NAMRTNet: Automatic Classification of Sleep Stages Based on Improved ResNet-TCN Network and Attention Mechanism
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
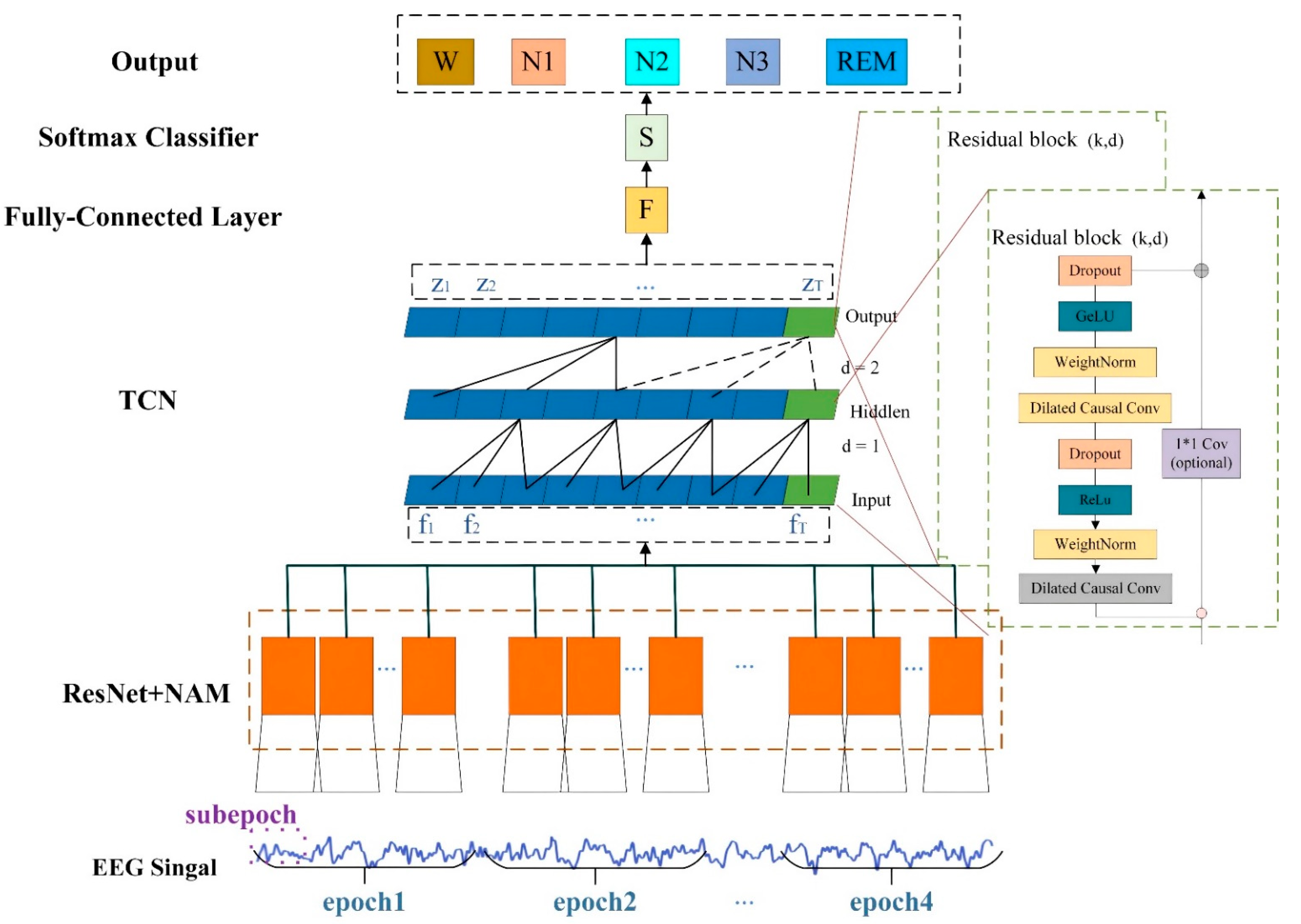
2.2. Model Overview
2.3. Multi-Sub-Epoch Feature Learning
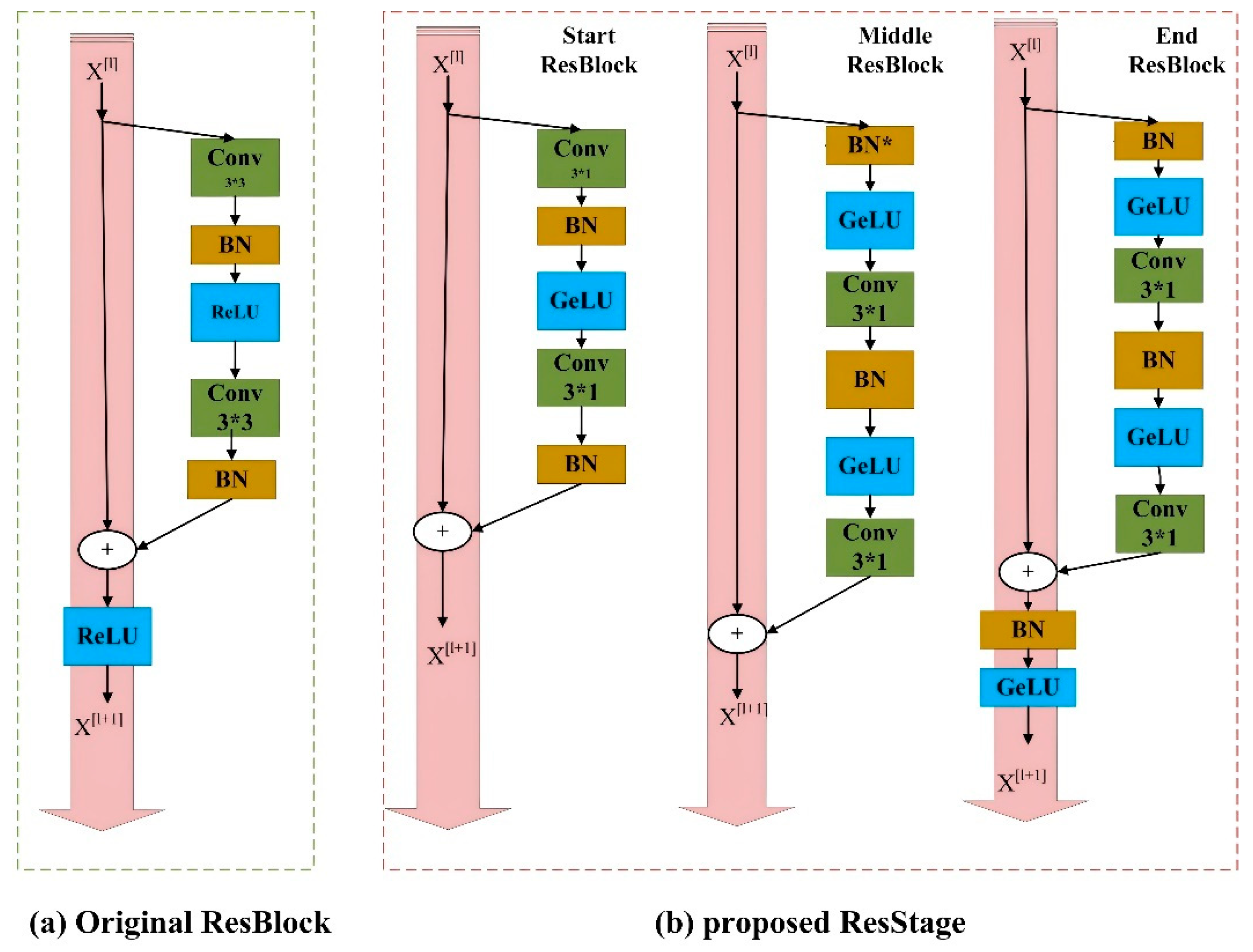
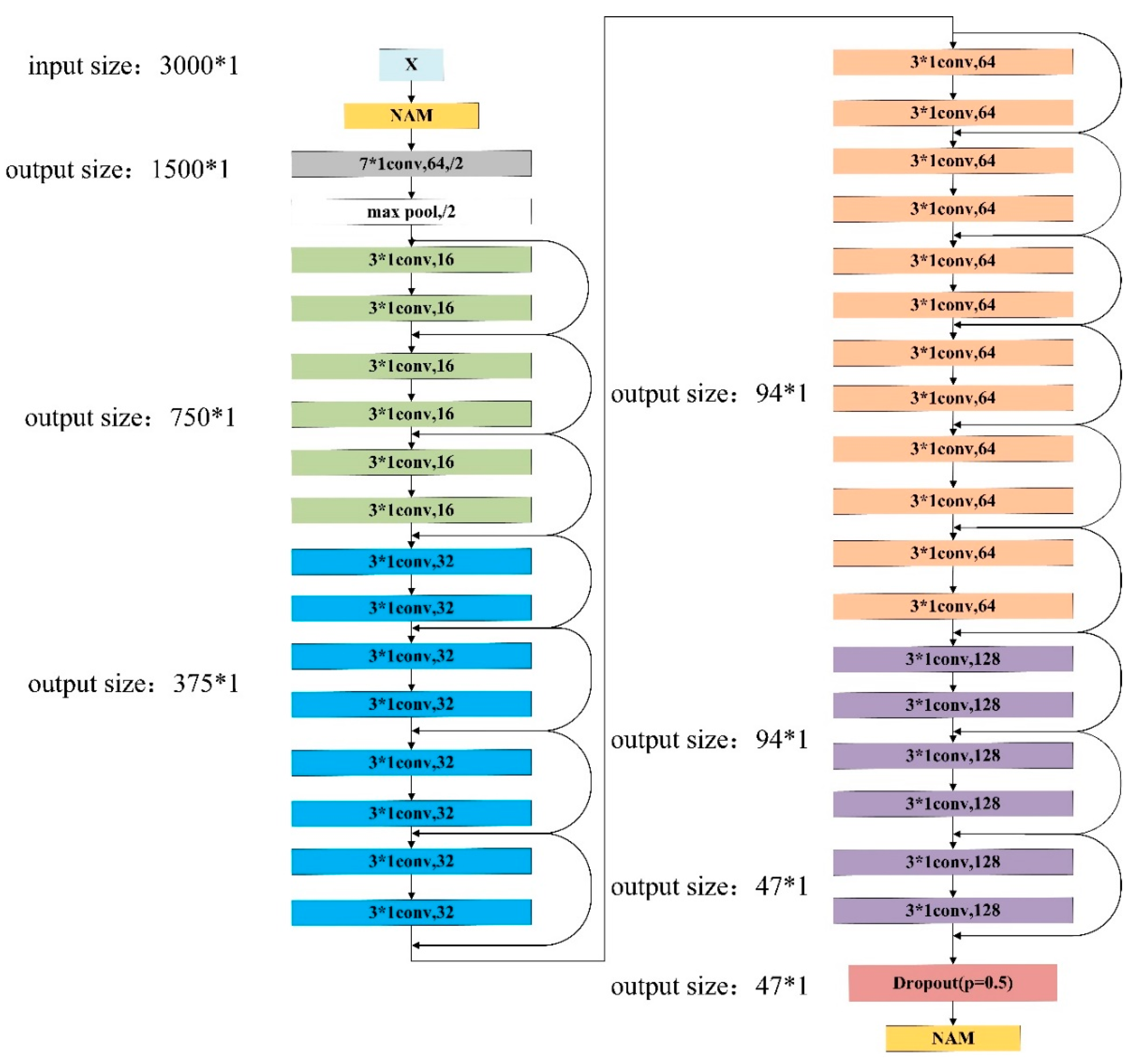
2.4. Improved ResNet 34
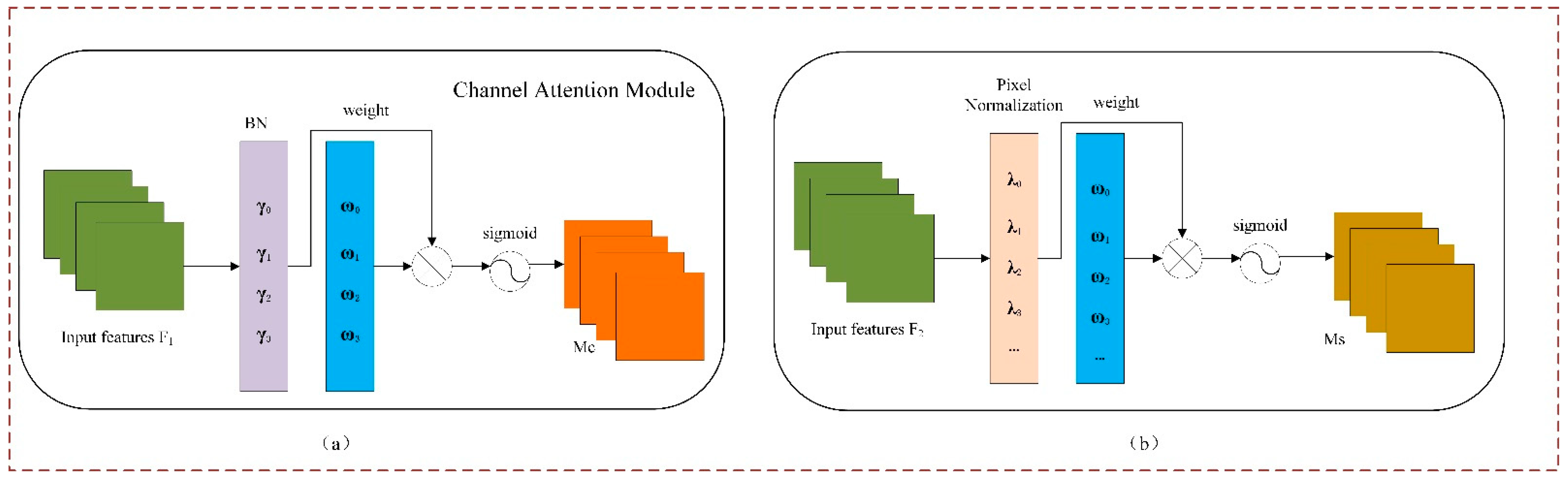
2.5. NAM Attention Mechanism
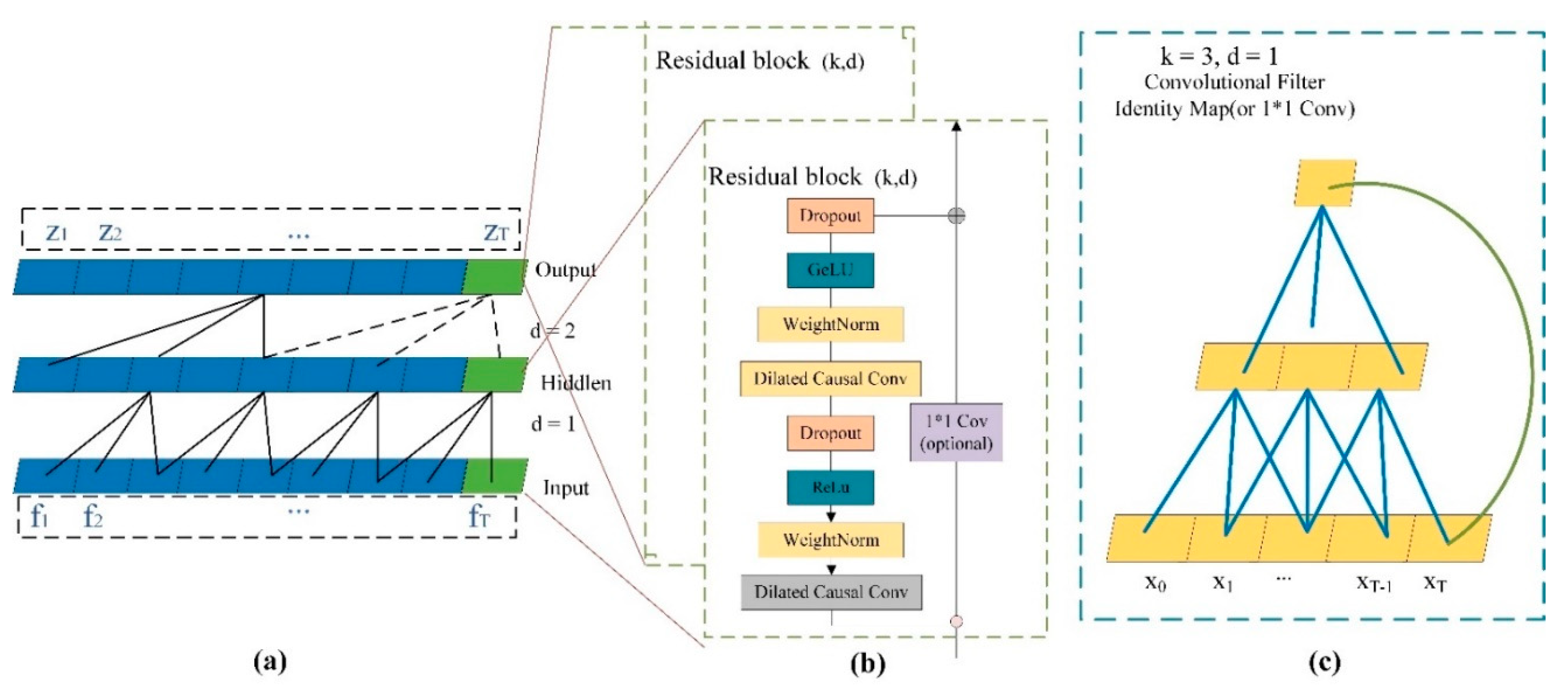
2.6. Long Time Series Dependencies
3. Results and Discussion
3.1. Evaluation Metrics
3.2. Parameters of the Optimizer
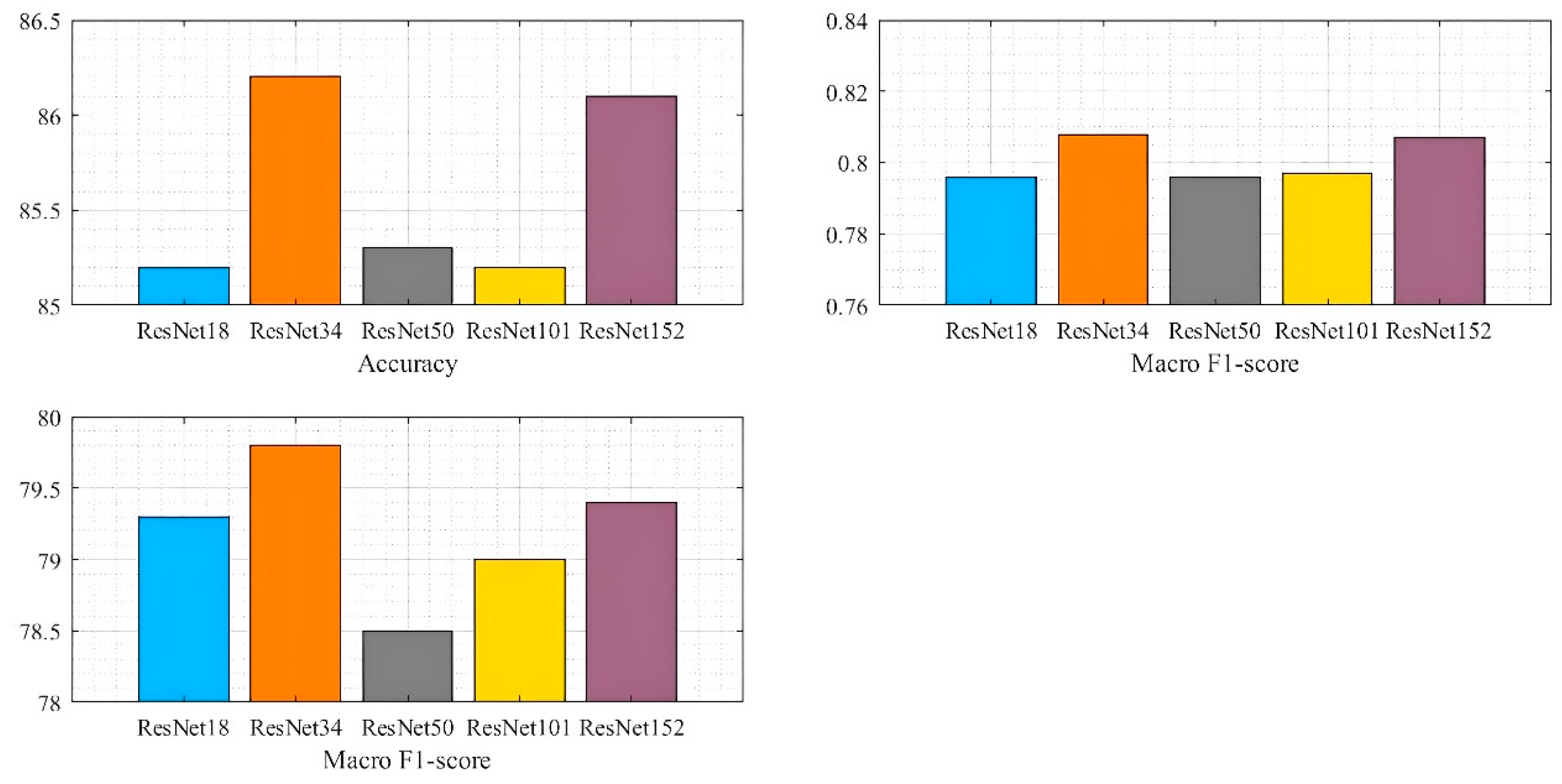
3.3. ResNet Layer Number Settings
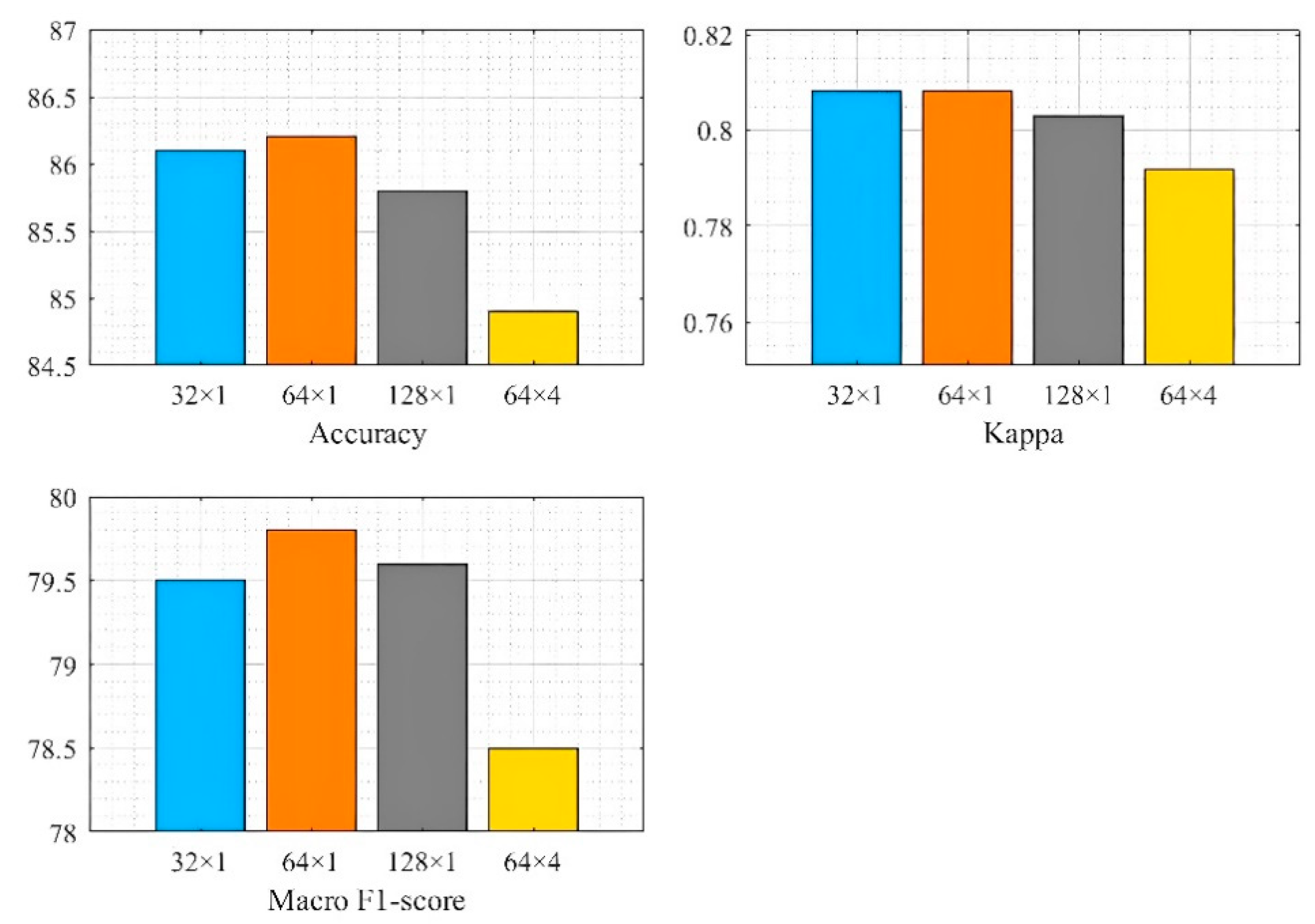
3.4. TCN Hidden Layer Setting
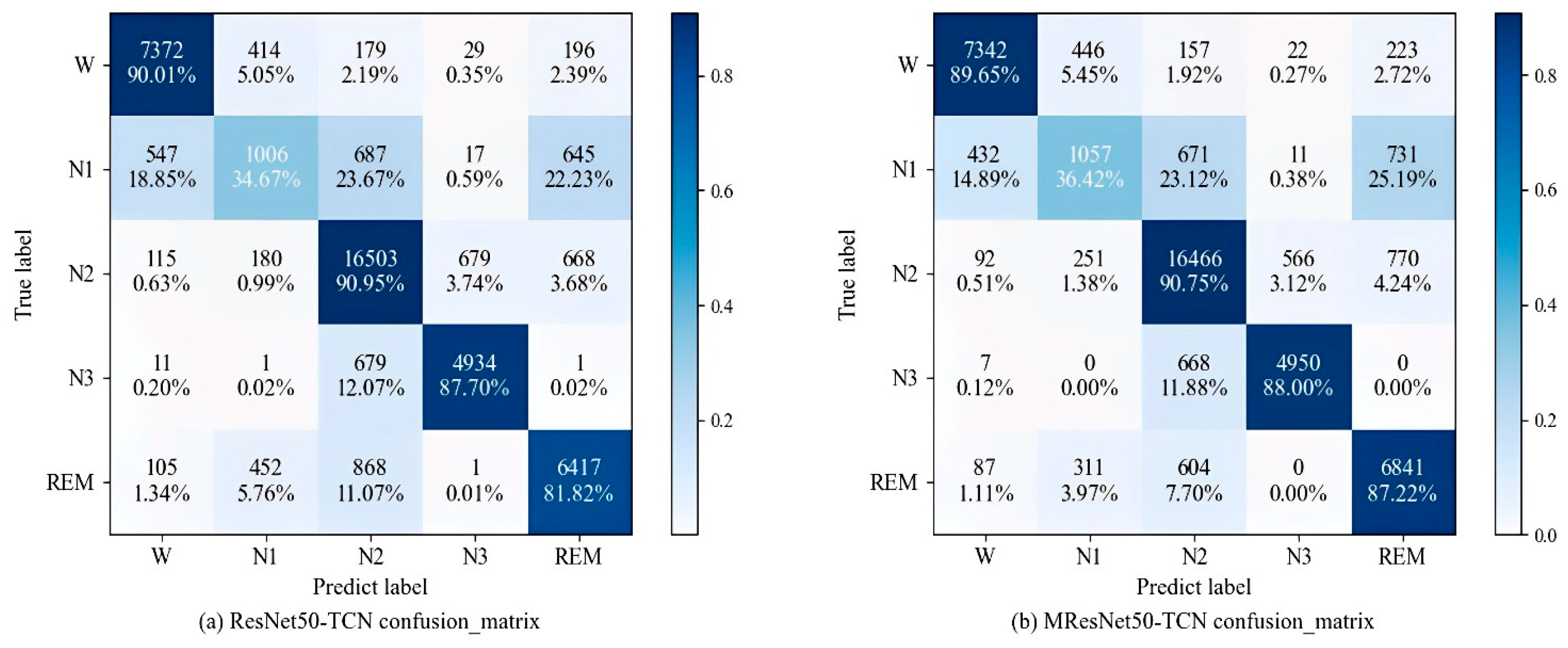
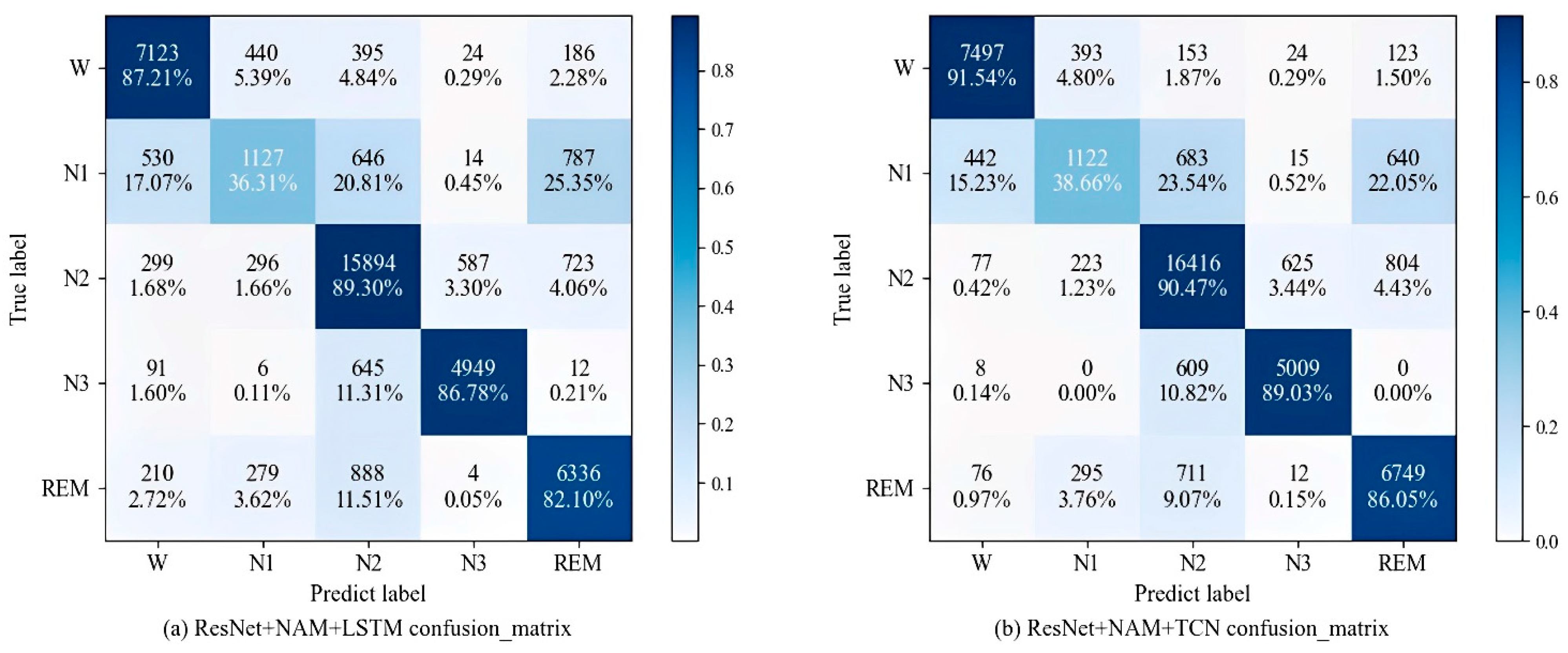
3.5. Comparative Experiments
3.6. K-Fold Crossover Experiment
3.7. Sleep Stage Scores
3.8. Comparison with the Forward Approach
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Luyster, F.S.; Strollo, P.J., Jr.; Zee, P.C.; Walsh, J.K. Boards of Directors of the American Academy of Sleep Medicine and the Sleep Research Society. Sleep A Health Imp. Sleep 2012, 35, 727–734. [Google Scholar] [CrossRef]
- Ruiz-Herrera, N.; Díaz-Román, A.; Guillén-Riquelme, A.; Quevedo-Blasco, R. Sleep Patterns during the COVID-19 Lockdown in Spain. Int. J. Environ. Res. Public Health 2023, 20, 4841. [Google Scholar] [CrossRef] [PubMed]
- Keenan, S.A. An overview of polysomnography. Handb. Clin. Neurophysiol. 2005, 6, 33–50. [Google Scholar]
- Stepnowsky, C.; Levendowski, D.; Popovic, D.; Ayappa, I.; Rapoport, D.M. Scoring accuracy of automated sleep staging from a bipolar electroocular recording compared to manual scoring by multiple raters. Sleep Med. 2013, 14, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.R.; Bhuiyan, M.I.H. Computer-aided sleep staging using complete ensemble empirical mode decomposition with adaptive noise and bootstrap aggregating. Biomed. Signal Process. Control. 2016, 24, 1–10. [Google Scholar] [CrossRef]
- Sharma, R.; Pachori, R.B.; Upadhyay, A. Automatic sleep stages classification based on iterative filtering of electroencephalogram signals. Neural Comput. Appl. 2017, 28, 2959–2978. [Google Scholar] [CrossRef]
- Hassan, A.R.; Bhuiyan, M.I.H. A decision support system for automatic sleep staging from EEG signals using tunable Q-factor wavelet transform and spectral features. J. Neurosci. Methods 2016, 271, 107–118. [Google Scholar] [CrossRef]
- Koley, B.; Dey, D. An ensemble system for automatic sleep stage classification using single channel EEG signal. Comput. Biol. Med. 2012, 42, 1186–1195. [Google Scholar] [CrossRef]
- Lajnef, T.; Chaibi, S.; Ruby, P.; Aguera, P.-E.; Eichenlaub, J.-B.; Samet, M.; Kachouri, A.; Jerbi, K. Learning machines and sleeping brains: Automatic sleep stage classification using decision-tree multi-class support vector machines. J. Neurosci. Methods 2015, 250, 94–105. [Google Scholar] [CrossRef]
- Zhu, G.; Li, Y.; Wen, P. Analysis and classification of sleep stages based on difference visibility graphs from a single-channel EEG signal. IEEE J. Biomed. Health Inform. 2014, 18, 1813–1821. [Google Scholar] [CrossRef]
- Lubin, A.; Johnson, L.C.; Austin, M.T. Discrimination among states of consciousness using EEG spectra. Psychophysiology 1969, 10, 593–601. [Google Scholar]
- Griffin, D.; Lim, J. Signal estimation from modified short-time Fourier transform. IEEE Trans. Acoust. Speech Signal Process. 1984, 32, 236–243. [Google Scholar] [CrossRef]
- Hazarika, N.; Chen, J.Z.; Tsoi, A.C.; Sergejew, A. Classification of EEG signals using the wavelet transform. Signal Process. 1997, 59, 61–72. [Google Scholar] [CrossRef]
- Huang, N.E. Hilbert-Huang Transform and Its Applications; World Scientific: Singapore, 2014. [Google Scholar]
- Khasawneh, N.; Fraiwan, M.; Fraiwan, L. Detection of K-complexes in EEG waveform images using faster R-CNN and deep transfer learning. BMC Med. Inform. Decis. Mak. 2022, 22, 297. [Google Scholar] [CrossRef]
- Tsinalis, O.; Matthews, P.M.; Guo, Y.; Zafeiriou, S. Automatic sleep stage scoring with single-channel EEG using convolutional neural networks. arXiv 2016, arXiv:1610.01683. [Google Scholar]
- Sors, A.; Bonnet, S.; Mirek, S.; Vercueil, L.; Payen, J.-F. A convolutional neural network for sleep stage scoring from raw single-channel EEG. Biomed. Signal Process. Control. 2018, 42, 107–114. [Google Scholar] [CrossRef]
- Sokolovsky, M.; Guerrero, F.; Paisarnsrisomsuk, S.; Ruiz, C.; Alvarez, S.A. Deep learning for automated feature discovery and classification of sleep stages. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 17, 1835–1845. [Google Scholar] [CrossRef]
- Li, F.; Yan, R.; Mahini, R.; Wei, L.; Wang, Z.; Mathiak, K.; Liu, R.; Cong, F. End-to-end sleep staging using convolutional neural network in raw single-channel EEG. Biomed. Signal Process. Control. 2020, 63, 102203. [Google Scholar] [CrossRef]
- Chambon, S.; Galtier, M.N.; Arnal, P.J.; Wainrib, G.; Gramfort, A. A deep learning architecture for temporal sleep stage classification using multivariate and multimodal time series. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 758–769. [Google Scholar] [CrossRef]
- Yulita, I.N.; Fanany, M.I.; Arymurthy, A.M. Fast Convolutional Method for Automatic Sleep Stage Classification. Health Inform. Res. 2018, 24, 170–178. [Google Scholar] [CrossRef]
- Berry, R.B.; Brooks, R.; Gamaldo, C.E.; Harding, S.M.; Marcus, C.; Vaughn, B.V. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications; American Academy of Sleep Medicine: Darien, IL, USA, 2015; pp. 1–7. [Google Scholar]
- Hsu, Y.-L.; Yang, Y.-T.; Wang, J.-S.; Hsu, C.-Y. Automatic sleep stage recurrent neural classifier using energy features of EEG signals. Neurocomputing 2013, 104, 105–114. [Google Scholar] [CrossRef]
- Supratak, A.; Dong, H.; Wu, C.; Guo, Y. DeepSleepNet: A model for automatic sleep stage scoring based on raw single-channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1998–2008. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.; Andreotti, F.; Cooray, N.; Chén, O.Y.; De Vos, M. SeqSleepNet: End-to-End Hierarchical Recurrent Neural Network for Sequence-to-Sequence Automatic Sleep Staging. Neural Syst. Rehabil. Eng. IEEE Trans. 2019, 27, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fan, R.; Liu, Y. Deep identity confusion for automatic sleep staging based on single-channel EEG. In Proceedings of the 2018 14th International Conference on Mobile Ad-Hoc and Sensor Networks (MSN), Shenyang, China, 6–8 December 2018; pp. 134–139. [Google Scholar]
- Huang, Y.; Liang, L.W. Joint sleep staging model based on pressure-sensitive sleep signal. IOP Conf. Ser. Mater. Sci. Eng. 2020, 740, 012159. [Google Scholar] [CrossRef]
- Gers, F.A.; Schmidhuber, J.; Cummins, F. Learning to forget: Continual prediction with LSTM. Neural Comput. 2000, 12, 2451–2471. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Luo, W.; Yu, F. Convolution- and Attention-Based Neural Network for Automated Sleep Stage Classification. Int. J. Environ. Res. Public Health 2020, 17, 4152. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Bai, S.; Kolter, J.Z.; Koltun, V. An empirical evaluation of generic convolutional and recurrent networks for sequence modeling. arXiv 2018, arXiv:1803.01271. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Lea, C.; Flynn, M.D.; Vidal, R.; Reiter, A.; Hager, G.D. Temporal convolutional networks for action segmentation and detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 156–165. [Google Scholar]
- Troncoso, A.; Salcedo-Sanz, S.; Casanova-Mateo, C.; Riquelme, J.C.; Prieto, L. Local models-based regression trees for very short-term wind speed prediction. Renew. Energy 2015, 81, 589–598. [Google Scholar] [CrossRef]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Phan, H.; Andreotti, F.; Cooray, N.; Chén, O.Y.; De Vos, M. DNN filter bank improves 1-max pooling CNN for single-channel EEG automatic sleep stage classification. In Proceedings of the 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 453–456. [Google Scholar]
- Vilamala, A.; Madsen, K.H.; Hansen, L.K. Deep convolutional neural networks for interpretable analysis of EEG sleep stage scoring. In Proceedings of the IEEE 27th International Workshop on Machine Learning for Signal Processing (MLSP), Tokyo, Japan, 25–28 September 2017; pp. 1–6. [Google Scholar]
- Mousavi, S.; Afghah, F.; Acharya, U.R. SleepEEGNet: Automated sleep stage scoring with sequence to sequence deep learning approach. PLoS ONE 2019, 14, e0216456. [Google Scholar] [CrossRef] [PubMed]
- Neng, W.; Lu, J.; Xu, L. Ccrrsleepnet: A hybrid relational inductive biases network for automatic sleep stage classification on raw single-channel eeg. Brain Sci. 2021, 11, 456. [Google Scholar] [CrossRef] [PubMed]
- Eldele, E.; Chen, Z.; Liu, C.; Wu, M.; Kwoh, C.K.; Li, X.; Guan, C. An attention-based deep learning approach for sleep stage classification with single-channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.; Chén, O.Y.; Koch, P.; Lu, Z.; McLoughlin, I.; Mertins, A.; De Vos, M. Towards more accurate automatic sleep staging via deep transfer learning. IEEE Trans. Biomed. Eng. 2020, 68, 1787–1798. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; Back, S.; Lee, S.; Park, D.; Kim, T.; Lee, K. Intra-and inter-epoch temporal context network (IITNet) using sub-epoch features for automatic sleep scoring on raw single-channel EEG. Biomed. Signal Process. Control. 2020, 61, 102037. [Google Scholar] [CrossRef]
- Supratak, A.; Guo, Y. TinySleepNet: An efficient deep learning model for sleep stage scoring based on raw single-channel EEG. In Proceedings of the 42nd Annual International Conference of the IEEE Engineering in Medicine Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 641–644. [Google Scholar]
| Dataset | W | N1 | N2 | N3 | REM | Total |
|---|---|---|---|---|---|---|
| Sleep-EDF | 8285 | 2804 | 17,799 | 5703 | 7717 | 42,308 |
| K-Folds | Overall Performance | Per-Class Performance | ||||||
|---|---|---|---|---|---|---|---|---|
| Acc | MF1 | κ | W | N1 | N2 | N3 | REM | |
| 10 | 83.7 | 76.4 | 0.77 | 90.7 | 35.8 | 88.9 | 90.4 | 86.8 |
| 15 | 84.8 | 77.9 | 0.793 | 90.3 | 39.9 | 88.9 | 86.3 | 83.9 |
| 20 | 86.2 | 79.8 | 0.808 | 92.0 | 45.5 | 89.4 | 88.6 | 83.5 |
| Method | Predicted | Per-Class Metrics | ||||||
|---|---|---|---|---|---|---|---|---|
| W | N1 | N2 | N3 | REM | PR | RE | F1 | |
| W | 7497 | 393 | 153 | 24 | 123 | 92.6 | 91.5 | 92.0 |
| N1 | 442 | 1122 | 683 | 15 | 640 | 55.2 | 38.7 | 45.5 |
| N2 | 77 | 223 | 16,416 | 625 | 804 | 88.4 | 90.5 | 89.4 |
| N3 | 8 | 0 | 609 | 5009 | 0 | 88.1 | 89.0 | 88.6 |
| REM | 76 | 295 | 711 | 12 | 6749 | 81.2 | 86.1 | 83.5 |
| Method | Overall Performance | Per-Class Performance | ||||||
|---|---|---|---|---|---|---|---|---|
| Acc | MF1 | κ | W | N1 | N2 | N3 | REM | |
| 1-max CNN [38] | 79.8 | 72.0 | 0.720 | - | - | - | - | - |
| VGG-FT [39] | 80.3 | - | - | - | - | - | - | - |
| SleepEEGNet [40] | 84.26 | 79.66 | 0.79 | 89.19 | 52.19 | 86.77 | 85.13 | 85.02 |
| CCRRSleepNet [41] | 84.29 | 79.81 | 0.78 | 89.01 | 51.73 | 87.25 | 88.20 | 82.86 |
| AttnSleep [42] | 84.3 | 77.7 | 0.776 | 85.4 | 50.9 | 88.8 | 86.4 | 86.5 |
| FT DeepSleepNet+ [43] | 84.4 | 78.8 | 0.781 | - | - | - | - | - |
| IITNet [44] | 84.6 | 79.0 | 0.782 | 81.0 | 50.5 | 88.2 | 86.9 | 87.2 |
| FT SeqSleepNet+ [43] | 85.2 | 79.6 | 0.789 | - | - | - | - | - |
| TinySleepNet [45] | 85.4 | 80.5 | 0.80 | 90.1 | 51.4 | 88.5 | 88.3 | 84.3 |
| proposed Method | 86.2 | 79.8 | 0.808 | 92.0 | 45.5 | 89.4 | 88.6 | 83.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Chen, C.; Meng, K.; Lu, L.; Cheng, X.; Fan, H. NAMRTNet: Automatic Classification of Sleep Stages Based on Improved ResNet-TCN Network and Attention Mechanism. Appl. Sci. 2023, 13, 6788. https://doi.org/10.3390/app13116788
Xu X, Chen C, Meng K, Lu L, Cheng X, Fan H. NAMRTNet: Automatic Classification of Sleep Stages Based on Improved ResNet-TCN Network and Attention Mechanism. Applied Sciences. 2023; 13(11):6788. https://doi.org/10.3390/app13116788
Chicago/Turabian StyleXu, Xuebin, Chen Chen, Kan Meng, Longbin Lu, Xiaorui Cheng, and Haichao Fan. 2023. "NAMRTNet: Automatic Classification of Sleep Stages Based on Improved ResNet-TCN Network and Attention Mechanism" Applied Sciences 13, no. 11: 6788. https://doi.org/10.3390/app13116788
APA StyleXu, X., Chen, C., Meng, K., Lu, L., Cheng, X., & Fan, H. (2023). NAMRTNet: Automatic Classification of Sleep Stages Based on Improved ResNet-TCN Network and Attention Mechanism. Applied Sciences, 13(11), 6788. https://doi.org/10.3390/app13116788





