1. Introduction
The synovial joint consists of contiguous bony surfaces covered by hyaline cartilage, connected by ligaments, and lined by a synovial membrane (SM); the capsule surrounds it. The SM includes two layers: the intima and the subintima [
1]. The intima represents the SM inner part and comprises one or two sheets of macrophages (Type A synoviocytes) and fibroblast-like synoviocytes (FLS) (Type B synoviocytes). Cells from the intima secrete the synovial fluid (SF), which provides articular cartilage lubrication, chondrocyte activity, and nutrition. In particular, Type B synoviocytes produce a glycosaminoglycan termed hyaluronan, which strongly contributes to the SF thick mucoid consistency [
2,
3]. The subintima is the SM outer part and comprises two-to-three layers of synoviocytes lying over loose connective tissue rich in fibroblasts.
Studies on SM have increased in previous years, showing progress in understanding the etiopathogenesis of common rheumatic diseases, such as osteoarthritis (OA) or rheumatoid arthritis (RA). In those pathologies, synovitis is directly responsible for several clinical symptoms and reflects the pathological progression [
4]. However, suitable in vitro models are still fundamental to fully investigate the factors contributing to inflammation [
5].
In vitro models facilitate the investigation and understanding of specific biological structures and processes. Conventional models rely on experimental cell cultures on monolayer or two-dimensional (2D) substrates. Generally, such culture models are used to perform preclinical analysis, but their predictive accuracy is poor since they cannot recapitulate the complexity of human tissues.
Studies in an appropriate animal model can help extrapolate in vitro data to clinical outcomes. However, animal sacrifice presents ethical issues and should be limited. Moreover, the differences between the animal model and the human body in terms of physiology and pathology may adversely impact the translation of the results achieved and the predictive power of drug efficiency [
6].
Novel biology and tissue engineering technologies have promoted the development of more sophisticated and robust in vitro models such as three-dimensional (3D) scaffolds, hydrogel-based formats, organoids, and organs-on-chips. Those models can better recapitulate the in vivo microenvironment, thus representing a promising alternative to the current state-of-the-art.
So far, the attempts at recapitulating the synovial tissue within in vitro models to study rheumatic diseases are limited and based on the inclusion of synovial fluid or synovial fibroblasts within 2D and 3D monoculture or co-culture models or the study of tissue explants [
6,
7]. However, poor physiological relevance and predictiveness of drug efficiency characterize these conventional models.
In recent years, more advanced solutions, like 3D microsystems, organoids, or microfluidic organ-on-chip approaches, were developed to recapitulate the synovium itself [
6,
7,
8,
9]. Though the use of multicompartment chips represents a noticeable improvement in the synovial joint in vitro modeling, these approaches still do not provide the possibility to replicate the synovium within a relevant-size three-dimensional engineered tissue, which would provide an interesting alternative to model the complexity of the in vivo environment in a representative fashion.
To this purpose, novel biofabrication methods like 3D printing could be combined with microscale technologies (i.e., soft lithography) and advanced biological systems, resulting in more complex culture systems tailored for specific applications [
10]. Those engineered platforms can provide additional degrees of flexibility and control over cell function and fate and, thus, build tissues that better emulate the dynamics of the in vivo conditions. Additionally, the models mentioned above allow for the monitoring of the pathophysiological conditions to study disease onset and progression and identify pathogenic factors, biomarkers, and potential therapies [
11].
Three-dimensional bioprinting technology enables the fabrication of structures with characteristics and functions as closely as possible to natural tissues by combining biomaterials, live cells, and active biomolecules in a bottom-up, layer-by-layer deposition through “Additive Manufacturing Technology.” Deposition may occur by different mechanisms such as extrusion, inkjet, or laser. In most cases, the created 3D tissue constructs require the action of ultraviolet (UV) light, chemical stimuli, or a heat source to achieve the desired mechanical properties, retain their shape, and provide the proper growth environment. Thanks to its high degree of control, 3D bioprinting has proved to be a crucial research technique for drug discovery, functional organ replacement, and regenerative medicine [
11,
12,
13].
In this study, we exploited volumetric extrusion bioprinting technology to fabricate a 3D-engineered construct composed of human fibroblast-like synoviocytes cells line (K4IM) embedded in a GelMA bioink. This study represents a proof of concept (PoC) to develop in the future a bioartificial SM and use it to identify the mechanisms involved in the onset and progression of many rheumatic diseases and to test novel therapeutic treatments.
2. Materials and Methods
2.1. Bioink Synthesis
GelMA was synthesized following the general protocol first reported by Van Den Bulcke et al. [
14]. Briefly, 10% weight/volume (
w/
v) Type B gelatin from bovine skin (Sigma Aldrich, St. Louis, MO, USA, gel strength 50–120 bloom) was dissolved into Dulbecco’s Phosphate Buffered Saline (DPBS, Sigma) at 50 °C for 1 h. Then, 4 mL every 100 mL of phosphate buffered saline (PBS) of Methacrylic Anhydride (MA, Sigma) was slowly added to the solution, with a 2 h reaction in dark at 40 °C, to introduce the methacryloyl groups to gelatin’s reactive amine and hydroxyl groups. The reaction was stopped by diluting the reaction mixture with DPBS. The resulting solution was dialyzed against deionized water with a cellulose membrane (12–14 kDa molecular weight cutoff, Sigma) for 1 week at 40 °C to remove low molecular weight impurities that could be cytotoxic. Finally, GelMA was freeze-dried. GelMA Degree of functionalization (DoF) was determined by using fluoraldehyde o-phthalaldehyde (OPA) reagent, which reacts with primary amines [
15]. In particular, GelMA was dissolved in PBS at the concentration of 2 mg/mL. Then, 300 μL of GelMA solution was mixed with 600 μL of fluoraldehyde o-phthalaldehyde reagent solution. Gelatin was used as a positive control. After 1 minute, fluorescence intensity was read by using Synergy™ HTX Multi-Mode Microplate Reader (BioTek, Winoosky, VT, USA) at 450 nm (Eexc = 360 nm). The DoF is then calculated as DoF = [1 − (Isample-Icontrol)]. The DoF value found was 70%.
Three different bioink formulations were evaluated as ink candidates, with a GelMA content of, respectively, 7.5, 10, and 12.5% w/v in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) photoinitiator concentration was kept constant at 0.25 w/v%.
To prepare the bioinks, 10 mg of LAP were first dissolved in 4 mL of culture medium. Then, the respective amount of lyophilized GelMA was gradually added to the solution until complete solubilization was observed. The process was performed in a heated bath at 37 °C to increase GelMA solubility and in a UV-protected container to avoid premature material crosslinking.
To sterilize the prepared ink formulations, the UV-protected containers were first placed in a heated bath at 60 °C. Subsequently, the different GelMA inks were filtered in a two-step approach using 0.45 and 0.22 micron polyethersulfone (PES) filters under laminar flow.
2.2. Bioink Characterizations
2.2.1. Swelling Test
GelMA’s degree of swelling was evaluated in distilled water at room temperature [
16]. 3D-printed samples were immersed in water for 2 days until the swollen samples reached an equilibrium, then removed, dried, and weighed. The swelling degree was evaluated applying the general formula: Swelling (%) = (Ws − Wi)/Wi × 100, where Ws = weight of swollen hydrogel and Wi = initial weight of hydrogel. The swelling experiments were performed at least three times. Differences between groups were analyzed by three-way ANOVA.
2.2.2. Rheological Measurement
Rheological measurements were performed through a rheometer (Physica MCR 302, Anton Paar, Graz, Austria) using a parallel plate configuration (φ = 15 mm) with a quartz bottom glass. For the experiments, a 200 μL sample was poured onto the plate and a UV-light at an intensity of 30 mWcm
−2 was placed under the quartz plate. The UV-light was switched on after 60 s to allow the system to stabilize before the polymerization onset. The obtained hydrogels were, thereafter, subjected to frequency sweep (frequency ω 0.1–100 rad/s with a constant γ = 1%) tests, from which the Young’s Modulus
E was evaluated [
17,
18]. All experiments were carried out at T = 37 °C. All rheological measurements were performed in triplicate.
2.2.3. Field Emission Scanning Electron Microscopy (FESEM)
The characterization of GelMA samples from a morphological point of view was performed by using a FESEM Zeiss Supra 40 (Oberkochen, Germany). To prepare the samples, 50 μL of GelMA were poured into a 96 well-plate and photopolymerized as previously described. Then, samples were incubated overnight with 200 µL of RPMI and lyophilized at −50 °C. Before characterization, the samples were coated with a film of Pt 5 nm thick [
19].
2.3. Structure Design and Printing Process Optimization
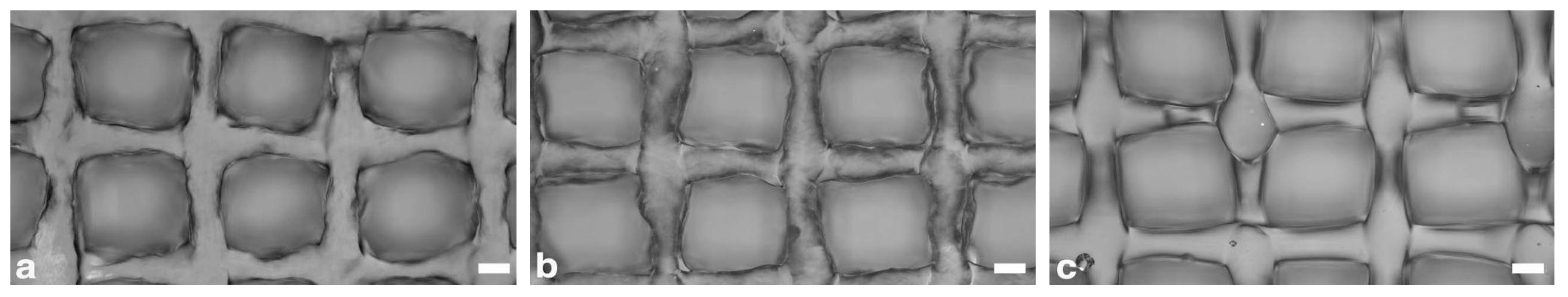
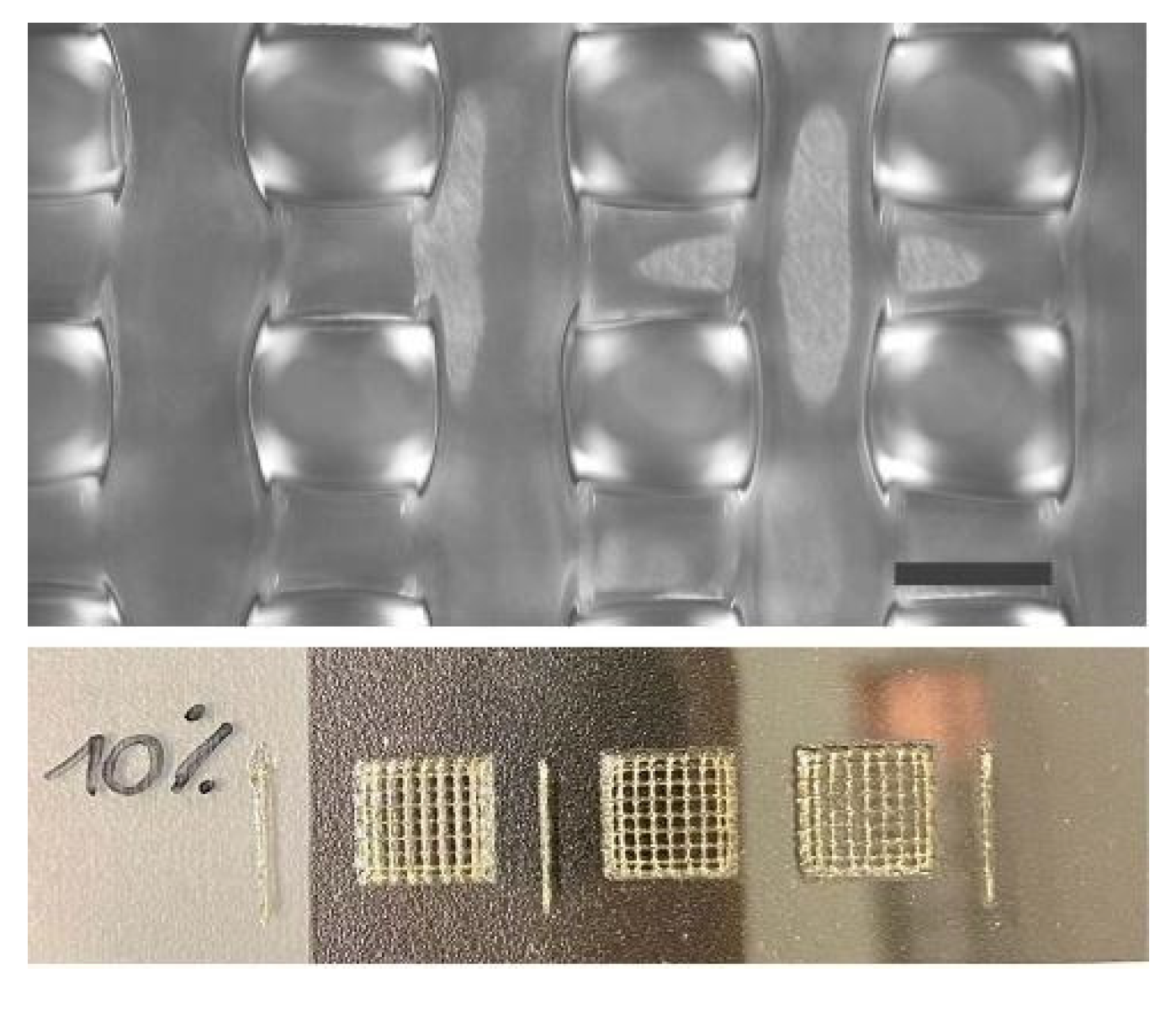
Three-dimensional scaffolds were designed through the BioCAD 1.1 software (REGENHU, Villaz-St-Pierre, Switzerland). The structures were characterized by a 6 × 6 mm square base, and a 0/90° grid pattern infill. The fiber diameter was set to 200 µm and the interfilament distance (center to center) to 1 mm, giving a theoretical pore size of 800 µm. A total of 16 layers were stacked for each scaffold, and after each layer deposition, a static UV irradiation was programmed using the software.
A first series of printing tests was performed on the different GelMA ink formulations (7.5, 10, and 12.5% w/v) prior to cell addition to optimize the process parameters.
All the prints were performed using a 3D Discovery Evolution platform (REGENHU, Villaz-St-Pierre, Switzerland). A volumetric dispensing technology was selected to provide improved control over the extruded flow rate, and thermal control options were applied to both the printhead and the substrate to exploit the high thermosensitivity of the ink.
After each deposited layer, a 15 s UV (wavelength 365 nm) irradiation was provided using the integrated light-curing kit to crosslink the deposited material and improve shape fidelity and stacking capabilities. A 200 µm cylindrical nozzle with a 12.7 mm length was used for all the tests, and the layer height was set to 100 µm, giving a final structure height of 1.6 mm.
Different printing parameter sets were tested to identify the optimal process conditions, and the fabricated structures were analyzed using a bright-field microscope (Eclipse 90i, Nikon Inc., Tokyo, Japan) to evaluate the correspondence of the internal microarchitecture to design parameters.
The tested parametric range for the ink formulations (GelMA 7.5, 10, and 12.5%
w/
v) are reported in
Table 1.
In a second step, the printing parameters optimization was repeated on the GelMA 10% w/v formulation, selected for the bioprinting experiments, with a 400 µm inner diameter and 25.4 mm length needle. These changes were decided to overcome some limitations encountered in the processability of the cell-laden bioinks with a 200 µm needle during preliminary bioprinting tests.
The printing parameters optimization followed the same procedure as before. The layer height was increased to 200 µm, and the number of layers was accordingly reduced to 8 to maintain a total construct height of 1.6 mm.
2.4. Immortalized Synoviocyte K4IM Cells
Prof. Murphy E. (School of Veterinary Medicine, University College, Dublin) kindly provided the immortalized human fibroblast-like synoviocyte cell line termed K4IM. It is a stable cell line obtained by immortalizing with SV40 T antigen (TAg) human synoviocytes from a patient undergoing meniscectomy. K4IM cells express intercellular adhesion molecules such as ICAM-1 (CD44, CD54) and CD95 (Fas), but not CD106 (vascular cell adhesion molecule 1; VCAM-1), platelet-derived growth factor (PDGF), and the receptors for interleukin1 (IL-1). They differ only in part from the parental wild-type phenotype [
20].
K4IM cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Sigma-Aldrich, St. Louis, MO, USA) supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum (FBS, Euroclone S.p.A., Milan, Italy).
2.5. 3D Bioprinting
The printing process was performed using a 3D Discovery Evolution platform (REGENHU, Villaz St. Pierre, Switzerland) integrated inside a Class II biosafety cabinet to prevent biological material contamination.
Three-dimensional cell-laden scaffolds were fabricated using the GelMA 10%
w/
v formulation as the basis for the bioink. The ink was prepared and sterilized according to the procedure reported in
Section 2.1. Then, K4IM cells with a density of 1.5 million/mL were resuspended within the biomaterial, previously kept at 37 °C to facilitate the process. The cells were carefully suspended in the biomaterial and dispensed into the cartridge, avoiding formation of air bubbles and cellular aggregates. Before printing, to prevent cell sedimentation effects, the bioink cartridge was briefly placed on ice to allow a first physical crosslinking of the bioink.
Subsequently, the cartridge was loaded within the volumetric extrusion printhead of the 3D Discovery Evolution, already kept cooled at 15 °C. The process was automatically performed within 12 well-plates, previously loaded on the cooled printing collector to maintain them at 6 °C for the whole process.
Square-based scaffolds of 6 × 6 mm with an infill pattern of 0/90°, a layer height of 200 µm, and a final height of 1.6 mm were printed using the optimized printing procedure. Twelve scaffolds could be fabricated using a single 2.5 mL cartridge, leading to a theoretical density of approximately 300.000 cells per scaffold.
The main process parameters are reported in
Table 2.
Immediately after printing, scaffolds were cultured with DMEM supplemented with 1% penicillin/streptomycin and 10% of FBS at 37 °C with 5% CO2. Scaffolds were then maintained in culture for up to 14 days with fresh medium change every 2 days. Scaffolds were collected on Days 0, 1 and 3 and Weeks 1 and 2 for biological tests.
2.6. Biological Assays
2.6.1. Live and Dead Assay
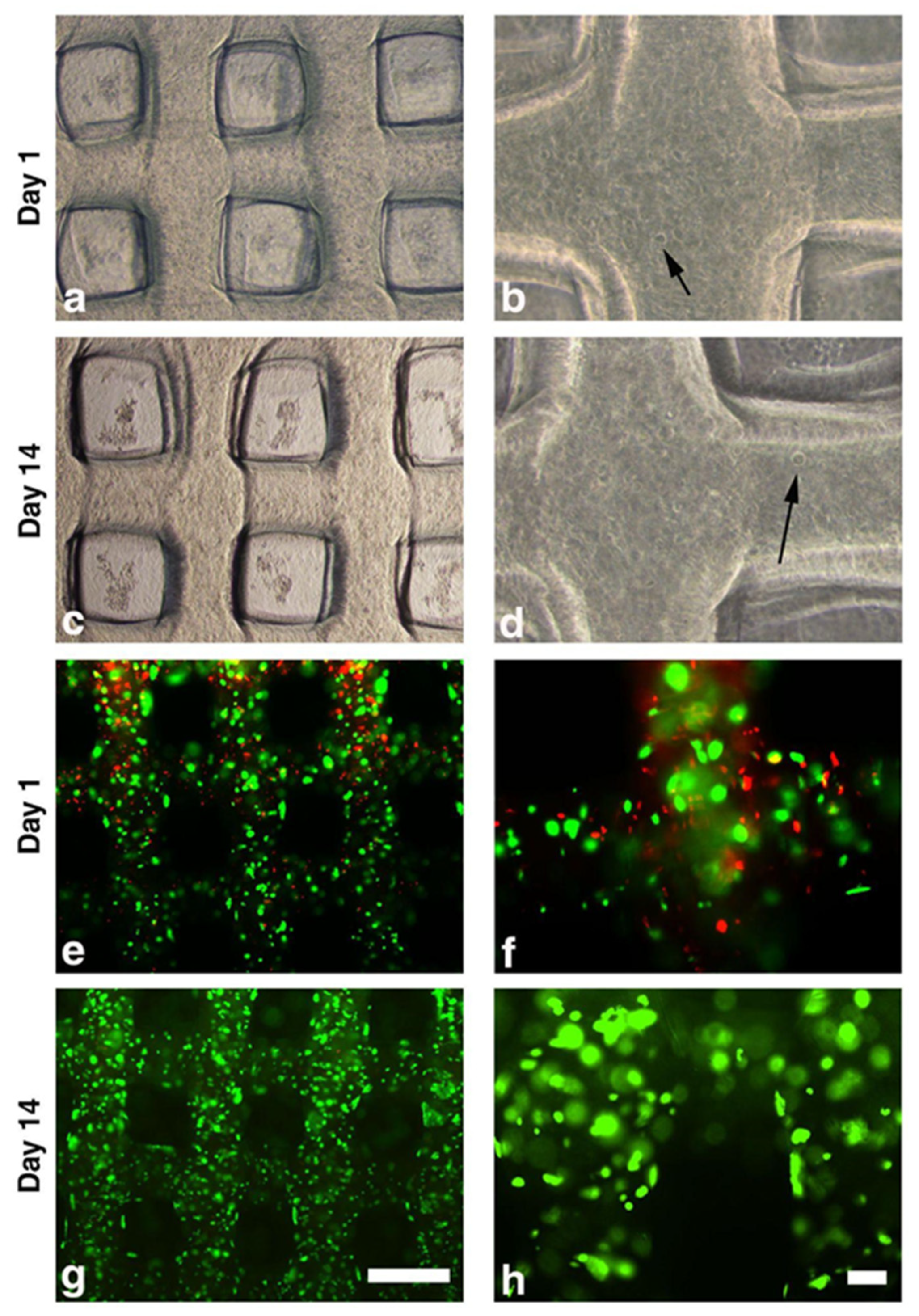
The Live and Dead assay was used to test cell viability (LIVE/DEAD™ Viability/Cytotoxicity Kit, ThermoFisher Scientific, Waltham, MA, USA) according to manufacturer instructions following the qualitative Fluorescence Microscopy Protocol. The test is based on the simultaneous determination of live (green) and dead (red) cells with two specific probes, calcein AM and ethidium homodimer (EthD-1), respectively. For each selected time point of culture (Day 1, Day 3, Day 7, and Day 14), a total number of four scaffolds from two sets of bioprinting processes (i.e., a duplicate for each bioprinting process) were used. Scaffolds were washed with phosphate-buffered saline (PBS) and incubated with ethidium homodimer 1 (4 µM) and calcein-AM (2 μM) for 30 min at 37 °C, 5% CO2. After two further washing steps, scaffolds were evaluated and images captured at 4× and 10× magnifications by a Nikon Eclipse Ni microscope equipped with NIS (Nikon Imaging Software 5.21.00) elements (Nikon Inc., Tokyo, Japan).
2.6.2. Cytotoxicity Assay
The quantification of cytotoxicity was evaluated in the cell supernatants using a colorimetric assay based on measurement of lactate dehydrogenase (LDH) activity (Cytotoxicity Detection Kit, Roche, Basel, Switzerland) according to the manufacturer instructions. A hundred microliters of supernatants from cultures at Day 1, Day 3, Day 7, and Day 14 were tested. Two scaffolds from two sets of bioprinting processes were used. The colorimetric detection of LDH activity in the supernatants was performed measuring the absorbance at 492–600 nm with a spectrophotometer (Infinite M200 TECAN, Männedorf, Switzerland) repeating the reading every 10 min for a total of 30 min.
2.7. Statistical Analysis
The GraphPad Prism for Windows (GraphPad Prism 5 software, San Diego, CA, USA) was used for statistical analysis. For the cytotoxicity assays, results are reported as a median and interquartile range (IQR) for continuous variables. The data were tested for normality using the D’Agostino & Pearson normality test and the two-way ANOVA with Bonferroni post-hoc correction test was used. A p value < 0.05 was considered as statistically significant.
4. Discussion
Synovial tissue plays a fundamental role in the immuno-inflammatory processes characteristic of OA and RA, the most common rheumatic diseases affecting joints [
21]. Therefore, understanding the mechanisms that regulate physiological and pathological synovium functions is crucial to shed light on the etiology and development of those diseases, and that might be helpful in discovering more effective therapies to minimize or counteract their progression. Recently, the use of synovial biopsies to investigate pathological mechanisms has been increasingly applied [
2], but reliability and culture condition issues limit their use. Recreating an in vitro synovial tissue 3D model would allow the evaluation of the critical pathways in a more biomimetic context and, in the meantime, represent a suitable alternative to 2D in vitro models for drug testing.
Our study represents a proof of concept (PoC) towards the possibility of fabricating through volumetric extrusion bioprinting a 3D engineered construct composed of GelMA embedded with a human synovial cell line. The construct could represent the first step towards the development of an in vitro synovial model.
To the best of our knowledge, a 3D bioprinted in vitro model of synovial tissue has no precedent in the literature. Previous in vitro attempts at recapitulating the synovium have been limited and primarily based on synovial fluid or synovial fibroblast monoculture or co-culture within 2D and 3D systems or tissue explants [
6,
7]. However, those models did not mimic the synovial structure accurately. More advanced applications, such as organoids or multicompartment organ-on-chip [
6,
7,
8,
9], significantly improved the in vitro model’s relevance. However, they still need to enable the fabrication of a relevant-size, three-dimensional engineered synovium. Only one study, by Lin et al. [
22], used 3D bioprinting technology by mixing RA-patients derived synoviocytes (MH7A) and vascular endothelial cells in a gelatin/alginate bioink. Nonetheless, their aim was to create a specific disease model to mimic the pathological characteristics of RA pannus vascular tissue [
23].
Interestingly, the authors have selected a grid structure design, as in our study. A grid structure favors cell well-being, guarantees nutrient transport, and promotes a new extracellular matrix (ECM) deposition. For these reasons the grid structure design is a well-established model used to bioprint and biofabricate a wide range of engineered tissues characterized by different internal microarchitectures (i.e., adipose tissue, liver, and neural tissue) [
24,
25,
26].
The use of hydrogels in bioprinting has recently emerged as a significant area of research. Hydrogels are the ideal materials for cell encapsulation due to their hydrated state that, on the other hand, provides challenges for generating high-fidelity constructs. Due to its rheological and mechanical features, we utilized GelMA, a widely diffused hydrogel biomaterial for 3D bioprinting. The high versatility of GelMA makes it suitable for recapitulating the soft tissue’s natural environment by enhancing stimuli responsiveness and integrating complex biochemical/biomechanical signals [
27].
GelMA presents, anyway, some drawbacks in printability due to its high temperature-dependent rheological properties that negatively impact the bioink processability, printing accuracy, and fidelity. To overcome these limitations, it is common to blend GelMA with different polymers or increase its concentration (above 15%). However, these approaches may be both detrimental to cell viability [
28,
29]. In this study, we selected a 10% GelMA concentration, which allows better biocompatibility but may present printability issues causing an up-to-twofold increase of the extruded filament size in comparison with the nozzle one or impaired stacking behavior due to discontinuities in the deposited filaments [
29,
30,
31,
32]. Therefore, we selected an Additive Manufacturing technology termed volumetric extrusion that allowed us to exert a finer control over GelMA material flow and printability. Furthermore, we implemented a dual gelation/crosslinking approach based on accurate thermal control of the printhead and the substrate, and layer-by-layer light irradiation, greatly improving material shape retention and stacking. As a result, we were able to fabricate relevant-sized 3D structures with microarchitectural features closely matching CAD design, with an average deviation of less than 1% in fiber diameter and pore size. At the same time, cell viability and functionality were proven not to be negatively affected by the bioprinting process in the selected conditions. However, volumetric extrusion also provides some limitations. In fact, cylindrical nozzles are preferred to conical ones to control the flow rate. This, combined with the need for longer needles to print inside a multi-well plate format, led to some restrictions regarding minimum fiber diameter. As observed during preliminary bioprinting tests, lower-size needles (200 µm inner diameter) resulted in low cell viability and critical failures in material deposition. Cell aggregates likely formed within the nozzle, leading to inhomogeneous material extrusion and frequent clogging phenomena. It would be challenging to fabricate structures with finer internal fibers. For this purpose, it is crucial to provide optimal conditions tailored for the specific bioink formulation. This is achieved not only by fine-tuning the bioprinting parameters but also by selecting the most suitable dispensing technology and process conditions.
The human synovial cell K4IM line that we used was embedded in the biomaterial and 3D bioprinted without detrimental effect on viability, showing that the bioprinted hydrogel provides a proper microenvironment for cell survival. We already demonstrated that this cell line might be a useful tool to test the effects of different biological compounds used in the treatment of OA [
33] and to evaluate different cellular pathways in OA studies [
34]. However, although the use of cell lines presents many advantages (i.e., easy to manipulate in culture; no ethical concerns associated with the use of animal and human tissue; reproducibility of results also obtained from different laboratories), it represents one of the major limitations of this study. Further research will be performed by using primary human cells, expecting to include also the macrophage component. Manferdini et al. compared human synoviocytes isolated from OA patients at two different passages (Passage 1 versus Passage 5) [
35]. They showed that synovial macrophages are present at Passage 1, but not at Passage 5. Although the K4IM cell line has a fibroblast-like phenotype without the macrophage component, in our study, it was a tool to prepare a synovial membrane 3D model in vitro suitable for further development to study different cellular pathways mechanisms, as well as the effect of pharmacological drugs in OA and other pathologies in which synovial membrane is involved.
Further studies will be needed based on our PoC to mimic more accurately the structure and cellular composition of the anatomical synovium, possibly based on the coordinated bioprinting of multiple materials and cell populations or on the integration of bioprinting and electrospinning/writing technologies to provide zonal differentiation through construct microarchitecture, as successfully proven by other studies [
36,
37].
5. Conclusions
The PoC study we presented is the first step to developing a 3D bioprinted in vitro model of synovial tissue. The use of a volumetric extrusion approach, combined with a dual gelification/crosslinking strategy based on accurate thermal control and layer-by-layer crosslinking, greatly enhanced the achievable shape fidelity and control over the scaffold internal microarchitectures. Moreover, the selected ink and process condition showed no detrimental effect on the embedded cells, which preserved their viability and metabolic activity for up to 14 days.
The reported approach could pave the way for the fabrication of more complex 3D bioprinted models, mimicking the structure and cellular composition of the synovial membrane accurately. These advanced 3D in vitro models could constitute a valuable tool to study the onset and progression of tissue-related pathologies and identify novel therapeutic treatments.