1. Introduction
Additive manufacturing, otherwise known as three-dimensional (3D) printing, is a rapidly growing technology whereby products are built on a layer-by-layer concept [
1,
2]. In many industrial fields, 3D printing has become central in product manufacturing since new shapes and prototypes can be realized after a suitable digital design.
In recent decades, several advances have enabled the 3D printing of biocompatible materials together with cells (bioinks) to produce complex 3D functional living-tissue-like constructs [
3,
4]. In this case, biocompatible materials such as hydrogels are used as printable elements instead of the plastic materials that are normally used for 3D printing. The bioink used to print cells consists of living cells, biocompatible biomaterials and/or active biomolecules [
5]. Due to these characteristics, bioprinting holds the promise of establishing systems that more closely mimic human in vivo microenvironments than current two-dimensional (2D) cell culture environments, filling the gap between 2D and animal models [
6,
7].
In the world, many researchers are working to adapt techniques and materials to the 3D bioprinting of cell models and tissues to be used in the field of tissue engineering and regenerative medicine, which is a key area in the biomedicine of the future. Nevertheless, while it is capable of creating new 3D cell models to answer many biological questions, the technology has remained inaccessible to most research labs due to the high price of commercially available bioprinters (USD 100,000–200,000) and low customization capacity, making it difficult for research groups to enter the field of bioprinting [
8].
Three-dimensional bioprinting is mainly based on three approaches: extrusion-, droplet- or laser-based bioprinting. Extrusion-based bioprinters, which are derived from inkjet printers, are the most commonly used type of printers for biological applications, and are designed for the high-precision deposition of bioinks [
9]. The main techniques in extrusion bioprinting use valve-based pneumatic or gear-driven actuator extrusion systems to drive cell suspensions out of a suitable needle tip.
The current professional bioprinters enable high-precision results by providing a sterile environment and cell-compatible printing conditions but with high overall costs. What is currently missing is a range of low-cost bioprinters to approach bioprinting techniques for simple applications.
Theoretically, 3D printing and bioprinting processes are based on the same printing principle: laying down materials or bioinks layer by layer in order to build a previously digitally designed 3D construct. Nevertheless, when bioinks are used, there are many problems to be considered; first of all, cell survival must be ensured in a bioprinting construct. For example, some researches have reported that the dispenser orifice size (150 μm) and the extrusion rate are fundamental parameters to consider for cell survival since they can control the mechanical stress to which the cells are subjected [
10].
To date, several studies have been carried out to try to adapt commercial 3D printers to biological purposes [
11,
12,
13,
14]. The use of commercial and low-cost 3D printers, modified for biological and medical research, allows for a great deal of freedom of maneuver to investigate specific objectives, including different printing methods, materials and the impact of environmental conditions on cells. However, to better evaluate the possibility of using commercial 3D printers instead of professional bioprinters for biological and medical applications, some features of the printing process have to be compared.
This study describes the implementation of a commercial 3D printer to obtain a 3D bioprinter and benchmarks its performance against a professional bioprinting tool taken as a reference. We also demonstrate that this bioprinter can be used to print a construct model of a vascularized medulloblastoma tumor (MB). MB is a cerebellar tumor that represents the most prevalent malignant brain tumor in children [
15,
16]. The possibility to intervene on the tumor through chemotherapy strongly depends on the presence and integrity of the blood–brain barrier (BBB), an endothelial structure that strictly controls the brain penetration of many molecules, including chemotherapy agents. The constructs we created using 3D bioprinting include the BBB that is present in the tumor in order to test the penetration of drugs through it.
We demonstrate the ability of our modified 3D printer to print the MB tumor model without affecting the cell vitality compared to the same printed model using a professional 3D bioprinter.
2. Materials and Methods
2.1. Modification of a Commercial Kentstrapper Verve 3D Printer in a BioVERVE Bioprinter
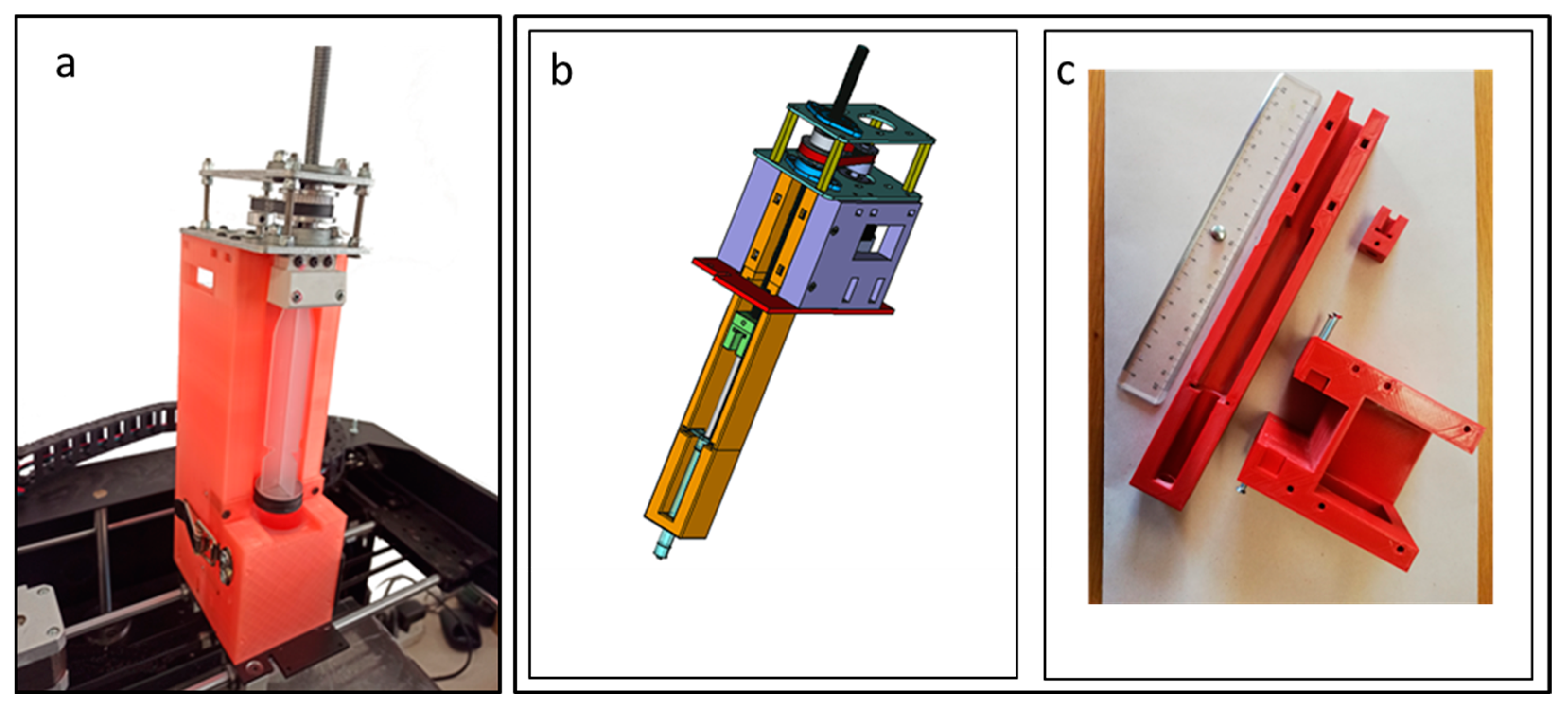
The aim of this work was to modify a commercial 3D printer (Kentstrapper Verve, Kentstrapper S.r.l., Florence, Italy) to create a bioprinter (BioVERVE) by replacing the default extruder used to print plastic filaments with a custom-made syringe extruder suitable for printing bioinks. The original syringe extruder was designed by Kentstrapper S.p.A. to accommodate a huge 60 mL luer-lock syringe and the ensemble of a step motor, gears, transmission belt, bearings and helical screw used to control the movement of the syringe plunger (
Figure 1a).
The 60 mL syringe extruder was used to verify the correct operation of the 3D printer in executing the G-code containing the print commands. Moreover, a first tuning of the syringe plunger movements was performed. The commands for the printer were provided as a G-code file. In particular, the correct parameter value for the M92 G-code (set axis step per millimeter) that determines how many turns of the step motor are needed to advance the syringe plunger of 1 mm was established. A value of 50 was found to be imposed as M92 parameter to print commercial silicone (i.e., 50 motor turns for 1 mm plunger advance).
The original Kentstrapper extruder was later modified to allocate a smaller 1 mL luer-lock syringe (more suitable for cell bioprinting purposes) and to reduce the overall size of the syringe extruder above the printer (
Figure 1b,c).
The new extruder design was largely inspired by the Kentstrapper’s design. The main modifications were related to the reduced size of a 1 mL syringe and to the reduction in the extruder size above the printer. In consideration of the small dimensions of the cell construct to be printed, the syringe was positioned below the rods sustaining the extruder for the
x-y motion, leaving just the step motor above the printer (
Figure 2).
Such syringe arrangement was chosen to realize an upper cover for the printer in order to protect the printing environment from outer contaminants.
The new adapter was entirely designed using FreeCAD (
https://www.freecadweb.org, (accessed on 20 September 2021)), a freeware and open-source parametric 3D modeler. The resulting model was subsequently sliced using Ultimaker Cura (V4.13, Ultimaker, Utrecht, the Netherlands—
https://github.com/Ultimaker/Cura/releases (accessed on 20 September 2021)), a freeware and open-source slicing application for 3D printers. The resulting G-code file was printed with a Kentstrapper Mavis 3D printer using polylactic acid (PLA) filaments. In order to print bioinks, the correct M92 G-code parameter value, which largely depends on the viscosity of the specific bioink used, the syringe nozzle diameter and, ultimately, the desired spatial resolution, was determined.
Several attempts were performed to establish the correct M92 value to be used in the bioprinting experiments, taking into account the ink viscosity. To this aim, conical 25 G nozzles were used to print a simple ring (5 mm diameter and 1 mm height), and several inks were tested (i.e., Cellink Xplore, Cellink Start and Cellink Laminink 411, Cellink Life Sciences, Göteborg, Sweden).
2.2. Printer Testing
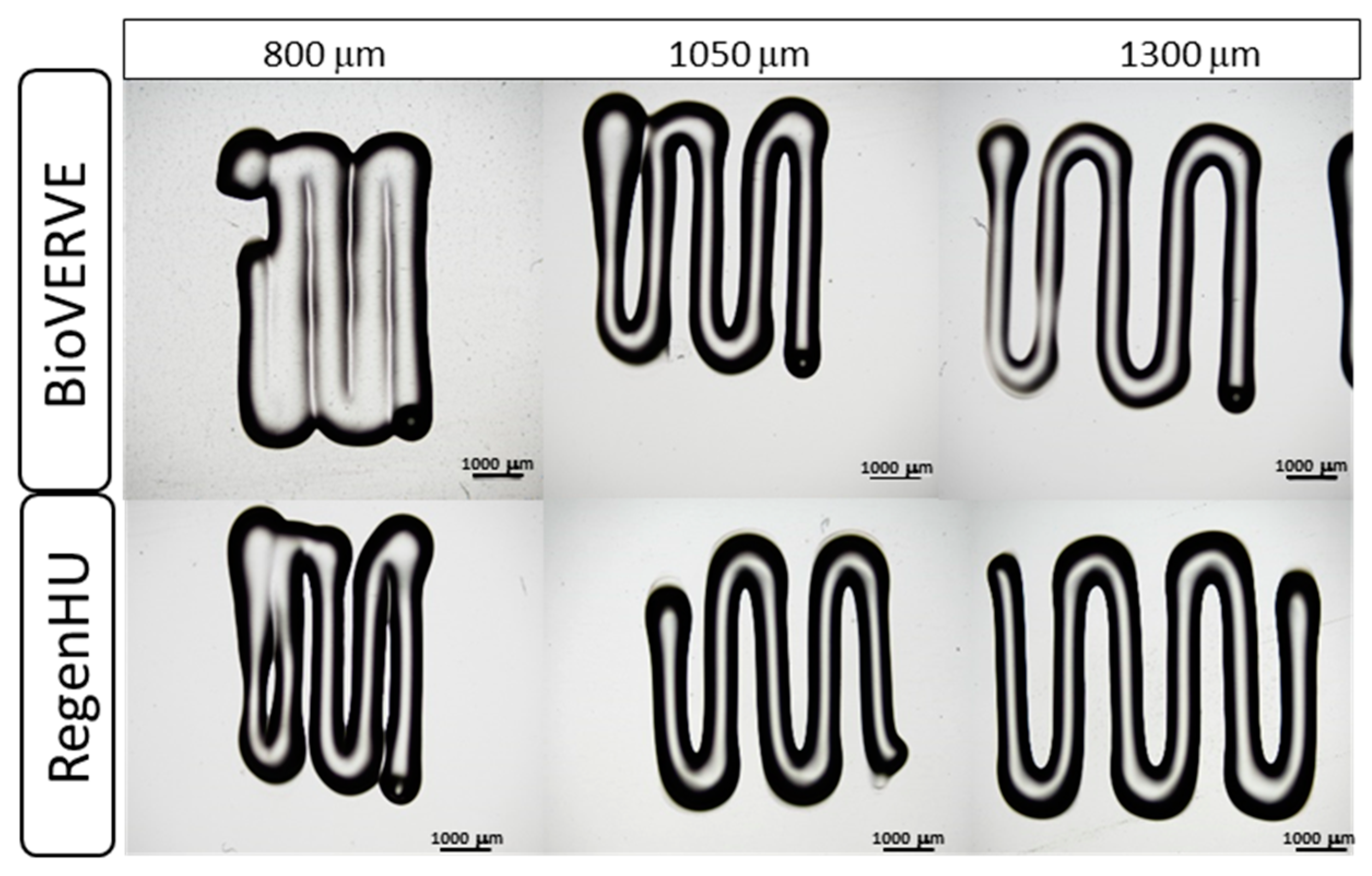
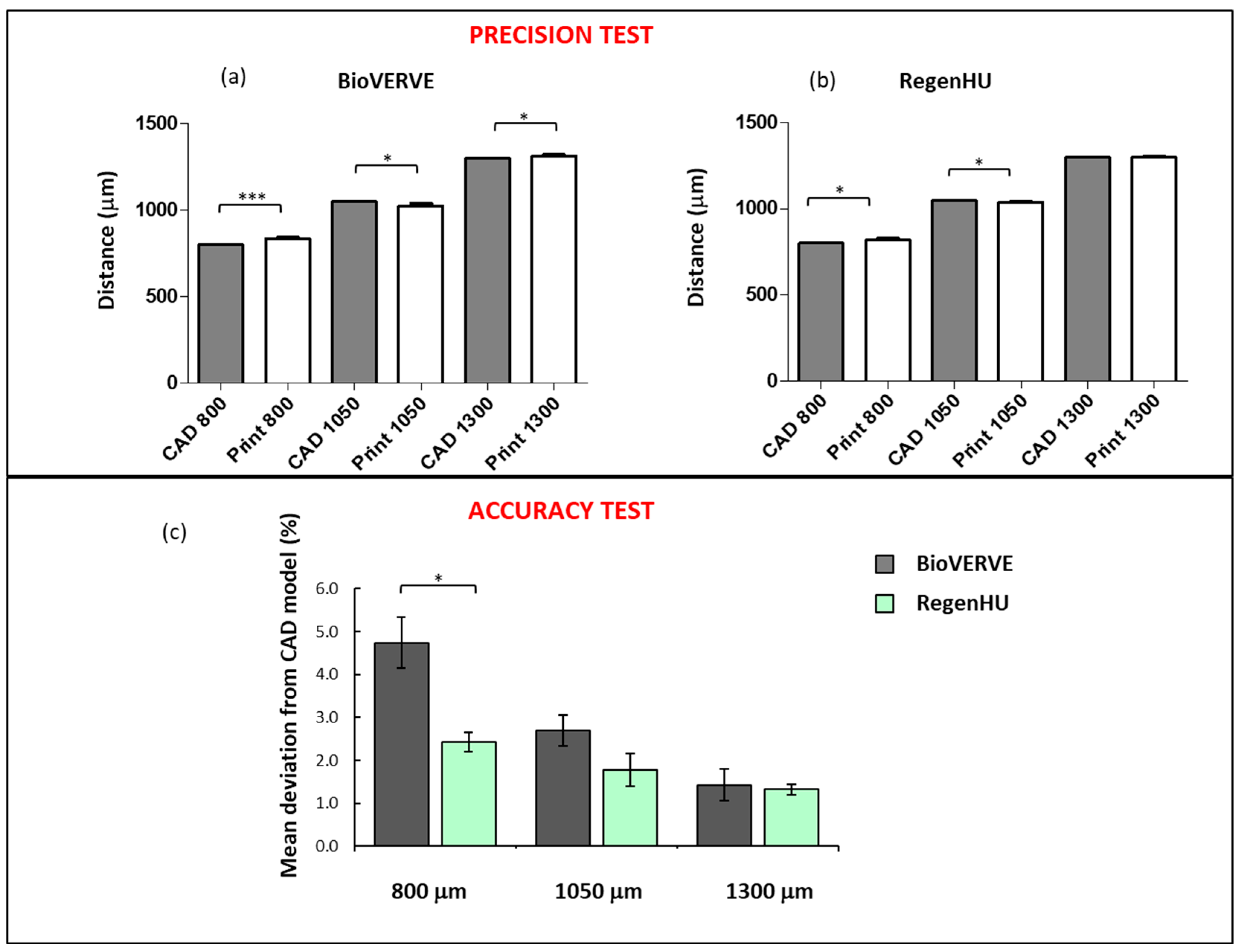
To evaluate the precision and accuracy of the printer (repeatability and fidelity to a specified CAD model, respectively), three Cellink Start hydrogel rectilinear line patterns consisting of five segments with varying distances (CAD model distances: 800, 1050 and 1300 μm) were printed using both the BioVERVE bioprinter and a professional 3D Discovery bioprinting platform (RegenHU, CH). Conical 25G nozzles and a velocity of 1 mm/sec were used to print the hydrogel with both bioprinters. The “UltiMaker Cura V4.13” software was used to design the CAD model for BioVERVE, while a dedicated CAD software (BioCAD, RegenHU, CH) was used to design the 3D structures to be printed via RegenHU. The three rectilinear patterns were printed and measured for each line distance. After printing, constructs were observed under an optical microscope, and the distance between the centers of each line segment, measured in three points along the line, was measured. Three experimental replicates were analyzed for each rectilinear line pattern printed with each bioprinter. A Nikon Eclipse 80i microscope (Nikon Instruments Europe B.V., Italy) was used to take images and the Nikon NIS-Element BR software (Nikon Instruments Europe B.V., Italy) was used to measure distances between lines in the BioVERVE constructs, while a Nikon Eclipse E880 microscope (Nikon Instruments Europe B.V., Italy) was used to take images and the Nikon NIS-Element BR software (Nikon Instruments Europe B.V., Italy) was used to measure distances between lines in the RegenHU constructs.
In the accuracy test, the mean percentage deviation (D%) of the line spacing distances of the printed model from the corresponding distances established in the CAD model was calculated as follows:
where d
CAD is the fixed distance in the specific CAD model and d
i represents the measured distances between the midpoints of the printed lines. The mean percentage deviations were compared using
t-test for both bioprinters.
2.3. Cell Culture and Cell Fluorescent Dye Labeling
Human medulloblastoma cell line (DAOY) was obtained from American Type Culture Collection (ATCC; Manassas, VA, USA) and routinely maintained in the complete growth medium Eagle’s Minimum Essential Medium (MEM) with 2 mM glutamine and 100 U penicillin/0.1 mg/mL streptomycin, supplemented with 10% fetal bovine serum.
Human endothelial cell line (HUVEC) was purchased from Lonza (Walkersville, MD, USA) and maintained in EGM-2 medium (Lonza) supplemented with EGMTM-2 SingleQuotsTM Kit (Lonza). Cells were cultured in standard CO2 incubators (90% humidity and 5% carbon dioxide) at 37 °C.
To visualize the arrangement of the DAOY and HUVEC cells within the model, the two cell lines were labeled using vital dyes. Briefly, DAOY and HUVEC cell lines were harvested using 0.05% trypsin, collected in suspension, counted and adjusted to a final concentration of 1 × 106 cells/mL. An amount of 5 μL/mL of either Vybrant DiI (red) or DiO (green) cell-labeling solution (Invitrogen, CA, USA) was added to each vial containing HUVEC or DAOY cells, respectively. The cell/dye mixture was then incubated for 15 min at 37 °C before being centrifuged at 200× g for 5 min (HUVEC) or at 130× g for 7 min (DAOY) to separate the supernatant containing residual dye. The remaining cell pellet was then washed twice in medium and used for the bioprinting.
2.4. Three-Dimensional Cell Bioprinting
The 3D in vitro MB cell model was printed using two different 3D extrusion bioprinters: a professional 3D Discovery platform (RegenHU, CH) accommodating four different print heads and a modified low-cost commercially available printer (VERVE printer, Kentstrapper S.p.A) with one print head (
Figure 2). Cellink Laminink 411 was purchased from Cellink (Cellink, USA) and used to print both cell lines at a cell density of 1 × 10
6 cells/mL. A 25G conical standard nozzle (Cellink, USA) was used. A tumor model characterized by a circle tumor core with a 5 mm diameter (DAOY, green cells), an endothelial ring (HUVEC, red cells) around the tumor core about 250 μm thick and a total height of about 2 mm was designed and printed. Cells were printed in sterile 12 well-plates (Corning Inc., New York, NY, USA) and crosslinked for 5 min using the Cellink Crosslinking Agent containing 50 mM calcium chloride (Cellink, Göteborg, Sweden).
2.5. Cell Viability Assays
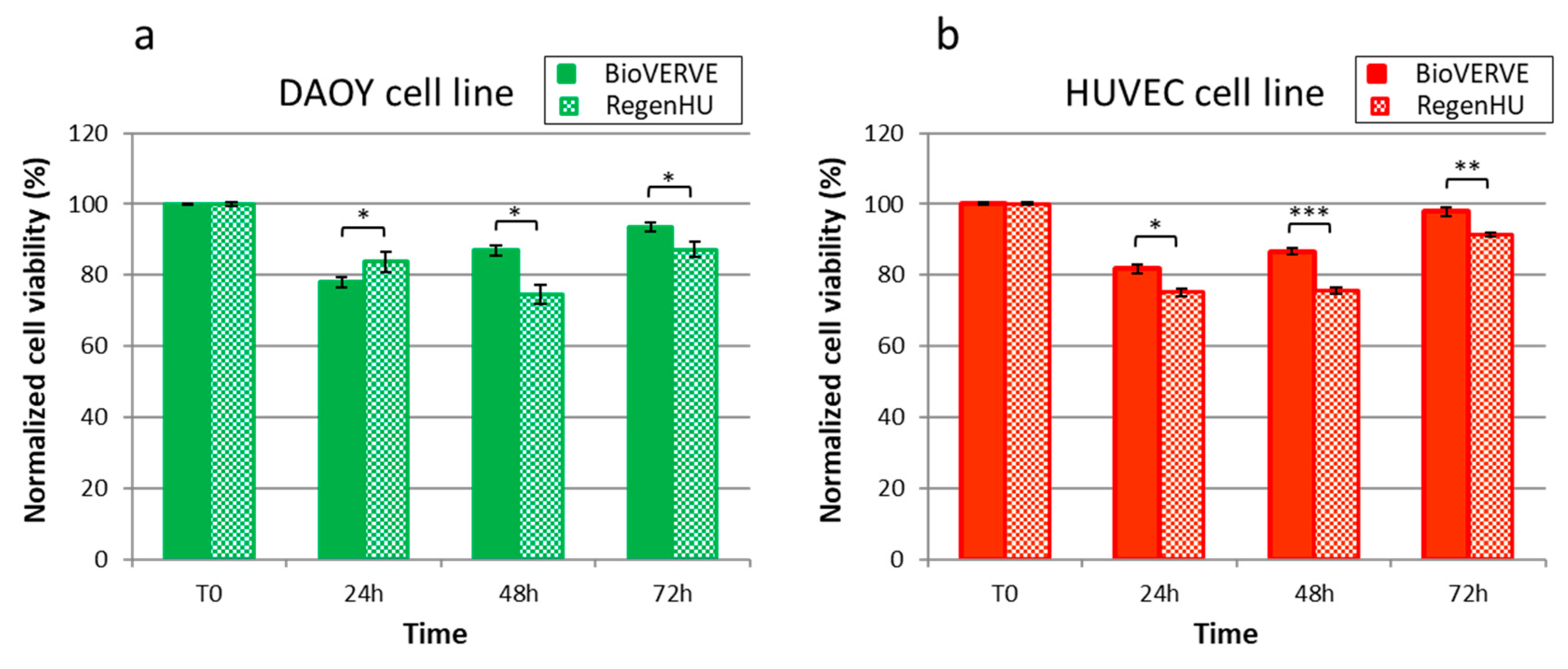
Three-dimensional bioprinted DAOY and HUVEC cells’ viability was assessed over time (24, 48 and 72 h post printing) by using LIVE/DEAD™ Cell Imaging Kit (488/570) (Thermo Fisher Scientific, Waltham, MA, USA), which allows for the visualization of live (green) and dead (red) cells. Briefly, the constructs were washed once with PBS, and Live/Dead reagent was added in a 1:1 ratio with phosphate-buffered saline (PBS) for 15 min at room temperature. Constructs were then observed with a Zeiss Axio Observer inverted fluorescent microscope using FITC and TRITC filters. Images were taken using the Zeiss ZEN PRO software, and live and dead cells lying in the same fields were manually counted. Three constructs were analyzed for each bioprinters, and at least 500 cells were counted/constructed. Cell viability was calculated as the percentage of live and dead cells on the total count and data were normalized to the unprinted cells’ viability (T0) for comparison.
2.6. RegenHU 3D Bioprinting
A dedicated CAD software (BioCAD, RegenHU, CH) was used to design the 3D structures. A high-precision microvalve-based dispensing technology (pneumatic printhead), used in contact mode with a dedicated 300 µm needle, was used to print the DAOY cells (tumor core), while a high-precision plunger dispenser was used to print the endothelial cells. For the pneumatic printhead, an extrusion pressure of 64 ± 1 MPa and a dosing distance of 0.1 mm were fixed to print the DAOY bioink, while for the plunger dispenser, a plunger velocity of 0.03 mm/s, corresponding to a flow rate velocity of 0.82 μL/s, was fixed to print the HUVEC bioink. Both print heads were kept at room temperature (RT) throughout the experiment. Sterility was guaranteed by the printing platform being integrated in a class II laminar flow hood.
2.7. BioVERVE 3D Bioprinting
The “UltiMaker Cura V4.13” software was used to design the model. The home-made BioVERVE bioprinter (
Figure 2) was equipped with a single 1 mL syringe extruder operated by a stepper. The tumor core (DAOY cells) and the endothelial ring (HUVEC cells) were printed sequentially by replacing the syringe, and the printing was performed at RT throughout the experiment.
For the BioVERVE bioprinter, an M92 value of E125 and a flux of 50% were fixed as printing conditions. Sterility was guaranteed with the BioVERVE bioprinter being housed in a class II laminar flow hood.
2.8. Statistical Analysis
All data were evaluated with GraphPad Prism 6 statistical software (GraphPad Software Inc., San Diego, CA, USA). Statistical significance between groups for most variables was determined using Student’s two-tailed t-test. p-values < 0.05 were considered to be statistically significant.
4. Conclusions
The main aim of our work was to adapt a 3D commercial printer designed for polymer prints to print bioink containing cells with a high performance. The need to use low-cost bioprinters is self-explanatory and increasingly evident. Three-dimensional bioprinting technology is gaining more and more traction and importance as a key technology for studying the effects of the extracellular environment on morphology, biology and cellular responses under particular conditions, but at the same time, it is an economically inaccessible technology for many laboratories.
In this work, we outlined a straightforward procedure for creating an affordable and highly accurate 3D bioprinter by modifying a readily available and inexpensive off-the-shelf 3D printer. Moreover, this system is well suited for customization and can be easily adapted by both basic and clinical research laboratories to investigate cellular interactions and/or for applications in tissue engineering.
The results reported in this article show how it would be possible, in the future, to have access to low-cost 3D bioprinters, expanding the accessibility of in vitro 3D bioprinted models to biomedical research. This type of system is ideal for performing experiments to study the effect of the 3D extracellular environment on cell biology and cellular responses to intrinsic and extrinsic agents acting on them. The low cost of the BioVERVE bioprinter and the acceptable performance in terms of printing resolution and cell survival, as benchmarked against the commercial reference obtained from RegenHU, make this bioprinter a good compromise between cost and performance.
Several works have reported different approaches to obtain a home-made extrusion bioprinter [
12,
14,
17,
18,
19]. In this study, we not only present the details of the home-made BioVERVE bioprinter, but also compare its performance with that of the professional RegenHU bioprinter. Of course, the professional RegenHU bioprinter has definitive advantages over the home-made BioVERVE, for example, greater simplicity in the model design via dedicated onboard CAD software and the possibility of combining multiple printing technologies (up to seven in the specific version at hand) in one multi-layer, multi-phase biomanufactured construct. However, minor significant differences are evident between the two bioprinters in terms of cell survival, thus suggesting that the printing process from both platforms affects cell integrity essentially in the same way. Furthermore, BioVERVE allowed us to obtain a good printing accuracy, which was close to that obtained with the RegenHU bioprinter, opening new opportunities for biomedical research. Implementing this type of 3D printer can open up new avenues for research, making these intricate machines more accessible for routine laboratory experimentation. By reducing the entry barrier, researchers can explore fresh perspectives and engage in a wider range of studies.