Using Ensemble OCT-Derived Features beyond Intensity Features for Enhanced Stargardt Atrophy Prediction with Deep Learning
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Imaging Dataset
2.2. Image Registration
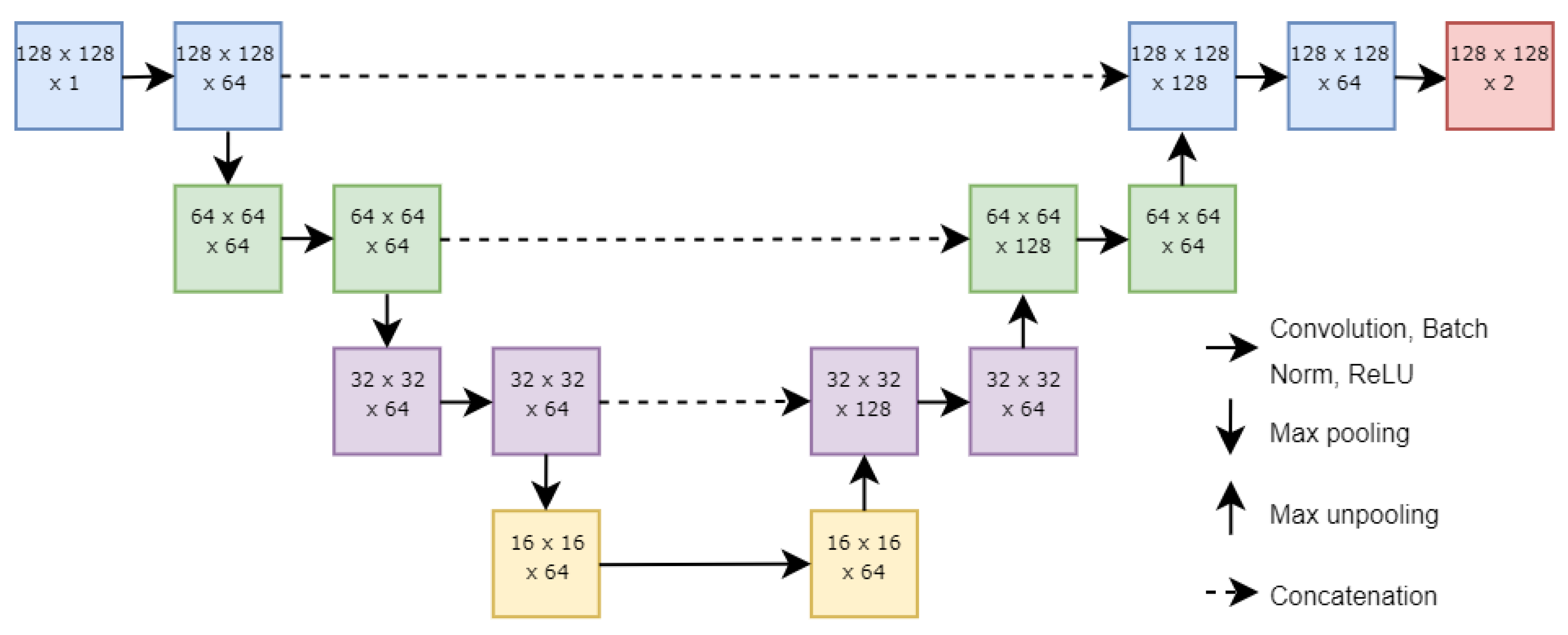
2.3. Neural Network Structure
2.4. Experimental Methods
3. Results
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Strauss, R.W.; Ho, A.; Muñoz, B.; West, S. The natural history of the progression of atrophy secondary to Stargardt disease (ProgStar) studies: Design and baseline characteristics: ProgStar report no. 1. Ophthalmology 2016, 123, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Schönbach, E.M.; Wolfson, Y.; Strauss, R.W.; Ibrahim, M.A.; Kong, X.; Muñoz, B.; Birch, D.G.; Cideciyan, A.V.; Hahn, G.-A.; Nittala, M.; et al. Macular sensitivity measured with microperimetry in Stargardt disease in the progression of atrophy secondary to Stargardt disease (ProgStar) study: Report no.7. JAMA Ophthalmol. 2017, 135, 696. [Google Scholar] [CrossRef] [PubMed]
- Strauss, R.W.; Muñoz, B.; Ho, A.; Jha, A.; Michaelides, M.; Mohand-Said, S.; Cideciyan, A.V.; Birch, D.; Hariri, A.H.; Nittala, M.G.; et al. Incidence of atrophic lesions in Stargardt disease in the progression of atrophy secondary to Stargardt disease (ProgStar) study: Report no. 5. JAMA Ophthalmol. 2017, 135, 687. [Google Scholar] [CrossRef] [PubMed]
- Strauss, R.W.; Muñoz, B.; Ho, A.; Jha, A.; Michaelides, M.; Cideciyan, A.V.; Audo, I.; Birch, D.G.; Hariri, A.H.; Nittala, M.G.; et al. Progression of Stargardt disease as determined by fundus autofluorescence in the retrospective progression of Stargardt disease study (ProgStar report no. 9). JAMA Ophthalmol. 2017, 135, 1232. [Google Scholar] [CrossRef]
- Ma, L.; Kaufman, Y.; Zhang, J.; Washington, I. C20-D3-vitamin A slows lipofuscin accumulation and electrophysiological retinal degeneration in a mouse model of Stargardt disease. J. Biol. Chem. 2010, 286, 7966–7974. [Google Scholar] [CrossRef]
- Kong, J.; Binley, K.; Pata, I.; Doi, K.; Mannik, J.; Zernant-Rajang, J.; Kan, O.; Iqball, S.; Naylor, S.; Sparrow, J.R.; et al. Correction of the disease phenotype in the mouse model of Stargardt disease by lentiviral gene therapy. Gene Ther. 2008, 15, 1311–1320. [Google Scholar] [CrossRef]
- Binley, K.; Widdowson, P.; Loader, J.; Kelleher, M.; Iqball, S.; Ferrige, G.; de Belin, J.; Carlucci, M.; Angell-Manning, D.; Hurst, F.; et al. Transduction of photoreceptors with equine infectious anemia virus lentiviral vectors: Safety and biodistribution of StarGen for Stargardt disease. Investig. Ophthalmol. Vis. Sci. 2013, 54, 4061. [Google Scholar] [CrossRef]
- Mukherjee, N.; Schuman, S. Diagnosis and management of Stargardt disease. In EyeNet Magazine; American Academy of Ophthalmology: San Francisco, CA, USA, 2014. [Google Scholar]
- Schmitz-Valckenberg, S.; Holz, F.; Bird, A.; Spaide, R. Fundus autofluorescence imaging: Review and perspectives. Retina 2008, 28, 385–409. [Google Scholar] [CrossRef]
- Huang, D.; Swanson, E.A.; Lin, C.P.; Schuman, J.S.; Stinson, W.G.; Chang, W.; Hee, M.R.; Flotte, T.; Gregory, K.; Puliafito, C.A.; et al. Optical coherence tomography. Science 1991, 254, 1178–1181. [Google Scholar] [CrossRef]
- Fujimoto, J.G.; Bouma, B.; Tearney, G.J.; Boppart, S.A.; Pitris, C.; Southern, J.F.; Brezinski, M.E. New technology for high-speed and high-resolution optical coherence tomography. Ann. N. Y. Acad. Sci. 1998, 838, 96–107. [Google Scholar] [CrossRef]
- Filho, M.A.B.; Witkin, A.J. Outer Retinal Layers as Predictors of Vision Loss. Rev. Ophthalmol. 2015, 15. [Google Scholar]
- Fujinami, K.; Zernant, J.; Chana, R.K.; Wright, G.A.; Tsunoda, K.; Ozawa, Y.; Tsubota, K.; Robson, A.G.; Holder, G.E.; Allikmets, R.; et al. Clinical and molecular characteristics of childhood-onset Stargardt disease. Ophthalmology 2015, 122, 326–334. [Google Scholar] [CrossRef]
- Fishman, G.A. Fundus flavimaculatus. A clinical classification. Arch. Ophthalmol. 1976, 94, 2061–2067. [Google Scholar] [CrossRef] [PubMed]
- Fujinami, K.; Lois, N.; Mukherjee, R.; McBain, V.A.; Tsunoda, K.; Tsubota, K.; Stone, E.M.; Fitzke, F.W.; Bunce, C.; Moore, A.T.; et al. A longitudinal study of Stargardt disease: Quantitative assessment of fundus autofluorescence, progression, and genotype correlations. Investig. Ophthalmol. Vis. Sci. 2013, 54, 8181–8190. [Google Scholar] [CrossRef] [PubMed]
- Lois, N.; Holder, G.E.; Bunce, C.; Fitzke, F.W.; Bird, A.C. Phenotypic subtypes of Stargardt macular dystrophy-fundus flavimaculatus. Arch. Ophthalmol 2001, 119, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Mishra, Z.; Wang, Z.; Sadda, S.R.; Hu, Z. Automatic Segmentation in Multiple OCT Layers for Stargardt Disease Characterization Via Deep Learning. Transl. Vis. Sci. Technol. 2021, 10, 24. [Google Scholar] [CrossRef]
- Kugelman, J.; Alonso-Caneiro, D.; Chen, Y.; Arunachalam, S.; Huang, D.; Vallis, N.; Collins, M.J.; Chen, F.K. Retinal boundary segmentation in Stargardt disease optical coherence tomography images using automated deep learning. Transl. Vis. Sci. Technol. 2020, 9, 12. [Google Scholar] [CrossRef]
- Charng, J.; Xiao, D.; Mehdizadeh, M.; Attia, M.S.; Arunachalam, S.; Lamey, T.M.; Thompson, J.A.; McLaren, T.L.; De Roach, J.N.; Mackey, D.A.; et al. Deep learning segmentation of hyperautofluorescent fleck lesions in Stargardt disease. Sci. Rep. 2020, 10, 16491. [Google Scholar] [CrossRef]
- Wang, Z.; Sadda, S.R.; Hu, Z. Deep learning for automated screening and semantic segmentation of age-related and juvenile atrophic macular degeneration. In Proceedings of the Medical Imaging 2019: Computer-Aided Diagnosis, San Diego, CA, USA, 17–20 February 2019. [Google Scholar]
- Zhao, P.; Branham, K.; Schlegel, D.; Fahim, A.T.; Jayasundera, K.T. Automated Segmentation of Autofluorescence Lesions in Stargardt Disease. Ophthalmol. Retin. 2022, 6, 1098–1104. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munchen, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Roy, A.G.; Conjeti, S.; Karri, S.P.K.; Sheet, D.; Katouzian, A.; Wachinger, C.; Navab, N. ReLayNet: Retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks. Biomed. Opt. Express 2017, 8, 3627–3642. [Google Scholar] [CrossRef]
- Fang, L.; Cunefare, D.; Wang, C.; Guymer, R.H.; Li, S.; Farsiu, S. Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search. Biomed. Opt. Express 2017, 8, 2732–2744. [Google Scholar] [CrossRef]
- Hu, K.; Hu, K.; Shen, B.; Zhang, Y.; Cao, C.; Xiao, F.; Gao, X. Automatic segmentation of retinal layer boundaries in OCT images using multiscale convolutional neural network and graph search. Neurocomputing 2019, 365, 302–313. [Google Scholar] [CrossRef]
- Kugelman, J.; Alonso-Caneiro, D.; Read, S.A.; Hamwood, J.; Vincent, S.J.; Chen, F.K.; Collins, M.J. Automatic choroidal segmentation in OCT images using supervised deep learning methods. Sci. Rep. 2019, 9, 13298. [Google Scholar] [CrossRef] [PubMed]
- Venhuizen, F.G.; van Ginneken, B.; Liefers, B.; van Grinsven, M.J.J.P.; Fauser, S.; Hoyng, C.; Theelen, T.; Sánchez, C.I. Robust total retina thickness segmentation in optical coherence tomography images using convolutional neural networks. Biomed. Opt. Express 2017, 8, 3292–3316. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhu, S. Visual interpretability for deep learning: A survey. Front. Inf. Technol. Electron. Eng. 2018, 19, 27–39. [Google Scholar] [CrossRef]
- Saha, S.; Wang, Z.; Sadda, S.; Kanagasingam, Y.; Hu, Z. Visualizing and understanding inherent features in SD-OCT for the progression of age-related macular degeneration using deconvolutional neural networks. Appl. AI Lett. 2020, 1, e16. [Google Scholar] [CrossRef]
- Hu, Z.; Wang, Z.; Sadda, S. Automated segmentation of geographic atrophy using deep convolutional neural networks. In Proceedings of the Medical Imaging 2018: Computer-Aided Diagnosis, Houston, TX, USA; 2018; Volume 10575, p. 1057511. [Google Scholar] [CrossRef]
- Stetson, P.F.; Yehoshua, Z.; Garcia Filho, C.A.A.; Nunes, R.P.; Gregori, G.; Rosenfeld, P.J. OCT minimum intensity as a predictor of geographic atrophy enlargement. Investig. Ophthalmol. Vis. Sci. 2014, 55, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; de Sisternes, L.; Chen, Q.; Leng, T.; Rubin, D.L. Automated geographic atrophy segmentation for SD-OCT images using region-based CV model via local similarity factor. Biomed. Opt. Express 2016, 7, 581–600. [Google Scholar] [CrossRef]
- Mishra, Z.; Ganegoda, A.; Selicha, J.; Wang, Z.; Sadda, S.R.; Hu, Z. Automated retinal layer segmentation using graph-based algorithm incorporating deep-learning-derived information. Sci. Rep. 2020, 10, 9541. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 4th International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Wang, Z.; Sadda, S.R.; Lee, A.; Hu, Z.J. Automated segmentation and feature discovery of age-related macular degeneration and Stargardt disease via self-attended neural networks. Sci. Rep. 2022, 12, 14565. [Google Scholar] [CrossRef]
- Anegondi, N.; Gao, S.S.; Steffen, V.; Spaide, R.F.; Sadda, S.R.; Holz, F.G.; Rabe, C.; Honigberg, L.; Newton, E.M.; Cluceru, J.; et al. Deep learning to predict geographic atrophy area and growth rate from multimodal imaging. Ophthalmol. Retin. 2023, 7, 243–252. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Ji, Z.; Niu, S.; Leng, T.; Rubin, D.L.; Yuan, S.; Chen, Q. An integrated time adaptive geographic atrophy prediction model for SD-OCT images. Med. Image Anal. 2021, 68, 101893. [Google Scholar] [CrossRef] [PubMed]
- Kuehlewein, L.; Hariri, A.H.; Ho, A.; Dustin, L.; Wolfson, Y.; Strauss, R.W.; Scholl, H.P.N.; Sadda, S.V.R. Comparison of manual and semiautomated fundus autofluorescence analysis of macular atrophy in stargardt disease phenotype. Retina 2016, 36, 1216–1221. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Siddiquee, M.R.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. Deep Learn. Med. Image Anal. Multimodal. Learn. Clin. Decis. Support 2018, 11045, 3–11. [Google Scholar] [CrossRef]
- Oktay, O.; Echlemper, J.; Le Folgoc, L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Li, M.; Chen, Y.; Ji, Z.; Xie, K.; Yuan, S.; Chen, Q.; Li, S. Image projection network: 3D to 2D image segmentation in OCTA images. IEEE Trans. Med. Imag. 2020, 39, 3343–3354. [Google Scholar] [CrossRef]
- Li, M.; Zhang, Y.; Ji, Z.; Xie, K.; Yuan, S.; Liu, Q.; Chen, Q. IPN-V2 and OCTA-500: Methodology and dataset for retinal image segmentation. arXiv 2020, arXiv:2012.07261. [Google Scholar]
- Chen, L. ARA-net: An attention-aware retinal atrophy segmentation network coping with fundus images. Front. Neurosci. 2023, 17, 1174937. [Google Scholar] [CrossRef]
- Lemmens, S.; Van Eijgen, J.; Van Keer, K.; Jacob, J.; Moylett, S.; De Groef, L.; Vancraenendonck, T.; De Boever, P.; Stalmans, I. Hyperspectral imaging and the retina: Worth the wave? Transl. Vis. Sci. Technol. 2020, 9, 9. [Google Scholar] [CrossRef]
- Sohrab, M.A.; Smith, R.T.; Fawzi, A.A. Imaging characteristics of dry age-related macular degeneration. In Seminars in Ophthalmology; Taylor & Francis: Boca Raton, FL, USA, 2011; Volume 26. [Google Scholar]






| ELM-EZ | EZ-IRPE | IRPE-ORPE | ORPE-C-S | ELM-IRPE | EZ ± 5 | |
|---|---|---|---|---|---|---|
| Ensemble | 0.824 | 0.804 | 0.803 | 0.706 | 0.830 | 0.803 |
| Mean Intensity | 0.196 | 0.126 | 0.344 | 0.259 | 0.349 | 0.260 |
| Standard Deviation | 0.558 | 0.694 | 0.199 | 0.443 | 0.303 | 0.430 |
| Maximum Intensity | 0.236 | 0.241 | 0.324 | 0.405 | 0.161 | 0.076 |
| Minimum Intensity | 0.425 | 0.089 | 0.297 | 0.179 | 0.327 | 0.077 |
| Median Intensity | 0.341 | 0.283 | 0.252 | 0.267 | 0.098 | 0.113 |
| Kurtosis | 0.771 | 0.240 | 0.592 | 0.293 | 0.451 | 0.219 |
| Skewness | 0.479 | 0.521 | 0.257 | 0.410 | 0.445 | 0.508 |
| Gray-Level Entropy | 0.247 | 0.276 | 0.413 | 0.016 | 0.677 | 0.237 |
| Thickness | 0.677 | 0.360 | 0.687 | 0.568 |
| ELM-EZ | EZ-IRPE | IRPE-ORPE | ORPE-C-S | ELM-IRPE | EZ ± 5 | |
|---|---|---|---|---|---|---|
| Ensemble | 0.977 | 0.973 | 0.974 | 0.964 | 0.980 | 0.971 |
| Mean Intensity | 0.921 | 0.834 | 0.873 | 0.902 | 0.845 | 0.854 |
| Standard Deviation | 0.902 | 0.958 | 0.885 | 0.897 | 0.782 | 0.881 |
| Maximum Intensity | 0.940 | 0.741 | 0.943 | 0.923 | 0.793 | 0.848 |
| Minimum Intensity | 0.920 | 0.677 | 0.894 | 0.889 | 0.912 | 0.792 |
| Median Intensity | 0.901 | 0.590 | 0.928 | 0.835 | 0.649 | 0.709 |
| Kurtosis | 0.971 | 0.842 | 0.933 | 0.887 | 0.858 | 0.823 |
| Skewness | 0.961 | 0.939 | 0.809 | 0.792 | 0.914 | 0.892 |
| Gray-Level Entropy | 0.947 | 0.895 | 0.853 | 0.808 | 0.948 | 0.686 |
| Thickness | 0.950 | 0.856 | 0.940 | 0.910 |
| ELM-EZ | EZ-IRPE | IRPE-ORPE | ORPE-C-S | ELM-IRPE | EZ ± 5 | |
|---|---|---|---|---|---|---|
| Ensemble | 0.809 | 0.828 | 0.801 | 0.696 | 0.823 | 0.791 |
| Mean Intensity | 0.327 | 0.213 | 0.312 | 0.458 | 0.421 | 0.180 |
| Standard Deviation | 0.684 | 0.606 | 0.294 | 0.377 | 0.498 | 0.411 |
| Maximum Intensity | 0.244 | 0.373 | 0.296 | 0.276 | 0.380 | 0.180 |
| Minimum Intensity | 0.427 | 0.060 | 0.152 | 0.328 | 0.301 | 0.074 |
| Median Intensity | 0.309 | 0.172 | 0.191 | 0.327 | 0.123 | 0.068 |
| Kurtosis | 0.640 | 0.376 | 0.528 | 0.321 | 0.405 | 0.172 |
| Skewness | 0.751 | 0.467 | 0.245 | 0.369 | 0.403 | 0.544 |
| Gray-Level Entropy | 0.624 | 0.530 | 0.405 | 0.082 | 0.606 | 0.280 |
| Thickness | 0.552 | 0.421 | 0.638 | 0.585 |
| ELM-EZ | EZ-IRPE | IRPE-ORPE | ORPE-C-S | ELM-IRPE | EZ ± 5 | |
|---|---|---|---|---|---|---|
| Ensemble | 0.970 | 0.973 | 0.973 | 0.964 | 0.977 | 0.970 |
| Mean Intensity | 0.860 | 0.818 | 0.929 | 0.925 | 0.848 | 0.911 |
| Standard Deviation | 0.927 | 0.927 | 0.798 | 0.836 | 0.898 | 0.867 |
| Maximum Intensity | 0.898 | 0.927 | 0.946 | 0.924 | 0.900 | 0.845 |
| Minimum Intensity | 0.852 | 0.920 | 0.929 | 0.878 | 0.837 | 0.892 |
| Median Intensity | 0.855 | 0.884 | 0.942 | 0.873 | 0.649 | 0.839 |
| Kurtosis | 0.964 | 0.811 | 0.911 | 0.872 | 0.801 | 0.663 |
| Skewness | 0.962 | 0.948 | 0.932 | 0.809 | 0.906 | 0.905 |
| Gray-Level Entropy | 0.898 | 0.885 | 0.824 | 0.830 | 0.920 | 0.723 |
| Thickness | 0.942 | 0.880 | 0.918 | 0.902 |
| ELM-IRPE | EZ ± 5 | p-Value | |
|---|---|---|---|
| All Features Dice Coefficient | 0.830 | 0.803 | 0.005 |
| Mean Intensity Dice Coefficient | 0.744 | 0.734 | 0.193 |
| p-value | <0.001 | <0.001 | |
| All Features Pixel Accuracy | 0.980 | 0.971 | 0.039 |
| Mean Intensity Pixel Accuracy | 0.967 | 0.968 | 0.046 |
| p-value | <0.001 | 0.011 |
| ELM-IRPE | EZ ± 5 | p-Value | |
|---|---|---|---|
| All Features Dice Coefficient | 0.823 | 0.791 | 0.003 |
| Mean Intensity Dice Coefficient | 0.762 | 0.700 | 0.395 |
| p-value | <0.001 | <0.001 | |
| All Features Pixel Accuracy | 0.977 | 0.970 | 0.076 |
| Mean Intensity Pixel Accuracy | 0.969 | 0.956 | 0.003 |
| p-value | 0.001 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishra, Z.; Wang, Z.; Sadda, S.R.; Hu, Z. Using Ensemble OCT-Derived Features beyond Intensity Features for Enhanced Stargardt Atrophy Prediction with Deep Learning. Appl. Sci. 2023, 13, 8555. https://doi.org/10.3390/app13148555
Mishra Z, Wang Z, Sadda SR, Hu Z. Using Ensemble OCT-Derived Features beyond Intensity Features for Enhanced Stargardt Atrophy Prediction with Deep Learning. Applied Sciences. 2023; 13(14):8555. https://doi.org/10.3390/app13148555
Chicago/Turabian StyleMishra, Zubin, Ziyuan Wang, SriniVas R. Sadda, and Zhihong Hu. 2023. "Using Ensemble OCT-Derived Features beyond Intensity Features for Enhanced Stargardt Atrophy Prediction with Deep Learning" Applied Sciences 13, no. 14: 8555. https://doi.org/10.3390/app13148555
APA StyleMishra, Z., Wang, Z., Sadda, S. R., & Hu, Z. (2023). Using Ensemble OCT-Derived Features beyond Intensity Features for Enhanced Stargardt Atrophy Prediction with Deep Learning. Applied Sciences, 13(14), 8555. https://doi.org/10.3390/app13148555






