Abstract
The standardization of data from medical studies and hospital information systems to a common data model such as the Observational Medical Outcomes Partnership (OMOP) model can help make large datasets available for analysis using artificial intelligence approaches. Commonly, automatic mapping without intervention from domain experts delivers poor results. Further challenges arise from the need for translation of non-English medical data. Here, we report the establishment of a mapping approach which automatically translates German data variable names into English and suggests OMOP concepts. The approach was set up using study data from the Hamburg City Health Study. It was evaluated against the current standard, refined, and tested on a separate dataset. Furthermore, different types of graphical user interfaces for the selection of suggested OMOP concepts were created and assessed. Compared to the current standard our approach performs slightly better. Its main advantage lies in the automatic processing of German phrases into English OMOP concept suggestions, operating without the need for human intervention. Challenges still lie in the adequate translation of nonstandard expressions, as well as in the resolution of abbreviations into long names.
Keywords:
OMOP; common data model; mapping; standardization; SNOMED CT; Germany; healthcare data; TF-IDF; HCHS 1. Introduction
Harmonizing large amounts of patients records in a medical context is a challenging process. To realize this, the medical information of a patient must be documented in a structured form, best through the inclusion of suitable data standards in electronic patient files. The use of data standards enables the possibility for rigid information networks, such as hospital information systems (HIS), to process the information they receive in the same way. This is a prerequisite for modern and complex tasks in medical informatics [1].
Data standards can be limited to certain topics or regions, and often differ in the complexity of their structure. For example, in the area of drug encoding, the ATC (Anatomical Therapeutic Chemical Classification System) standard is primarily used in the European context, whereas RxNorm (Medical Prescription Normalized) is the standard in the US American context. The ICD-10 (International Classification of Diseases) code offers another example. It is often used as a local version of the ICD-10 standard published by the WHO in a respective country. Thus, the ICD-10-FR used in France has semantic differences to the ICD-10-GM, which is used in Germany. The transmission of information between individual information systems can differ. Additionally, the different data standards can be structured with varying degrees of complexity. The ICD-10 standard uses a taxonomic structure, whereas SNOMED CT (SNOMED Clinical Terms)—a standard used, for example, in the USA for coding diagnoses and procedures—has an ontological structure. Taxonomic data standards form classes and, thus, structure the data hierarchically. Ontologies are built on concepts, which are related. Therefore, hierarchical but also non-hierarchical, i.e., logical, relations can be displayed [2]. Many individual data standards served their purpose in the past years, but, increasingly, the inclusion of different data standards within an HIS leads to digital processes being considerably slowed down. In recent years, various approaches have been explored to address these data management challenges regarding semantic interoperability.
One approach that allows for a high degree of semantic interoperability is the use of a common data model (CDM). One well-known CDM is the Observational Medical Outcomes Partnership (OMOP) model of the Observational Health Data Sciences and Informatics (OHDSI) community. Local data are assigned to a standardized concept, which in turn is based on a data standard. The OMOP model is a relational data model. The clinical data area of the OMOP model stores patient data, whereas the standardized vocabularies build a large metadata repository within the OMOP model that includes several data standards such as ICD-10, SNOMED CT, or LOINC (Logical Observation Identifiers Names and Codes), a well-established standard for laboratory measurements. The vocabularies have to be actively included during the mapping process in order to standardize the local data and enable semantic interoperability. For example, the OMOP standard in the Condition_occurrence domain is SNOMED CT. Institutions that instead use ICD-10 for coding diagnoses can map their local ICD-10 codes to the respective SNOMED CT equivalent via the standardized vocabularies. This approach generates a high degree of semantic interoperability.
The standardization of medical data harbors great potential as it provides the basis to create large medical datasets through either data pooling or federated learning for analysis using notoriously data-hungry artificial intelligence methods [3]. Efforts are already underway to map German medical data to the OMOP CDM by applying the new Episode Domain to German Cancer Registry data [4], implementing OMOP at eight German hospitals [5], using HL7 FHIR to integrate German registry data into OMOP [6], mapping German infection-control-related data across openEHR, FHIR, and OMOP [7], creating a concept to transfer German drug [8] and procedure data [9] to OMOP, as well as by the establishment of an ETL (Extraction, Transformation, Loading)-process to OMOP for all German university hospitals [10]. However, challenges arise for one from medical data that does not adhere to any established data standard. This is frequently the case for study data, which is often tailored towards a specific research goal, and especially within questionnaire data. Furthermore, problems occur from the need for translation of non-English to English, which becomes necessary if specific vocabularies are not available in a certain language.
OHDSI provides a set of software tools to help prepare ETLs of structured data from common terminologies, vocabularies, and coding schemes called WhiteRabbit and Rabbit-In-A-Hat. To help map source codes, preferably from standard terminologies, to OMOP the OHDSI program offers USAGI [11]. It has, for example, recently been used to map clinical studies by condition to OMOP [12]. Usagi performs similarity mapping using term frequency-inverse document frequency (TF-IDF). TF-IDF is a statistical measure that represents term importance. It is a popular method often used by search engines. However, TF-IDF similarities are based merely on similar occurrences of keywords. Model training as in machine learning applications is not possible. Also, USAGI does not provide an option to translate non-English codes to English but suggests using Google Translate.
Recently, a deep-learning-based approach to terminology mapping to OMOP called TOKI has been published [13]. In contrast to TF-IDF, TOKI uses embedding-based semantic similarities where words are embedded into a semantic space defined primarily through their co-occurrence within text corpora. This makes individual word vectors easily comparable. TOKI reports a greater than ten percent improved matching accuracy compared to USAGI. Unfortunately, it does not provide a translation function and the source code of TOKI is not publicly available.
Furthermore, an NLP-based software solution called CLAMP exists [14]. It comprises a graphical user interface (GUI) to build customized NLP pipelines of sequential NLP tasks including tokenization, sentence boundary detection, part-of-speech tagger, named entity recognition, and others. It approaches clinical concept extraction as a supervised named entity recognition (NER) task and has recently been used to map COVID-19 signs and symptoms from clinical text to OMOP concepts [15]. NLP-based methods for the mapping of clinical text to OMOP such as CLAMP and related are being promoted by the OHDSI Natural Language Processing Working Group [16].
In this study, we use the TF-IDF method similarly to USAGI to map medical data from the Hamburg City Health Study (HCHS) to OMOP concepts. The HCHS is a large, population-based cohort study of 45,000 participants from the general population of Hamburg, Germany [17]. Participants undergo 18 examinations primarily targeting major organ system functions and structures including extensive imaging examinations. Additionally, before, during, and after the baseline visit validated self-report questionnaires asking for a variety of lifestyle and environmental conditions and habits are filled out.
2. Materials and Methods
2.1. Data
The methodology was developed and tested using two datasets.
2.1.1. HCHS Dataset
The Hamburg City Health Study (HCHS) is an ongoing single-center, prospective, observational, population-based cohort study of randomly selected residents of the metropolitan region of Hamburg, Germany, between the age of 45 and 74 years, aiming to investigate the development of chronic diseases [17]. The study is registered at clinicaltrials.gov, NCT03934957.
The dataset includes all variable names from the study variable manual for examination data from the hospital information system (n = 1033) as well as from the questionnaires (n = 1649), which had been provided as csv files from the study variable manual.
To evaluate the mapping pipeline, 100 random HCHS variable names were selected and mapped to standard concepts of the OMOP standardized vocabularies by a domain expert for mapping of cancer registry data to OMOP and CDM implementer with 3 years of experience (Supplementary Table S1). The terms were manually translated into English using Google Translate. Afterwards, the web application Athena [18] was used as a reference for the search for suitable OMOP concepts.
For method refinement, each 10 variable names were selected from the examination and the questionnaire dataset.
2.1.2. Anesthesiology Dataset
A total of 10 variable names were selected from questionnaire data from the VIDIAC study on videolaryngoscopic intubation and difficult airway classification [19].
A total of 10 variable names were selected from device data readouts of a monitoring system for hemodynamic parameters.
2.2. Methods
2.2.1. Mapping to OMOP Concepts
The mapping pipeline involves a series of sequential tasks. Initially, medical abbreviations in the German input terms are replaced by using a dictionary of 874 common German medical abbreviations and their long names [20]. This is followed by translation to English using HuggingFace transformer version 4.25.1 with model ‘Helsinki-NLP/opus-mt-de-en’ [21]. The translated data are then subjected to the removal of stop words and punctuation, lowercase conversion, and lemmatization via spaCy version 3.3.1 using model ‘en_core_web_lg’ to generate keywords [22]. Thereafter, the Scikit-learn library version 0.24.2 [23] is utilized to generate TF-IDF vectors for keywords and OMOP SNOMED CT (and LOINC) concepts, including their synonyms. The vectors are then used to compute the cosine similarity between words, identifying the top similar concepts for each variable.
2.2.2. Graphical User Interface
The JavaScript web framework Vue.JS version 3.2.37 was used for programming the prototype, as Vue.JS is easy to learn with knowledge of JavaScript, TypeScript, and HTML and can be used to implement single- and multipage web applications. To retrieve the data from the backend the extension Apollo in version 3.6.9 was used.
For mobile app development Framework7.io was used. Framework7 is a free and open-source mobile HTML framework that allows you to develop hybrid mobile apps for iOS and Android. It can potentially be integrated directly into Vue via an extension. To implement the Swipe-GUI the swipeableCards plugin (https://github.com/elzahaby/swipeableCards) was used.
To evaluate the usability of created GUIs, expert user testings and interviews were conducted with physicians and a computer scientist from the Department of Anesthesiology as well as from the Institute for Applied Medical Informatics. Both senior scientists and residents with little or no scientific experience were represented. A total of 8 experts participated in the interview (female = 2, male = 6).
The answers to the interview questions were collected electronically using the web-based application LimeSurvey. The questionnaire contained 42 questions, 28 of which were free-text fields. For GUI-related questions of the interview see Supplementary Table S2.
3. Results
3.1. Concept of Mapping Pipeline
The aim of the project was to create a workflow that helps map data that does not currently adhere to a common data standard such as health study data. Large parts of these data are commonly not mapped to a standardized vocabulary such as ICD-10, RxNorm, or ATC. Therefore, mapping of these data to a common data model such as OMOP cannot be performed in an automated fashion. The current process is to use the USAGI tool which suggests OMOP concepts based on the relevance of keywords within OMOP concept names using TF-IDF. When dealing with non-English data this is preceded by manual translation using a publicly available internet service such as Google Translate as well as some form of manual simplification of especially long phrases from questionnaire data (Figure 1 top). Our approach eliminates the need for human–computer interaction during the process of translation, keyword preprocessing, and concept matching (Figure 1 bottom) as needed in the current process using USAGI.
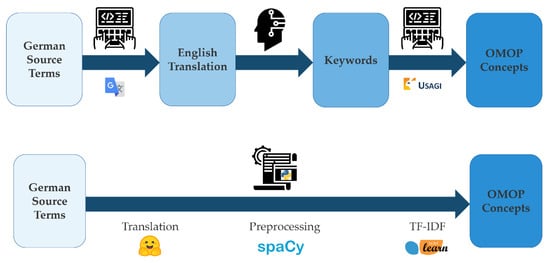
Figure 1.
Top: current workflow of mapping German study data to the OMOP common data model; Bottom: new semiautomated approach.
The workflow is realized in Python and uses publicly available tools from Hugging Face (https://huggingface.co), spaCy (https://spacy.io), and scikit-learn (https://scikit-learn.org).
Firstly, common German medical abbreviations (e.g., ‘KHK’—‘Koronare Herzkrankheit’, Engl. ‘Coronary Artery Disease’) are automatically transformed into their long names using a custom dictionary before translation using Hugging Face (Figure 2). The so-obtained English expressions are automatically preprocessed to remove common words (‘stop words’ like ‘a’, ‘the’, etc.) and punctuation; to remove inflections such as plurals, verb tense, etc. (‘lemmatization’); and are then put into lowercase using spaCy. These remaining keywords are used for concept matching via TF-IDF from scikit-learn. The algorithm is publicly available and fully customizable, and may serve as the foundation for similar tasks in different contexts.
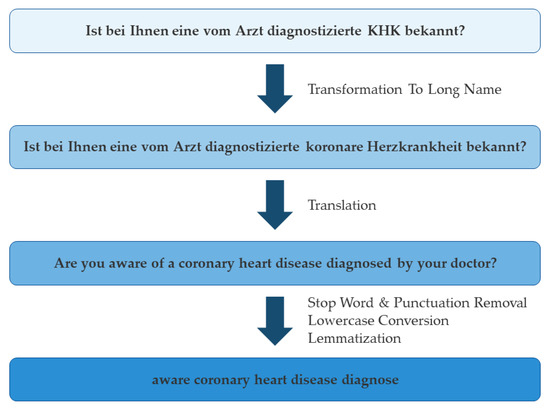
Figure 2.
Example of the translation and preprocessing workflow.
One major challenge lies in delivering a meaningful, automated translation of non-English medical expressions into English. The workflow relies on a popular transformer model trained on the OPUS-MT open-source parallel corpus [24].
However, medical expressions can be particularly challenging. For one, the training data of common translation models contains only a few medical texts. Therefore, less frequent medical terms may be unknown to the model. For example, ‘Luftnot’ translates to ‘air need’ rather than ‘shortness of breath’. Therefore, current efforts of the community aim to establish a text corpus and model specifically for medical text [25,26,27,28,29].
Additionally, medical expressions, particularly in Germany, often make use of Latin expressions (e.g., ‘Ulcus cruris’, Engl. ‘leg ulcer’). The use of uncommon abbreviations (e.g., ‘RR’ for ‘riva rocci’ which represents ‘blood pressure’) and of colloquialisms or layman’s terms (e.g., ‘Schaufensterkrankheit’, ‘offenes Bein’ for leg ulcer) hamper the translation task even further.
Medical expressions, which fail an appropriate translation from German to English, subsequently fail to generate useful OMOP concept suggestions at the end of the workflow.
3.2. Code Refinement
The initially created workflow performed inferiorly to the manual method using USAGI. Especially, medical synonyms such as ‘coronary arteriosclerosis’ instead of ‘coronary artery disease’ were not recognized.
To refine the mapping algorithm to improve suggestion results, we selected a small set of variable names to evaluate results in detail and modify code to improve them. The original code (ORI) underwent three rounds of refinement: Firstly, we applied minor changes to cosine similarity score calculation and package use (MIN). Thereafter, a major change was applied to also use OMOP concept synonyms for term matching (SYN). Finally, duplicate concepts from synonym matching were removed from the list of results (DUP).
A comparison of the final results from our algorithm with the standard USAGI method showed comparable results in 12 of 20 cases. In 4 of 20 cases it delivered improved results; in 2 of 20 cases the results were worse. In 2 of 20 cases the translation itself failed to return meaningful expressions which could then be mapped to OMOP concepts.
The different cases are represented in Table 1. The identified concepts are color-coded qualitatively based on if they are a good match to the German source term (dark green—good match, light green—acceptable match, orange—poor match, red—no match). For a full list of results for all 20 terms see Supplementary Table S3.

Table 1.
Comparison of mapping results during code refinement with USAGI.
For a thorough evaluation of our refined mapping algorithm, we applied it to all HCHS examination and questionnaire variable names (Figure 3, Supplementary Table S4). When considering the top similarity score for the best concept suggestion for each term, we observe that examination terms extracted from the hospital information system perform better than those from questionnaires (p-value = 1.717 × 10−11, Welch Two Sample t-test), with mean similarity scores for examinations of 0.6717 (first quartile: 0.5486, median: 0.6572, and third quartile: 0.7781), whereas for questionnaires of 0.6509 (first quartile: 0.5173, median: 0.6293, and third quartile: 0.7636).
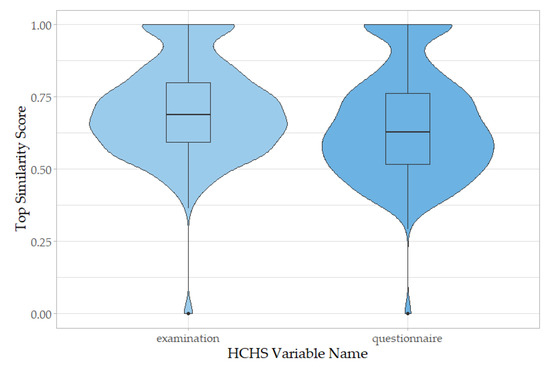
Figure 3.
Distribution of top similarity scores of suggested OMOP concepts for all HCHS variable names. Variable names from the HCHS examinations dataset (left) map better based on keyword to concept similarity score than those from the HCHS questionnaires dataset (right). (p-value = 1.717 × 10−11, Welch Two Sample t-test).
A total of 0.96% of examination variable names did not have any mapping suggestions (mean similarity score = 0), as did 0.75% of questionnaire variable names.
In a random subset of variable names, the algorithm identified the same concept as one of its top five concept suggestions as an OMOP mapping expert in 56 percent of cases (50 of 90) (Supplementary Table S1, highlighted concepts).
3.3. Application to Independent Datasets
In order to evaluate the performance of our algorithm when mapping other datasets, we applied the refined code to a small dataset of variable names from an anesthesiology study as well as to abbreviated variables from anesthesiology measurements. The first set of study variables is used to measure the direct applicability of our code to other datasets of similar structure. The abbreviated set was chosen for one to evaluate the mappability of abbreviations (mostly via synonyms), as well as the use of an expanded list of OMOP concepts including both SNOMED CT and LOINC terms and their effect on mapping performance.
The first task of providing OMOP concept suggestions for anesthesiology study names was similar to the task the algorithm was designed for. However, some of the medical terms in this methodological study may be more uncommon than those in the HCHS public health cohort study. Here, the algorithm was able to find a suitable OMOP concept in five of ten cases (Supplementary Table S5). In one case, ambiguous spelling of the term causes failure, as ‘Thyreomentaler Abstand’ translates to ‘thyreomental distance’ but not ‘thyromental distance’ for concept 4142891. In another case, the term ‘Maskenbeatmung unmöglich’ translates to ‘mask breathe impossible’, which relates to the concept ‘controlled ventilation’ (4074665). This logical relation is, however, not known by the algorithm. The term ‘Kehlkopf/Atemwegstrauma’ which translates to ‘larynx/respiratory trauma’ should be associated with the concept ‘injury of larynx’ (4053585, synonym: laryngeal trauma). However, it does not appear in the algorithm’s top five concept suggestions, probably because of the differing word combination of the synonymous words larynx/laryngeal and trauma/injury. In another case, the translated verb ‘tracheotomize’ does not lead to an association with the noun tracheotomy for which multiple potentially suitable concepts exist (‘incision of trachea’ (4168133), ‘Tracheostomy, emergency procedure by transtracheal approach’ (4208093). In one case (‘Simplified Airway Risk Index’), no fitting OMOP concept could be identified.
When comparing the algorithm performance using an OMOP concept library only containing LOINC terms or only SNOMED CT terms or a combined library, no negative effects when using the combined library could be observed.
The second task using anesthesiology-related measurements illustrated the challenge of mapping abbreviated medical terms to concepts (Supplementary Table S6). Here, seven of ten abbreviations failed to map to the correct concept, as these abbreviations neither occur in the OMOP concept name nor in its concept synonyms. In contrast, when using the correct long names as provided by a medical domain expert all of these standard measurements could be mapped correctly.
3.4. Graphical User Interface for Semiautomated Concept Mapping
Because of the complexity of the mapping process, the need for human interaction, preferably by domain experts, to select the appropriate concept in the target CDM to ensure high-quality mappings persists. Intelligent user interfaces facilitate this human–computer interaction. Designed as the front end of an automated, intelligent premapping microservice that could be integrated into a metadata repository [30], it focusses on what has been the focus of science for years. In many areas, especially in the health sector, the requirements for usability are well defined. In the European Medical Device Regulation, usability and the proof of testing (e.g., for the risk of incorrect operation) is of high importance. The clear presentation of relevant information, prevention of operational errors through intuitive and consistent operation, and a high level of user ergonomics are defined in ISO/IEC 62366-1:2015 [31].
We designed three exemplary graphical user interfaces (GUIs) to display descriptive information for each mapping item (e.g., study variable name) as well as for the interaction during the mapping of information elements.
The Floating-Action-Button (FAB)-GUI consists of two consecutive pages (Figure 4). First, the user is provided with an overview of the data elements to be processed in tabular form. After clicking on the table row, a page with information about the individual element is displayed, including the element name and a short description. Up to four of the most-likely entries found in the LOINC or SNOMED CT database are displayed. In addition, the four most probable classes are offered for selection with blue icons. By clicking on one of the terms, the mapping suggestion is selected by the user. At the bottom of the second page, information on the input dataset is displayed which can be expanded. The goal of this GUI is to direct the user to a single element via a selection page where the necessary information about the selected concept is presented.
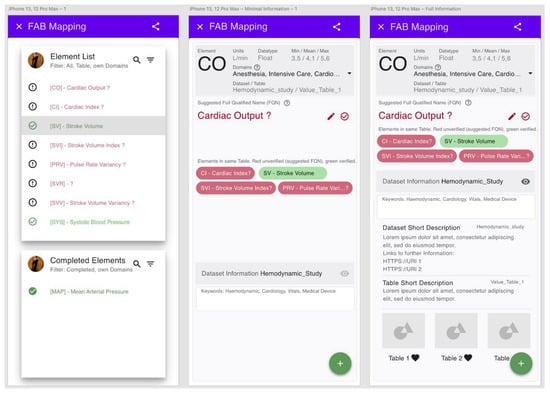
Figure 4.
Example of the Floating-Action-Button graphical user interface (FAB-GUI). Left: first page with tabular view of mappable items. Middle: second page with mapping suggestions for a selected item. Right: second page with expanded dataset description.
Alternatively, the Table-GUI shows a table with all elements to be mapped on one page. For each table element the mapping suggestions appear directly below the information about the element. When you click on a mapping suggestion, the element mapped with it disappears from the list. As information elements the user is offered the same elements as in the FAB mapping.
Finally, a Swipe-GUI was developed which displays only one element at a time as a swipeable card from a stack of cards. The user can swipe the card left or right to perform an assignment to one of two mapping suggestions. The information elements on the card are intentionally limited. When you tap on a suggested concept, it expands to show the information similar to the other GUIs.
To evaluate the performance of the different GUIs, user testing among a small group of potential users was carried out. The participants were given test versions on a smart phone, displaying mapping suggestions for the anesthesiology dataset. Afterwards an online questionnaire was completed. The questions were mostly in free-text form and aimed at capturing the individual user experience rather than delivering a quantitative comparison of the different GUI concepts, as the way of interaction with the application strongly depends on the habits of the individual user.
Overall, the analysis of user responses showed that the FAB-GUI and the Table-GUI were received about equally well, with the Table-GUI perceived as faster when dealing with a large list of elements. The concept of the Swipe-GUI was well received by the majority of participants.
4. Discussion
We present an approach for mapping nonstandardized German medical data to the OMOP common data model. The established workflow handles German data from translation to concept suggestion without the need for human–computer interaction. This can be of advantage particularly when dealing with large datasets and frequent mapping tasks. The performance of our algorithm is comparable to the manual method, in that it suggests the right OMOP concept as frequently.
However, there is still room for improvement: For one, a major challenge lies in the correct translation of clinical expressions, especially from unstructured data. Here, a great need exists for a comprehensive multilingual medical text corpus as the basis for improved language models. Although some efforts are being made in individual countries, there seems to be the need for a concerted effort to include a comprehensive collection of languages in this corpus. For example, a project called the European Clinical Case Corpus (E3C, https://e3c.fbk.eu) has generated a freely available multilingual corpus in English, French, Italian, Spanish, and Basque but is not applicable to German texts. And, German efforts to create a medical text corpus for natural language processing as part of the Medical Informatics Initiative are currently predominantly limited to the German language itself.
Additional to the translation of non-English terms to English, the project has highlighted problems with the identification of word synonyms. For example, the algorithm struggles with the equivalence of verbs, nouns, and adjectives of the same word stem (larynx vs. laryngeal; tracheotomize vs. tracheotomy), as well as words with similar meaning (injury vs. trauma). To improve results, a semantic web with word embeddings representing semantic distances could be utilized which would account for such semantic similarities [32,33,34].
Furthermore, the comprehensiveness of concept synonyms within the OMOP catalogue could be reviewed. Especially for standard medical abbreviations, shortcomings have become apparent. However, abbreviations are inherently ambiguous. For example, the abbreviation CO can stand for carbon monoxide or cardiac output, and PR may mean progesterone receptor or pulse rate. Therefore, it seems that some form of integration of domain knowledge is inevitable. For example, a large dictionary of 858 thousand medical acronyms and abbreviations [35] could be integrated in the algorithm, combined with the need for user–computer interaction to select the domain-specific appropriate long name.
Problems also arise from the words and phrases chosen in these unstructured datasets, with the use of nonstandard expressions and layman’s terms affecting translation and subsequent mapping. Also, the relation between individual variables (such as questions within a questionnaire) has an impact. As each variable name is processed independently, a semantic reference of a question to the one before needs to be avoided under all circumstances. Therefore, the early involvement of a domain expert for data standardization during study design should be considered.
Overall, the presented mapping tool shows a feasible approach for the automation of specific mapping tasks. Its code is publicly available and customizable, and could be integrated into a metadata repository. Besides a user-friendly graphical user interface, additional functions could be added, such as the ability to select specific OMOP vocabularies or domains.
5. Conclusions
For the first time, our study describes a semiautomated mapping process for nonstandardized German data to English OMOP concepts. Our publicly available tool using TF-IDF suggests concepts after automated translation, and can be embedded into a user-friendly graphical user interface. Improvements in translation tools for medical text will lead to improved mapping tools such as ours in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app13148159/s1.
Author Contributions
Conceptualization: S.N.; methodology/software: A.C., C.G., P.B. and H.H.; validation: J.C. and S.N.; formal analysis: A.C.; data curation: A.C.; writing—original draft preparation: S.N.; writing—review and editing: R.T. and S.B.; visualization: S.N.; supervision: F.Ü.; project administration: S.N. All authors have read and agreed to the published version of the manuscript.
Funding
The participating institutes and departments from the University Medical Center Hamburg-Eppendorf all contributed with individual and scaled budgets to the overall funding. We acknowledge financial support from the Open Access Publication Fund of UKE—Universitätsklinikum Hamburg-Eppendorf and DFG—German Research Foundation. The HCHS is additionally funded by the euCanSHare grant agreement [grant number 825903-euCanSHare H2020]; the Foundation Leducq [grant number 16 CVD 03] and the innovative medicine initiative [Grant Number 116074]. The HCHS is further supported by Deutsche Gesetzliche Unfallversicherung (DGUV); Deutsches Krebsforschungszentrum (DKFZ); Deutsches Zentrum für Herz-Kreislauf-Forschung (DZHK); Deutsche Stiftung für Herzforschung; Seefried Stiftung; Bayer; Amgen, Novartis; Schiller; Siemens; Topcon, Unilever; and by donations from the ’Förderverein zur Förderung der HCHS e.V.‘ and TePe® (2014). Sponsor funding has in no way influenced the content or management of this study.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The computer code generated for this project is available at https://github.com/UKEIAM/de.uke.iam.automapping). All hyperlinked websites of tools and repositories were last accessed and accessible on 12 July 2023.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Institute of Medicine (US) Committee on Data Standards for Patient Safety. Patient Safety: Achieving a New Standard for Care; Aspden, P., Corrigan, J.M., Wolcott, J., Erickson, S.M., Eds.; National Academies Press (US): Washington, DC, USA, 2004; ISBN 978-0-309-09077-3. [Google Scholar]
- Haendel, M.A.; Chute, C.G.; Robinson, P.N. Classification, Ontology, and Precision Medicine. N. Engl. J. Med. 2018, 379, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, N.; Peng, Y.; Wolfien, M.; Zoch, M.; Sedlmayr, M. OMOP CDM Can Facilitate Data-Driven Studies for Cancer Prediction: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 11834. [Google Scholar] [CrossRef]
- Carus, J.; Nürnberg, S.; Ückert, F.; Schlüter, C.; Bartels, S. Mapping Cancer Registry Data to the Episode Domain of the Observational Medical Outcomes Partnership Model (OMOP). Appl. Sci. 2022, 12, 4010. [Google Scholar] [CrossRef]
- Maier, C.; Lang, L.; Storf, H.; Vormstein, P.; Bieber, R.; Bernarding, J.; Herrmann, T.; Haverkamp, C.; Horki, P.; Laufer, J.; et al. Towards Implementation of OMOP in a German University Hospital Consortium. Appl. Clin. Inform. 2018, 9, 54–61. [Google Scholar] [CrossRef]
- Fischer, P.; Stöhr, M.R.; Gall, H.; Michel-Backofen, A.; Majeed, R.W. Data Integration into OMOP CDM for Heterogeneous Clinical Data Collections via HL7 FHIR Bundles and XSLT. Stud. Health Technol. Inform. 2020, 270, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, E.; Thun, S. From OpenEHR to FHIR and OMOP Data Model for Microbiology Findings. Stud. Health Technol. Inform. 2021, 281, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, I.; Zoch, M.; Wilhelm, M.; Sedlmayr, M.; Bathelt, F. Transfer of Clinical Drug Data to a Research Infrastructure on OMOP—A FAIR Concept. Stud. Health Technol. Inform. 2021, 287, 63–67. [Google Scholar] [CrossRef]
- Reinecke, I.; Kallfelz, M.; Sedlmayr, M.; Siebel, J.; Bathelt, F. Evaluation and Challenges of Medical Procedure Data Harmonization to SNOMED-CT for Observational Research. Stud. Health Technol. Inform. 2022, 294, 405–406. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Henke, E.; Reinecke, I.; Zoch, M.; Sedlmayr, M.; Bathelt, F. An ETL-process design for data harmonization to participate in international research with German real-world data based on FHIR and OMOP CDM. Int. J. Med. Inform. 2023, 169, 104925. [Google Scholar] [CrossRef]
- USAGI for Vocabulary Mapping. Available online: https://www.ohdsi.org/analytic-tools/usagi/ (accessed on 17 April 2023).
- Liu, H.; Carini, S.; Chen, Z.; Phillips Hey, S.; Sim, I.; Weng, C. Ontology-based categorization of clinical studies by their conditions. J. Biomed. Inform. 2022, 135, 104235. [Google Scholar] [CrossRef]
- Kang, B.; Yoon, J.; Kim, H.Y.; Jo, S.J.; Lee, Y.; Kam, H.J. Deep-learning-based automated terminology mapping in OMOP-CDM. J. Am. Med. Inform. Assoc. 2021, 28, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Soysal, E.; Wang, J.; Jiang, M.; Wu, Y.; Pakhomov, S.; Liu, H.; Xu, H. CLAMP—A toolkit for efficiently building customized clinical natural language processing pipelines. J. Am. Med. Inform. Assoc. 2018, 25, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Abu-El-Rub, N.; Gray, J.; Pham, H.A.; Zhou, Y.; Manion, F.J.; Liu, M.; Song, X.; Xu, H.; Rouhizadeh, M.; et al. COVID-19 SignSym: A fast adaptation of a general clinical NLP tool to identify and normalize COVID-19 signs and symptoms to OMOP common data model. J. Am. Med. Inform. Assoc. 2021, 28, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- OHDSI Natural Language Processing Working Group. Available online: https://www.ohdsi.org/web/wiki/doku.php?id=projects:workgroups:nlp-wg (accessed on 17 April 2023).
- Jagodzinski, A.; Johansen, C.; Koch-Gromus, U.; Aarabi, G.; Adam, G.; Anders, S.; Augustin, M.; der Kellen, R.B.; Beikler, T.; Behrendt, C.-A.; et al. Rationale and Design of the Hamburg City Health Study. Eur. J. Epidemiol. 2020, 35, 169–181. [Google Scholar] [CrossRef]
- Athena. Available online: https://athena.ohdsi.org (accessed on 17 April 2023).
- Kohse, E.K.; Siebert, H.K.; Sasu, P.B.; Loock, K.; Dohrmann, T.; Breitfeld, P.; Barclay-Steuart, A.; Stark, M.; Sehner, S.; Zöllner, C.; et al. A model to predict difficult airway alerts after videolaryngoscopy in adults with anticipated difficult airways—The VIDIAC score. Anaesthesia 2022, 77, 1089–1096. [Google Scholar] [CrossRef]
- Medizinische Abkürzungen. Available online: https://www.bionity.com/de/lexikon/Medizinische_Abk%C3%BCrzungen.html (accessed on 17 April 2023).
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M.; et al. Transformers: State-of-the-Art Natural Language Processing. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations; Association for Computational Linguistics, Online, 16–20 November 2020; pp. 38–45. [Google Scholar]
- spaCy 2: Natural Language Understanding with Bloom Embeddings, Convolutional Neural Networks and Incremental Parsing. Available online: https://sentometrics-research.com/publication/72/ (accessed on 17 April 2023).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Tiedemann, J.; Thottingal, S. OPUS-MT—Building open translation services for the World. In Proceedings of the 22nd Annual Conference of the European Association for Machine Translation; European Association for Machine Translation, Lisboa, Portugal, 3–5 November 2020; pp. 479–480. [Google Scholar]
- Yang, X.; Chen, A.; PourNejatian, N.; Shin, H.C.; Smith, K.E.; Parisien, C.; Compas, C.; Martin, C.; Costa, A.B.; Flores, M.G.; et al. A large language model for electronic health records. NPJ Digit. Med. 2022, 5, 194. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, X.; Hou, Y.; Li, G.; Wang, H.; Xu, H.; Xiang, Y.; Tang, B. Multimodal Data Matters: Language Model Pre-Training Over Structured and Unstructured Electronic Health Records. IEEE J. Biomed. Health Inform. 2022, 27, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Naseem, U.; Dunn, A.G.; Khushi, M.; Kim, J. Benchmarking for biomedical natural language processing tasks with a domain specific ALBERT. BMC Bioinform. 2022, 23, 144. [Google Scholar] [CrossRef]
- Frei, J.; Frei-Stuber, L.; Kramer, F. GERNERMED++: Transfer Learning in German Medical NLP. arXiv 2022, arXiv:2206.14504. [Google Scholar] [CrossRef]
- Roller, R.; Seiffe, L.; Ayach, A.; Möller, S.; Marten, O.; Mikhailov, M.; Alt, C.; Schmidt, D.; Halleck, F.; Naik, M.; et al. A Medical Information Extraction Workbench to Process German Clinical Text. arXiv 2022, arXiv:2207.03885. [Google Scholar] [CrossRef]
- Kadioglu, D.; Breil, B.; Knell, C.; Lablans, M.; Mate, S.; Schlue, D.; Serve, H.; Storf, H.; Ückert, F.; Wagner, T.; et al. Samply.MDR—A Metadata Repository and Its Application in Various Research Networks. Stud. Health Technol. Inform. 2018, 253, 50–54. [Google Scholar] [PubMed]
- ISO/IEC 62366-1; Medical devices—Part 1: Application of Usability Engineering to Medical Devices. International Electrotechnical Commission: Geneva, Switzerland, 2015.
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient Estimation of Word Representations in Vector Space. arXiv 2013, arXiv:1301.3781. [Google Scholar] [CrossRef]
- Zhang, J.; Kowsari, K.; Harrison, J.H.; Lobo, J.M.; Barnes, L.E. Patient2Vec: A Personalized Interpretable Deep Representation of the Longitudinal Electronic Health Record. IEEE Access 2018, 6, 65333–65346. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Q.; Bai, H.; Liu, C.; Liu, W.; Zhang, Y.; Jiang, L.; Xu, H.; Wang, K.; Zhou, Y. EHR2Vec: Representation Learning of Medical Concepts From Temporal Patterns of Clinical Notes Based on Self-Attention Mechanism. Front. Genet. 2020, 11, 630. [Google Scholar] [CrossRef]
- Medical Abbreviations. Available online: https://www.allacronyms.com/medical/abbreviations (accessed on 17 April 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).