Object Detection for Brain Cancer Detection and Localization
Abstract
1. Introduction
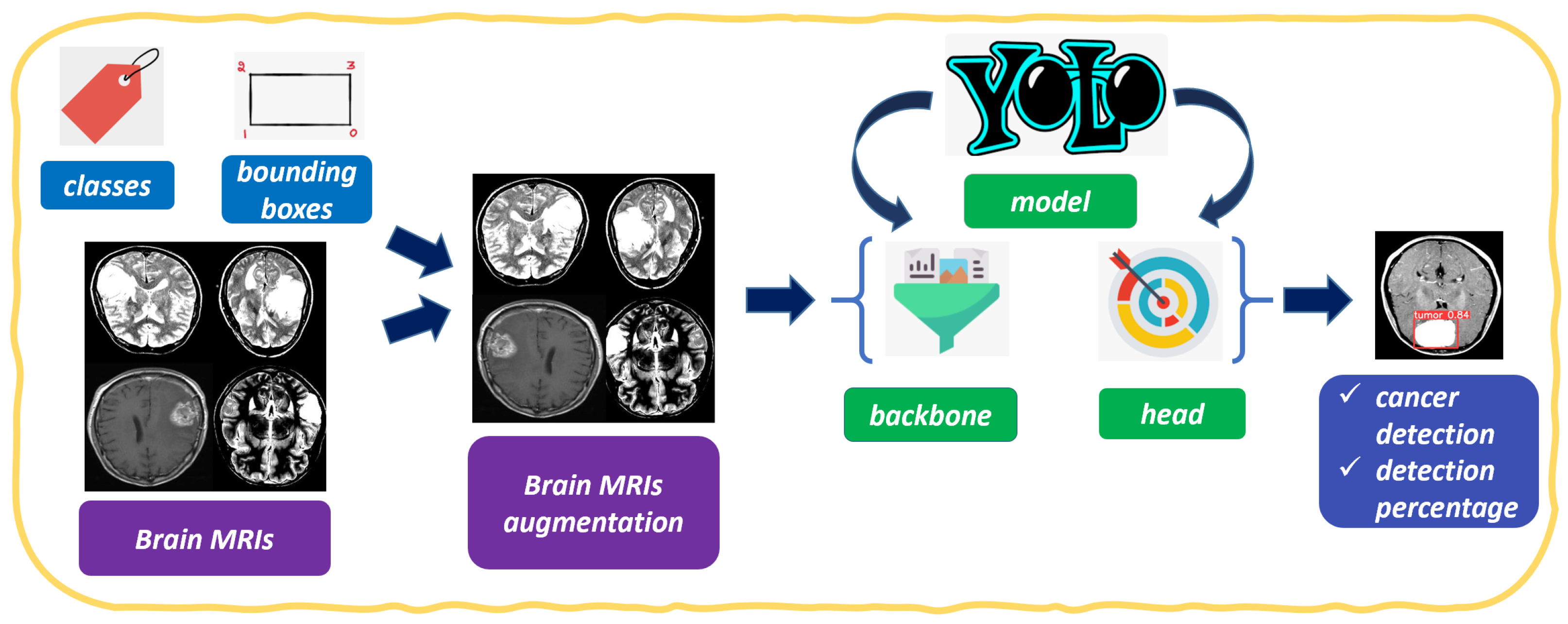
2. The Method
- Anchor-free detection. YOLOv8 does away with the use of anchor boxes, which were a key component of previous YOLO models. Anchor boxes are pre-defined bounding boxes that are used to classify objects in an image. However, anchor boxes can be restrictive, and they can make it difficult for the model to learn to detect objects that are not well-represented by the anchor boxes. YOLOv8 uses a new technique called anchor-free detection, which allows the model to learn to detect objects of any size and shape.
- C3 convolutions. YOLOv8 uses a new type of convolution called C3 convolutions. C3 convolutions are more efficient than traditional convolutions, and they allow the model to learn more complex features.
- Mosaic augmentation. YOLOv8 uses a new type of data augmentation called mosaic augmentation. Mosaic augmentation creates a new image by stitching together four randomly cropped images. This helps the model to learn to generalize to different object appearances and different lighting conditions.
- Real-time detection: YOLO is known for its real-time object detection capabilities. In the context of medical image classification, this means that YOLO can quickly and efficiently identify abnormalities, lesions, or specific medical conditions in real-time, allowing for faster diagnosis and treatment planning.
- Object localization: YOLO is designed to not only classify objects but also precisely localize them within the image. In medical imaging, this localization can be crucial for identifying the exact location and extent of abnormalities, aiding medical professionals in making accurate diagnoses.
- Handling multiple classes: YOLO can handle multiple classes or categories simultaneously. In medical image classification, this is advantageous as it allows the model to detect and classify various medical conditions or abnormalities within a single image.
- Single-pass approach: YOLO follows a single-pass approach, making predictions in one pass through the neural network. This design makes YOLO faster and more computationally efficient compared to some other object detection models, making it suitable for large-scale medical image datasets.
- Transfer learning: YOLO can leverage pre-trained models on large image datasets (e.g., ImageNet) for feature extraction and then fine-tune the model on medical image datasets. This transfer learning approach allows the model to learn relevant features from general images and then adapt them to medical images, even with limited labeled medical data.
- Generalizability: YOLO has shown promising generalization capabilities across different domains and tasks. This is essential in medical image classification, as medical datasets can vary in terms of image quality, patient demographics, and equipment, and a model that can generalize well is desirable.
- Ongoing research and development: YOLO is an actively researched object detection architecture, and advancements in its design and training methodologies continue to improve its performance. This ongoing research ensures that adopting YOLO in medical image classification can benefit from the latest advancements in computer vision and deep learning.
- Classify the image as cancerous;
- Detect the area (i.e., localize) related to brain cancer;
- Assign a probability of the cancer presence.
3. Experimental Analysis
3.1. The Dataset
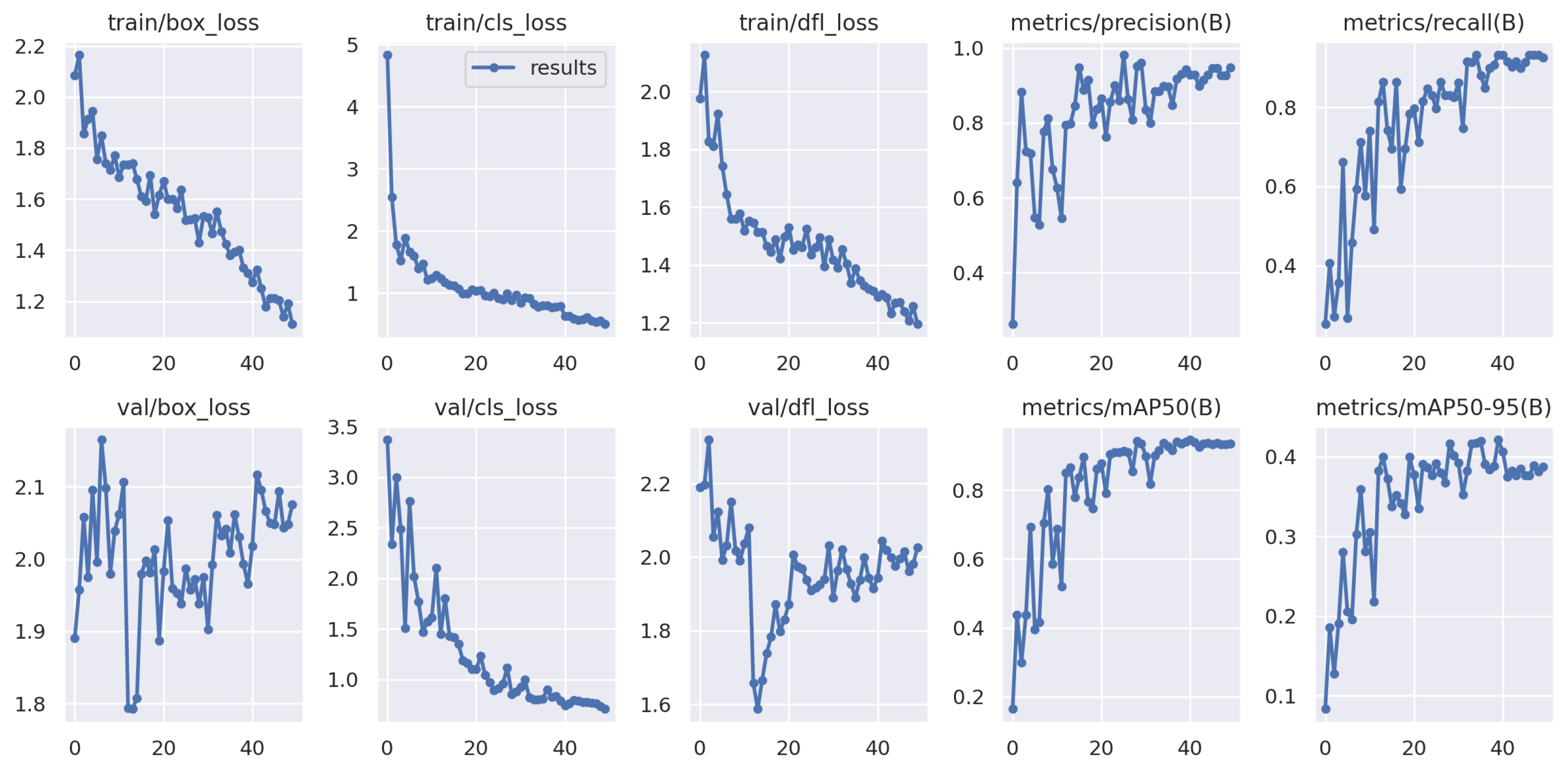
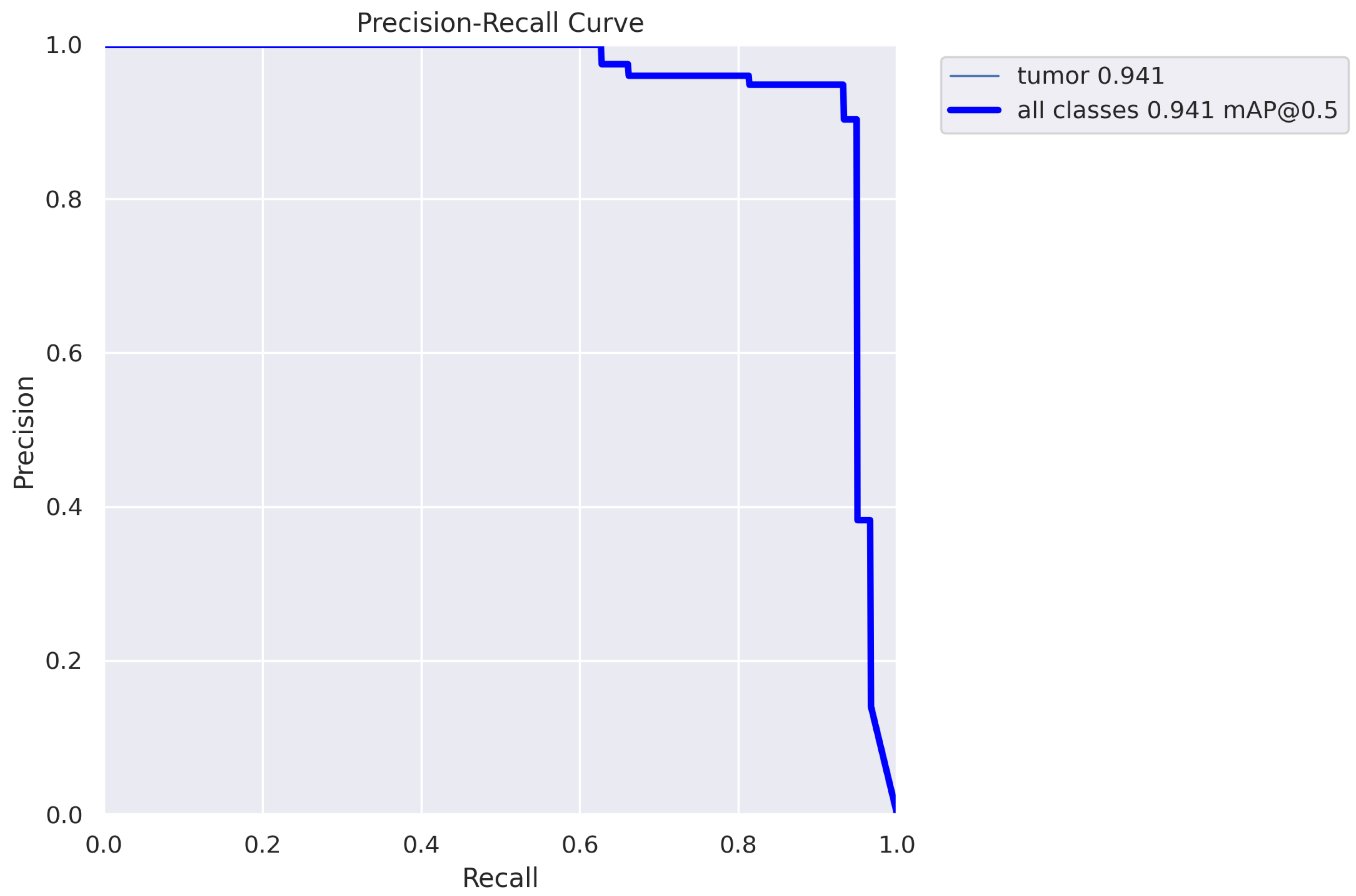
3.2. The Results
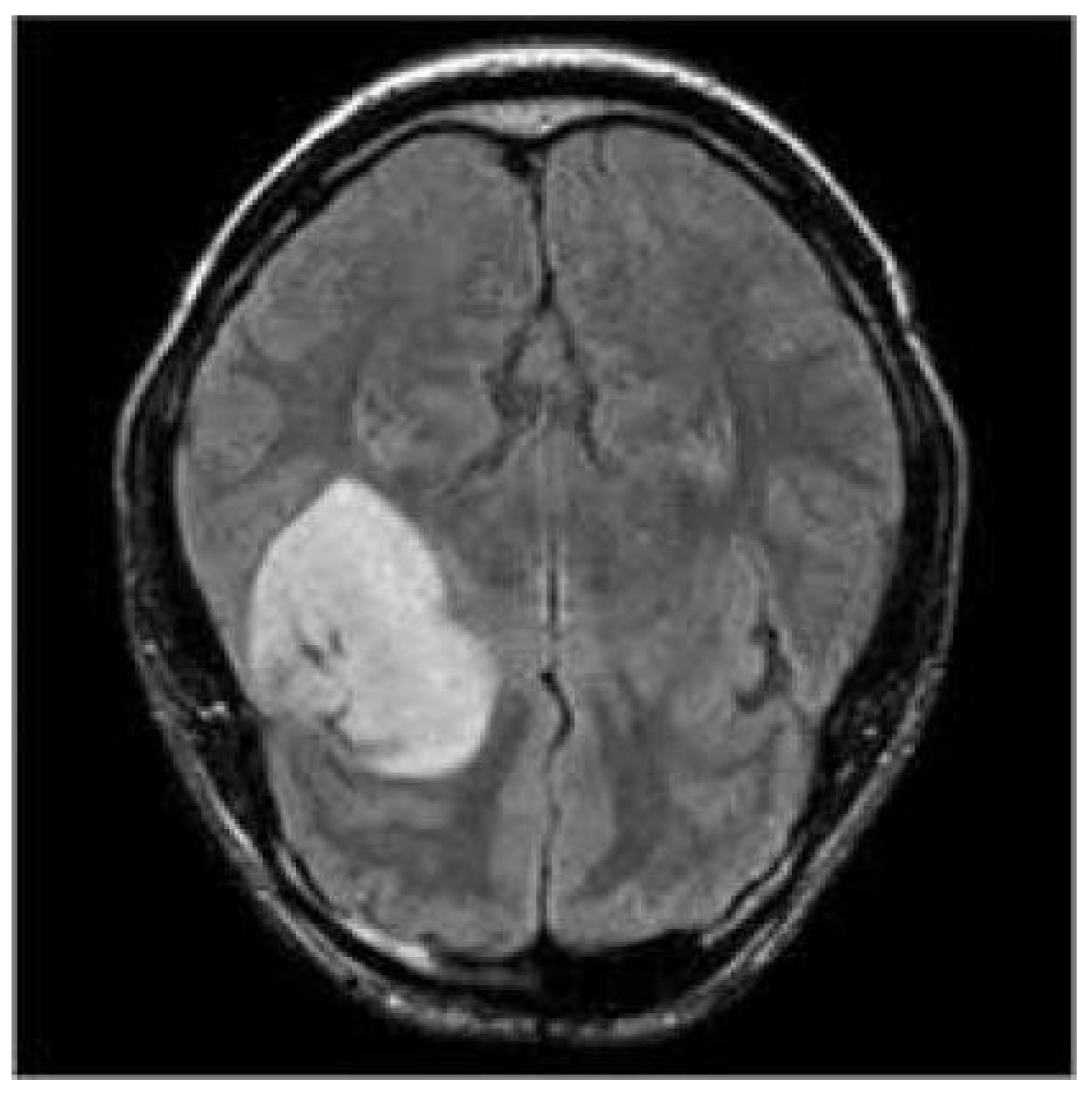
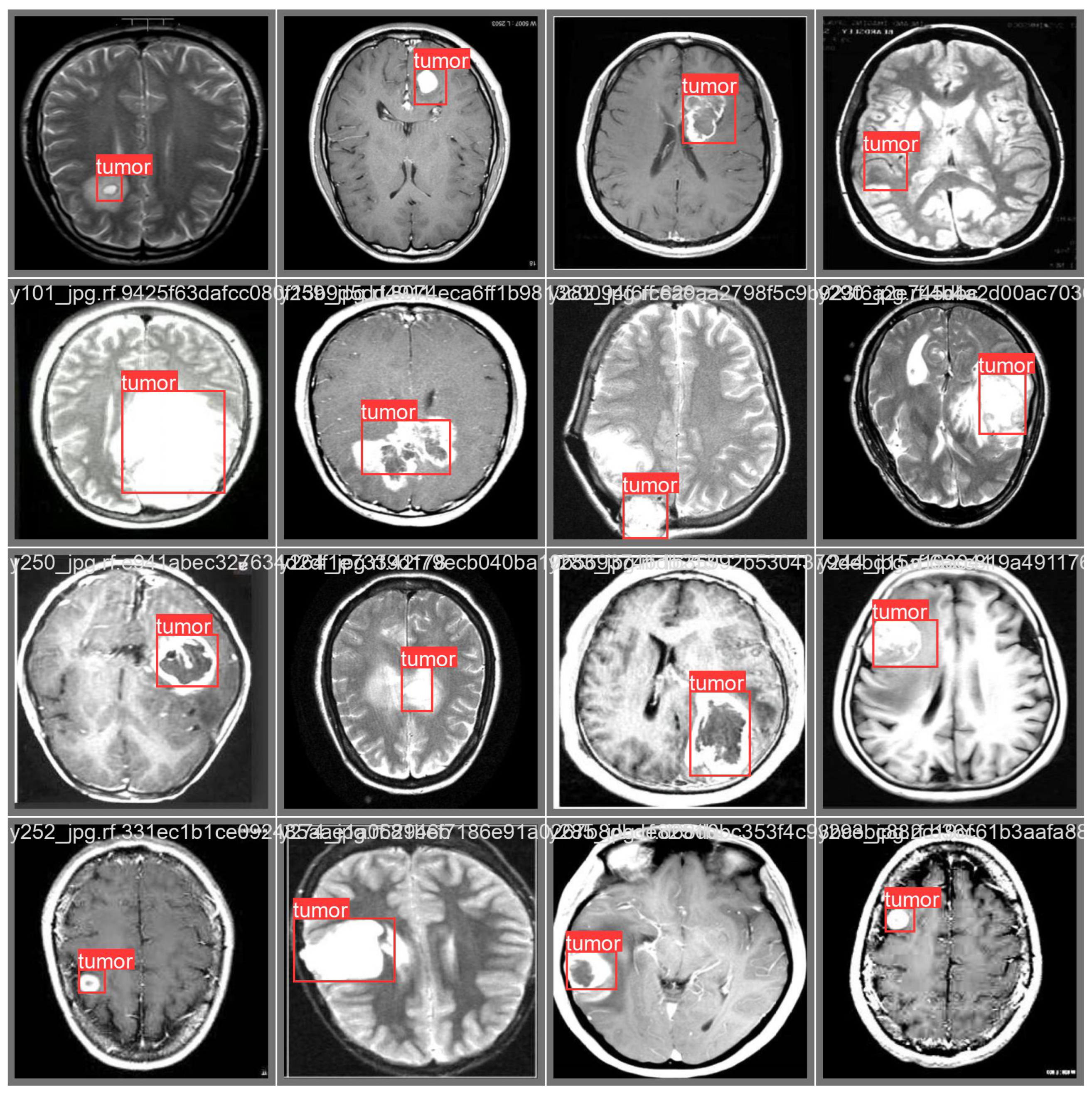
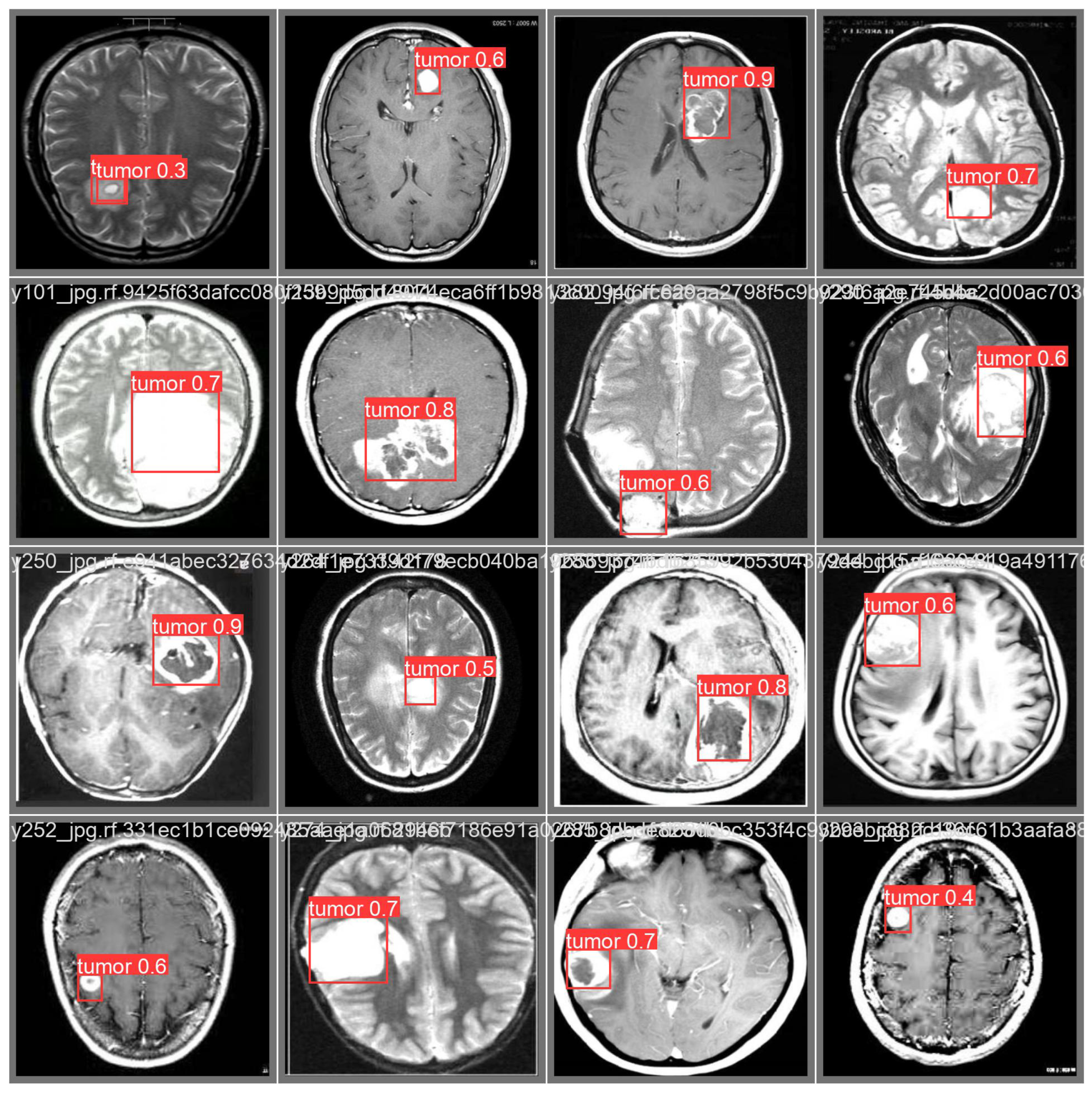
3.3. Prediction Examples
4. Related Work
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Di Giammarco, M.; Martinelli, F.; Mercaldo, F.; Santone, A. High Grade Brain Cancer Segmentation by means of Deep Learning. Procedia Comput. Sci. 2022, 207, 1633–1640. [Google Scholar] [CrossRef]
- Weitzner, M.A.; Meyers, C.A.; Gelke, C.K.; Byrne, K.S.; Levin, V.A.; Cella, D.F. The Functional Assessment of Cancer Therapy (FACT) Scale. Development of a brain subscale and revalidation of the general version (FACT-G) in patients with primary brain tumors. Cancer 1995, 75, 1151–1161. [Google Scholar] [CrossRef]
- Gleeson, H.K.; Shalet, S.M. The impact of cancer therapy on the endocrine system in survivors of childhood brain tumours. Endocr.-Relat. Cancer 2004, 11, 589–602. [Google Scholar] [CrossRef]
- Hauptmann, M.; Byrnes, G.; Cardis, E.; Bernier, M.O.; Blettner, M.; Dabin, J.; Engels, H.; Istad, T.S.; Johansen, C.; Kaijser, M.; et al. Brain cancer after radiation exposure from CT examinations of children and young adults: Results from the EPI-CT cohort study. Lancet Oncol. 2023, 24, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Di Giammarco, M.; Iadarola, G.; Martinelli, F.; Mercaldo, F.; Ravelli, F.; Santone, A. Explainable Deep Learning for Alzheimer Disease Classification and Localisation. In Proceedings of the International Conference on Applied Intelligence and Informatics; Springer: Cham, Switzerland, 2022; pp. 129–143. [Google Scholar]
- Fomchenko, E.I.; Holland, E.C. Stem cells and brain cancer. Exp. Cell Res. 2005, 306, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Mercaldo, F.; Di Giammarco, M.; Apicella, A.; Di Iadarola, G.; Cesarelli, M.; Martinelli, F.; Santone, A. Diabetic retinopathy detection and diagnosis by means of robust and explainable convolutional neural networks. Neural Comput. Appl. 2023, 35, 17429–17441. [Google Scholar] [CrossRef]
- Tandel, G.S.; Biswas, M.; Kakde, O.G.; Tiwari, A.; Suri, H.S.; Turk, M.; Laird, J.R.; Asare, C.K.; Ankrah, A.A.; Khanna, N.; et al. A review on a deep learning perspective in brain cancer classification. Cancers 2019, 11, 111. [Google Scholar] [CrossRef]
- Martinelli, F.; Mercaldo, F.; Santone, A. Water Meter Reading for Smart Grid Monitoring. Sensors 2023, 23, 75. [Google Scholar] [CrossRef]
- Martinelli, F.; Mercaldo, F.; Santone, A. Smart Grid monitoring through Deep Learning for Image-based Automatic Dial Meter Reading. In Proceedings of the 2022 IEEE International Conference on Big Data (Big Data), Osaka, Japan, 17–20 December 2022; pp. 4534–4542. [Google Scholar]
- Mercaldo, F.; Martinelli, F.; Santone, A. A proposal to ensure social distancing with deep learning-based object detection. In Proceedings of the 2021 International Joint Conference on Neural Networks (IJCNN), Shenzhen, China, 18–22 July 2021; pp. 1–5. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Jiang, P.; Ergu, D.; Liu, F.; Cai, Y.; Ma, B. A Review of Yolo algorithm developments. Procedia Comput. Sci. 2022, 199, 1066–1073. [Google Scholar] [CrossRef]
- Sanchez, S.; Romero, H.; Morales, A. A review: Comparison of performance metrics of pretrained models for object detection using the TensorFlow framework. IOP Conf. Ser. Mater. Sci. Eng. 2020, 844, 012024. [Google Scholar]
- Sah, S.; Shringi, A.; Ptucha, R.; Burry, A.M.; Loce, R.P. Video redaction: A survey and comparison of enabling technologies. J. Electron. Imaging 2017, 26, 051406. [Google Scholar] [CrossRef]
- Hurtik, P.; Molek, V.; Hula, J.; Vajgl, M.; Vlasanek, P.; Nejezchleba, T. Poly-YOLO: Higher speed, more precise detection and instance segmentation for YOLOv3. Neural Comput. Appl. 2022, 34, 8275–8290. [Google Scholar] [CrossRef]
- Horak, K.; Sablatnig, R. Deep learning concepts and datasets for image recognition: Overview 2019. In Proceedings of the Eleventh International Conference on Digital Image Processing (ICDIP 2019), Guangzhou, China, 10–13 May 2019; Volume 11179, pp. 484–491. [Google Scholar]
- YOLO. YOLO Dataset. 2022. Available online: https://universe.roboflow.com/yolo-hz3ua/yolo-fj4s3 (accessed on 17 July 2023).
- Isselmou, A.E.K.; Zhang, S.; Xu, G. A novel approach for brain tumor detection using MRI images. J. Biomed. Sci. Eng. 2016, 9, 44. [Google Scholar] [CrossRef]
- Badran, E.F.; Mahmoud, E.G.; Hamdy, N. An algorithm for detecting brain tumors in MRI images. In Proceedings of the 2010 International Conference on Computer Engineering and Systems (ICCES), Cairo, Egypt, 30 November–2 December 2010; pp. 368–373. [Google Scholar]
- Sharma, K.; Kaur, A.; Gujral, S. Brain tumor detection based on machine learning algorithms. Int. J. Comput. Appl. 2014, 103, 7–11. [Google Scholar] [CrossRef]
- Ramteke, R.; Monali, Y.K. Automatic medical image classification and abnormality detection using K-Nearest Neighbour. Int. J. Adv. Comput. Res. 2012, 2, 190–196. [Google Scholar]
- Xuan, X.; Liao, Q. Statistical structure analysis in MRI brain tumor segmentation. In Proceedings of the 2007 Fourth International Conference on Image and Graphics, ICIG 2007, Chengdu, China, 22–24 August 2007; pp. 421–426. [Google Scholar]
- Gadpayleand, P.; Mahajani, P. Detection and classification of brain tumor in MRI images. Int. J. Emerg. Trends Electr. Electron. 2013, 23, 2320–9569. [Google Scholar]
- Jafari, M.; Shafaghi, R. A hybrid approach for automatic tumor detection of brain MRI using support vector machine and genetic algorithm. Glob. J. Sci. Eng. Technol. 2012, 3, 1–8. [Google Scholar]
- Amin, S.E.; Mageed, M. Brain tumor diagnosis systems based on artificial neural networks and segmentation using MRI. In Proceedings of the 2012 8th International Conference on Informatics and Systems (INFOS), Giza, Egypt, 14–16 May 2012; pp. 15–25. [Google Scholar]
- Reis, D.; Kupec, J.; Hong, J.; Daoudi, A. Real-Time Flying Object Detection with YOLOv8. arXiv 2023, arXiv:2305.09972. [Google Scholar]
- Paul, S.; Ahad, D.M.T.; Hasan, M.M. Brain Cancer Segmentation Using YOLOv5 Deep Neural Network. arXiv 2022, arXiv:2212.13599. [Google Scholar]
- Selvy, P.T.; Dharani, V.; Indhuja, A. Brain tumour detection using deep learning techniques. Int. J. Sci. Res. Comput. Sci. Eng. Inf. Technol. 2019, 169, 175. [Google Scholar] [CrossRef]
- Alsubai, S.; Khan, H.U.; Alqahtani, A.; Sha, M.; Abbas, S.; Mohammad, U.G. Ensemble deep learning for brain tumor detection. Front. Comput. Neurosci. 2022, 16, 1005617. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, S.; Rezayi, S.; Keshavarz, H.; Niakan Kalhori, S.R. MRI-based brain tumor detection using convolutional deep learning methods and chosen machine learning techniques. BMC Med. Inform. Decis. Mak. 2023, 23, 16. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Kim, J.S.; Park, D.K.; Whangbo, T. Automated Detection of Gastric Lesions in Endoscopic Images by Leveraging Attention-based YOLOv7. IEEE Access 2023. [Google Scholar] [CrossRef]
- Masood, M.; Nazir, T.; Nawaz, M.; Javed, A.; Iqbal, M.; Mehmood, A. Brain tumor localization and segmentation using mask RCNN. Front. Comput. Sci. 2021, 15, 156338. [Google Scholar] [CrossRef]
- Dipu, N.M.; Shohan, S.A.; Salam, K.A. Brain Tumor Detection Using Various Deep Learning Algorithms. In Proceedings of the 2021 International Conference on Science & Contemporary Technologies (ICSCT), Dhaka, Bangladesh, 5–7 August 2021; pp. 1–6. [Google Scholar]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. An ensemble learning approach for brain cancer detection exploiting radiomic features. Comput. Methods Programs Biomed. 2020, 185, 105134. [Google Scholar] [CrossRef]

| Layer | From | n | Params | Module | Arguments |
|---|---|---|---|---|---|
| 0 | −1 | 1 | 928 | modules.Conv | [3, 32, 3, 2] |
| 1 | −1 | 1 | 18,560 | modules.Conv | [32, 64, 3, 2] |
| 2 | −1 | 1 | 29,056 | modules.C2f | [64, 64, 1, True] |
| 3 | −1 | 1 | 73,984 | modules.Conv | [64, 128, 3, 2] |
| 4 | −1 | 2 | 197,632 | modules.C2f | [128, 128, 2, True] |
| 5 | −1 | 1 | 295,424 | modules.Conv | [128, 256, 3, 2] |
| 6 | −1 | 2 | 788,480 | modules.C2f | [256, 256, 2, True] |
| 7 | −1 | 1 | 1,180,672 | modules.Conv | [256, 512, 3, 2] |
| 8 | −1 | 1 | 1,838,080 | modules.C2f | [512, 512, 1, True] |
| 9 | −1 | 1 | 656,896 | modules.SPPF | [512, 512, 5] |
| 10 | −1 | 1 | 0 | upsampling.Upsample | [None, 2, ‘nearest’] |
| 11 | [−1, 6] | 1 | 0 | modules.Concat | [1] |
| 12 | −1 | 1 | 591,360 | modules.C2f | [768, 256, 1] |
| 13 | −1 | 1 | 0 | upsampling.Upsample | [None, 2, ‘nearest’] |
| 14 | [−1, 4] | 1 | 0 | modules.Concat | [1] |
| 15 | −1 | 1 | 148,224 | modules.C2f | [384, 128, 1] |
| 16 | −1 | 1 | 147,712 | modules.Conv | [128, 128, 3, 2] |
| 17 | [−1, 12] | 1 | 0 | modules.Concat | [1] |
| 18 | −1 | 1 | 493,056 | modules.C2f | [384, 256, 1] |
| 19 | −1 | 1 | 590,336 | modules.Con | [256, 256, 3, 2] |
| 20 | [−1, 9] | 1 | 0 | modules.Concat | [1] |
| 21 | −1 | 1 | 1,969,152 | modules.C2f | [768, 512, 1] |
| 22 | [15, 18, 21] | 1 | 2,116,822 | modules.Detect | [2, [128, 256, 512]] |
| Step | Image | Labels | Precision | Recall | Specificity | mAP_0.5 | mAP_0.5:0.95 |
|---|---|---|---|---|---|---|---|
| Training | 60 | 59 | 0.948 | 0.926 | 0.931 | 0.935 | 0.388 |
| Testing | 60 | 59 | 0.943 | 0.932 | 0.938 | 0.941 | 0.421 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mercaldo, F.; Brunese, L.; Martinelli, F.; Santone, A.; Cesarelli, M. Object Detection for Brain Cancer Detection and Localization. Appl. Sci. 2023, 13, 9158. https://doi.org/10.3390/app13169158
Mercaldo F, Brunese L, Martinelli F, Santone A, Cesarelli M. Object Detection for Brain Cancer Detection and Localization. Applied Sciences. 2023; 13(16):9158. https://doi.org/10.3390/app13169158
Chicago/Turabian StyleMercaldo, Francesco, Luca Brunese, Fabio Martinelli, Antonella Santone, and Mario Cesarelli. 2023. "Object Detection for Brain Cancer Detection and Localization" Applied Sciences 13, no. 16: 9158. https://doi.org/10.3390/app13169158
APA StyleMercaldo, F., Brunese, L., Martinelli, F., Santone, A., & Cesarelli, M. (2023). Object Detection for Brain Cancer Detection and Localization. Applied Sciences, 13(16), 9158. https://doi.org/10.3390/app13169158







