba, A.; Telea, A. Automated Digital Hair Removal by Threshold Decomposition and Morphological Analysis. In Proceedings of the Mathematical Morphology and Its Applications to Signal and Image Processing: 12th International Symposium, ISMM 2015, Reykjavik, Iceland, 27–29 May 2015; Proceedings 12. Springer: Cham, Switzerland, 2015. [Google Scholar]
- Transfer Learning Using Pretrained Network—MATLAB & Simulink. Available online: mathworks.com (accessed on 9 July 2023).
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Net-works. Adv. Neural Inf. Process. Syst. 2012, 25. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Redmon, J. Darknet: Open Source Neural Networks in C. Available online: https://pjreddie.com/darknet (accessed on 9 July 2023).
- Wang, W.; Li, Y.; Zou, T.; Wang, X.; You, J.; Luo, Y. A Novel Image Classification Approach via Dense-MobileNet Models. Mob. Inf. Syst. 2020, 2020, 7602384. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A.; Liu, W.; et al. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar] [CrossRef]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Phoenix, AZ, USA, 12–17 February 2016; Volume 31. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar] [CrossRef]
- Shazia, A.; Xuan, T.Z.; Chuah, J.H.; Usman, J.; Qian, P.; Lai, K.W. A comparative study of multiple neural network for detection of COVID-19 on chest X-ray. EURASIP J. Adv. Signal Process. 2021, 2021, 50. [Google Scholar] [CrossRef]
- Teachable Machine. Available online: https://teachablemachine.withgoogle.com (accessed on 9 July 2023).
- Algorithm & Component Reference for Azure Machine Learning Designer. Available online: https://learn.microsoft.com/en-us/azure/machine-learning/component-reference/component-reference?view=azureml-api-2 (accessed on 9 July 2023).
- Innovate Faster with Enterprise-Ready Generative AI. Available online: https://cloud.google.com/vertex-ai (accessed on 9 July 2023).
- What Is a REST API? Available online: https://www.redhat.com/en/topics/api/what-is-a-rest-api (accessed on 9 July 2023).
- No-Fuss AI for Your App: Meet Salesforce Einstein. Available online: https://www.toptal.com/salesforce/easy-ai-cloud-salesforce-einstein (accessed on 9 July 2023).
- Giving Vertex AI, the New Unified ML Platform on Google Cloud, a Spin. Available online: https://towardsdatascience.com/giving-vertex-ai-the-new-unified-ml-platform-on-google-cloud-a-spin-35e0f3852f25 (accessed on 9 July 2023).
- What Is a Salesforce Sandbox? Available online: https://www.salesforceben.com/salesforce-sandbox/ (accessed on 9 July 2023).
- Shrivastava, M. Learning Salesforce Einstein; Packt Publishing Ltd.: Birmingham, UK, 2017; ISBN 1787125289, 9781787125285. [Google Scholar]
- Lars Malmqvist Architecting AI Solutions on Salesforce: Design Powerful and Accurate AI-Driven State-of-the-Art Solutions Tailor-Made for Modern Business Demands; Packt Publishing Ltd.: Birmingham, UK, 2021; ISBN 1801072299, 9781801072298.
- Einstein Vision and Language Model Builder: User Guide. Available online: https://quip.com/z6a4AlCUw8n3 (accessed on 9 July 2023).
- Salesforce AI Research. Available online: https://blog.salesforceairesearch.com/ (accessed on 9 July 2023).
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Menegola, A.; Tavares, J.M.; Fornaciali, M. Towards Automated Melanoma Screening: An Overview of Emerging Image Processing and Machine Learning Techniques. Arch. Dermatol. Res. 2017, 309, 679–696. [Google Scholar]
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), Hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2018, arXiv:1710.05006. [Google Scholar]
- Mendonça, T.; Marques, J.S. Comparison of Segmentation Methods for Melanoma Diagnosis in Dermoscopy Images. IEEE Trans. Med. Imaging 2013, 32, 742–753. [Google Scholar]
- Han, S.S.; Park, G.H.; Lim, W.; Kim, M.S.; Na, J.I.; Park, I.; Chang, S.E. Deep neural networks show an equivalent and often superior performance to dermatologists in onychomycosis diagnosis: Automatic construction of onychomycosis databases by region-based convolutional deep neural network. PLoS ONE 2017, 12, e0183913. [Google Scholar] [CrossRef]
- Brinker, J.; Hekler, A.; Hauschild, A.; Berking, C.; Schilling, B.; Enk, A.; Haferkamp, S.; Karoglan, A.; von Kalle, C.; Weichenthal, M.; et al. Comparing artificial intelligence algorithms to 157 German dermatologists: The melanoma classification benchmark. Eur. J. Cancer 2019, 111, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Hossin, M.; Sulaiman, M.N. A Review on Evaluation Metrics for Data Classification Evaluations. Int. J. Data Min. Knowl. Manag. Process 2015, 5, 1–11. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Kaur, R.; GholamHosseini, H.; Sinha, R.; Lindén, M. Melanoma Classification Using a Novel Deep Convolutional Neural Network with Dermoscopic Images. Sensors 2022, 22, 1134. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Kamrul Hasan, M.; Ahmad, M. Skin Lesion Classification Using Convolutional Neural Network for Mela-noma Recognition. In Proceedings of the International Joint Conference on Advances in Computational Intelligence, Birulia, Bangladesh, 20–21 November 2020; Uddin, M.S., Bansal, J.C., Eds.; Algorithms for Intelligent Systems. Springer: Singapore, 2021. [Google Scholar] [CrossRef]
- Rezaoana, N.; Hossain, M.S.; Andersson, K. Detection and Classification of Skin Cancer by Using a Parallel CNN Model. In Proceedings of the 2020 IEEE International Women in Engineering (WIE) Conference on Electrical and Computer Engineering (WIECON-ECE), Bhubaneswar, India, 26–27 December 2020; pp. 380–386. [Google Scholar] [CrossRef]
- Tahir, M.; Naeem, A.; Malik, H.; Tanveer, J.; Naqvi, R.A.; Lee, S.-W. DSCC_Net: Multi-Classification Deep Learning Models for Diagnosing of Skin Cancer Using Dermoscopic Images. Cancers 2023, 15, 2179. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).