1. Introduction
In today’s rapidly advancing era of technology and automation, the healthcare industry is actively exploring innovative solutions to enhance patient care and improve diagnostic practices. One area that holds immense potential is the automated analysis of medical diagnostic videos. By leveraging deep learning techniques, these videos can be processed and analyzed in a manner that significantly enhances the efficiency and accuracy of diagnosis. Deep learning has emerged as a powerful tool for analyzing medical videos, enabling automated tasks such as classification, detection, and segmentation. The application of deep learning models, such as convolutional neural networks (CNNs) and long short-term memory (LSTM) networks, has demonstrated remarkable success in extracting meaningful information from medical videos. These models have been employed to address various challenges, including the classification of different types of medical videos and the segmentation of specific anatomical structures.
There are two types of medical diagnostic videos: ultrasound and endoscopy. Cardiac ultrasound is known as echocardiography, and we refer to echocardiography as a third type of medical diagnostic video for the rest of this research work. Several researchers [
1] have exploited traditional and machine learning techniques to analyze medical videos, such as echocardiographic videos. The analysis tasks include segmentation of left ventricle (LV), myocardium [
2], and anterior mitral leaflet [
3] from echocardiographic videos. The researchers also apply machine learning techniques for classifying informative frames from endoscopy videos [
4]. Other traditional techniques such as naive Bayesian [
5], sliding windows Gauss–Seidel [
6], and the polar active contour model [
7] are explored to perform different operations on ultrasound videos to make diagnosis easy for medical practitioners. However, such research articles are out of the scope of this research. We have only reviewed the application of deep learning techniques for medical video analysis.
This article presents a detailed review of medical diagnostic video types and deep learning methods applied for their analysis. We have several contributions. First, to the best of our knowledge, this is the first work that comprehensively provides details about the study of all diagnostic videos. Second, we have thoroughly reviewed many research articles on deep-learning-based medical video analysis. Third, current publications are discussed along with their datasets, purpose, performance, methods, and limitations. Fourthly, we identified which deep learning models would be suitable for classifying, segmenting, and detecting clinical videos. Last, we discussed challenges and future research directions for fellow researchers and practitioners.
The paper is structured as follows:
Section 2 and
Section 3 discuss the methodology and taxonomy followed for this study.
Section 4 defines and explains the different deep learning techniques used for extracting spatial and temporal data from medical videos.
Section 5 discusses publicly available datasets for medical diagnostic videos.
Section 6,
Section 7 and
Section 8 give a detailed literature review of deep learning techniques applied to echocardiography, endoscopy, and ultrasound videos. The literature review for each video type is divided into three subsections: Classification, Segmentation, and Detection. In
Section 9, we discuss and provide our opinion on methods used for medical video analysis. In
Section 10 and
Section 11, the challenges, future implications, and conclusions for deep learning and medical video analysis are provided.
2. The Methodology Followed for This Study
2.1. Research Gap and Scope of the Review
The current reviews [
8,
9] on medical image analysis using deep learning focus primarily on the application of convolutional neural networks (CNNs) to static medical images, such as CT scans, MRI, and X-ray images. These reviews discuss various deep learning architectures, such as U-Net, DenseNet, and ResNet, and their application to medical image analysis tasks such as segmentation, classification, and detection. However, these reviews do not discuss the analysis of spatial and temporal features in medical images. Spatial features refer to the arrangement and distribution of pixels in an image, while temporal features refer to changes that occur over time in a sequence of images, such as in videos or dynamic medical imaging modalities such as ultrasound and endoscopy.
While CNNs can be applied to temporal medical images, such as in video segmentation and classification tasks, their ability to capture spatiotemporal features is limited. To address this limitation, researchers have developed various deep learning architectures, such as 3D CNNs, that can capture both spatial and temporal features simultaneously. Thus, the current reviews on medical image analysis using deep learning mainly focus on static medical images and do not discuss the analysis of spatial and temporal features.
The focus of this article is to provide a comprehensive review of the application of deep learning techniques for the analysis of medical diagnostic videos, i.e., ultrasound and endoscopy. The article reviews the deep learning techniques used for extracting spatial and temporal data from these videos and discusses publicly available datasets for medical diagnostic videos. This article also includes a detailed literature review of deep learning techniques applied to each type of medical diagnostic video for classification, segmentation, and detection tasks. Finally, this article discusses the challenges, future implications, and conclusions for deep learning and medical video analysis.
2.2. Search Strategy
A comprehensive literature search was undertaken using the databases of PubMed, ACM Digital Library, IEEE Xplorer, Elsevier, and Springer. We mainly focused on the research articles published from 2016 to 2022 but included a few research papers from earlier years. For article search, we used the following keyword combinations:
Deep learning and medical diagnostic videos. This keyword combination resulted in 180 research papers.
Deep learning and echocardiography videos. This keyword combination resulted in 120 research papers.
Deep learning and endoscopy videos. This keyword combination resulted in 250 research papers.
Deep learning and ultrasound videos. This keyword combination resulted in 200 research papers.
Deep learning and polyp detection OR classification OR segmentation. This keyword combination resulted in 160 research papers.
Deep learning and cardiac phases OR LV classification OR detection OR segmentation. This keyword combination resulted in 150 research papers.
Deep learning and informative frames OR classification OR detection OR segmentation in ultrasound data. This keyword combination resulted in 200 research papers.
2.3. Inclusion and Exclusion Criteria
Inclusion criteria were as follows:
Studies reporting performance of deep learning models for the analysis of medical diagnostic videos. Review or survey articles which cover the topic of deep learning applied to medical diagnostic videos. Research papers reporting running time, AUC, sensitivity, diagnostic accuracy, and specificity or papers with adequate information to calculate these data. Studies published between 2016 and 2022. Exclusion criteria were as follows:
Papers reporting results only on image data. Papers published before 2016. Papers which have explored machine learning but not deep learning or only explored traditional approaches. Conference papers which are not Scopus-indexed.
2.4. Search Result and Study Selection
First, we selected around 350 papers based on the relevance of their titles to our topic. The title and abstract of the retrieved articles were then screened for relevance by all the authors independently. The decision of inclusion and exclusion was taken on the above criteria. Following this approach, full-text reviews of the relevant studies were completed. Disagreements about the study’s relevance were settled by consensus after screening and following a full-text review.
2.5. Data Extraction
For each study, we extracted the following data: primary details such as first author and year of publication, then the purpose of the study such as classification or segmentation. Afterwards, details about the method applied and the data used were extracted, such as the format of the video, resolution, and size of the dataset. Lastly, we extracted the results regarding AUC, sensitivity, specificity, etc.
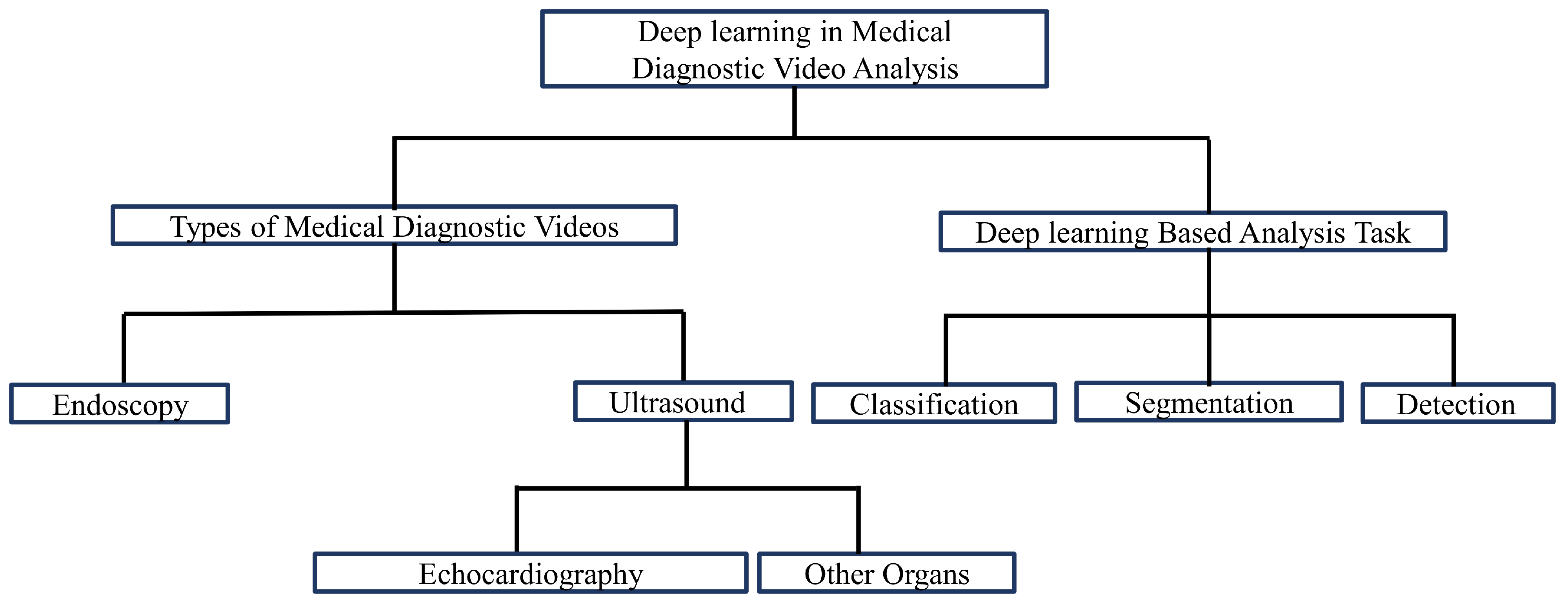
3. Taxonomy of Deep-Learning-Based Medical Video Analysis
In this section, we discuss the organization of the review of recent contributions in the field of deep-learning-based medical video analysis. The taxonomy followed in this literature review is shown in
Figure 1. In the following subsections, we explain each block of our taxonomy in detail.
3.1. Types of Medical Videos
Echocardiography, endoscopy, and ultrasound procedures generate three types of videos for diagnosing diseases, such as the presence of polyps in body organs and cardiomyopathy. There are also a few other types of videos, such as surgery and training videos, that are used for analysis and teaching purposes. As this study only deals with diagnostic videos, such videos are out of the scope of our research. Further details about these videos are given in the following subsections.
(1) Echocardiography: Echocardiography is a noninvasive and affordable imaging technology that helps doctors diagnose a heart’s pumping strength, blood flow, the presence of a tumor, the functionality of valves, and the physiology of the heart [
10]. Echocardiography uses sound waves by passing a probe called a transducer over the chest. Anatomical structures of the heart are displayed on the screen of the echocardiography machine as sequential frames over time. For echocardiography videos, temporal resolution plays a vital role. Temporal resolution is the time from starting one frame to the next; it shows the ultrasound system’s ability to capture structures with rapid movements, such as the cardiac cycle. Low temporal resolution can cause data loss, resulting in an incorrect diagnosis. Another challenge medical practitioners face is poor video quality caused by improper probe handling and loss of signals.
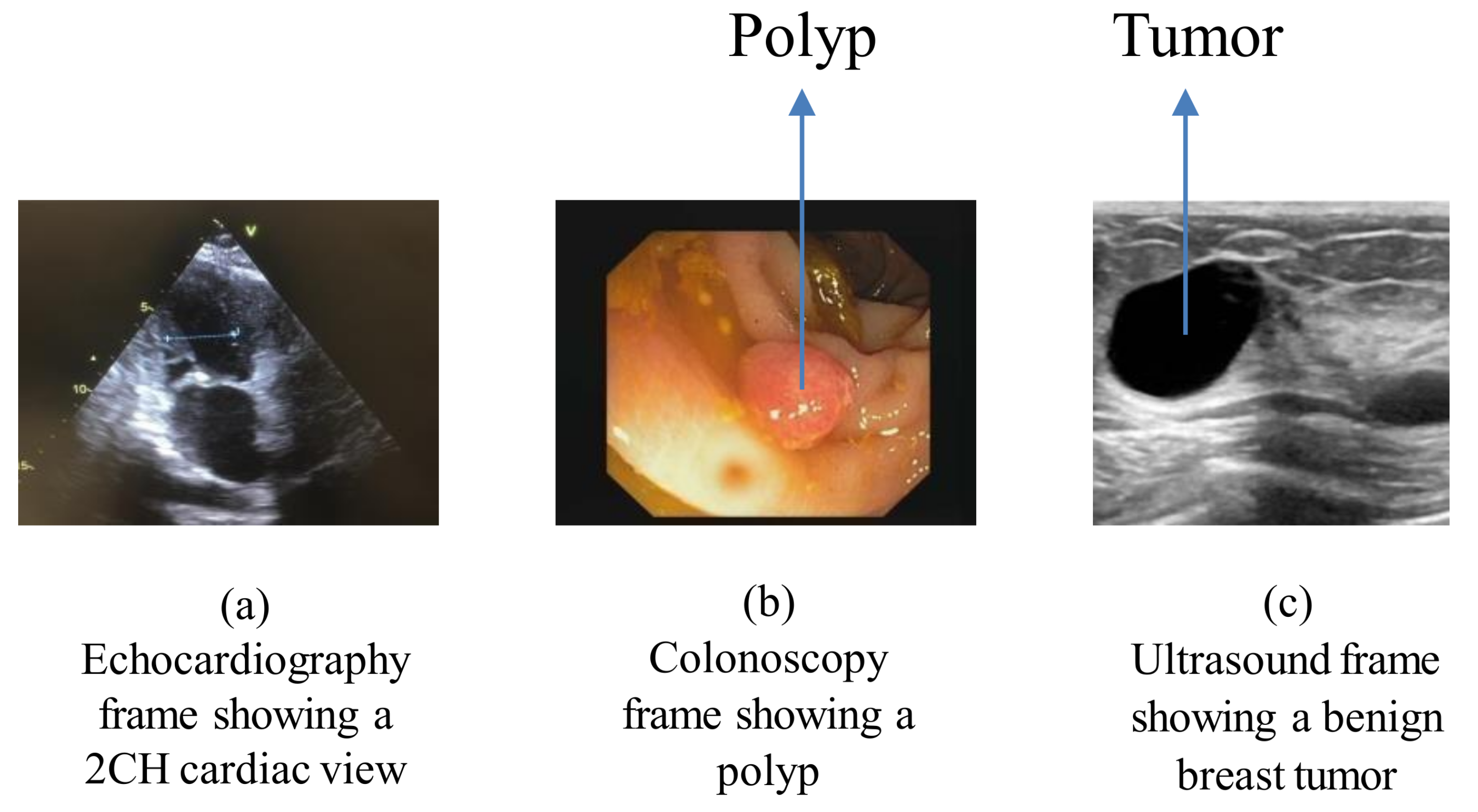
Figure 2a depicts the echocardiography frame showing a two-chamber view of the heart.
(2) Endoscopy: Endoscopy is a procedure in which an endoscope is passed inside the human body to examine an organ, such as the intestine, colon, or stomach. Endoscopy is the preferred test for various types of cancer, polyps, and lesion detection [
11]. The technique for capturing endoscopy and echocardiography videos is different; hence, the results are dissimilar. The endoscopy videos are colorful and usually of good quality compared with the echocardiography videos. The presence of interference, such as food and gastric juices, in the digestive system makes it challenging for medical staff to analyze the regions of interest. The presence of uninformative frames in endoscopy videos makes the diagnosis process time-consuming and inefficient in terms of computational resources.
Figure 2b depicts the endoscopy frame showing a polyp in the colon area of the body.
(3) Ultrasound: Ultrasound is extensively used for diagnosing the causes of pain, swelling, and infection in any part of the body (e.g., kidney, liver, and gallbladder) in a cost-effective manner [
12]. Ultrasound imaging uses high-frequency sound waves and can be performed on various organs to examine them internally. Ultrasound can also guide surgeons during surgery regarding the area of interest. The relatively low-energy acoustic waves used during ultrasound imaging cannot penetrate thick layers of human tissue; for example, in obese people, the liver and other vital abdominal organs can lie four to five centimeters below the surface. Such cases present more physical strain on sonographers and radiologists.
Figure 2c depicts the ultrasound frame showing a benign breast tumor.
3.2. Deep-Learning-Based Analysis Tasks
(1) Classification: Classification is a process that involves assigning a class label to the input data [
13,
14]. Mostly, a classification task allocates a single label to the input. However, sometimes it involves predicting the probability across two or more class labels. The classification model assumes that the classes are mutually exclusive in these cases. The research articles that have explored deep-learning-based classification techniques for echocardiography, endoscopy, and ultrasound are reviewed in
Section 6.1,
Section 7.1 and
Section 8.1.
(2) Segmentation: Image segmentation is a process in which the input is broken down into segments, which helps reduce the image’s complexity to make further processing or analyzing the image easier and simpler [
15]. Segmentation, in other words, is a method of assigning labels to pixels. All pixels belonging to the same class have a common label assigned to them. The research articles that have explored deep-learning-based segmentation techniques for echocardiography, endoscopy, and ultrasound are reviewed in
Section 6.2,
Section 7.2 and
Section 8.2.
(3) Detection: Object detection is a technique that identifies a particular object’s location in an image or video [
16]. Object detection can be used to count objects in a scene and determine and track their precise locations while accurately labeling them. The research articles that have explored deep-learning-based detection techniques for echocardiography, endoscopy, and ultrasound are reviewed in
Section 6.3,
Section 7.3 and
Section 8.3.
4. An Overview of Deep Learning Techniques
The following are a few commonly used deep learning techniques in the medical field.
4.1. Convolutional Neural Networks
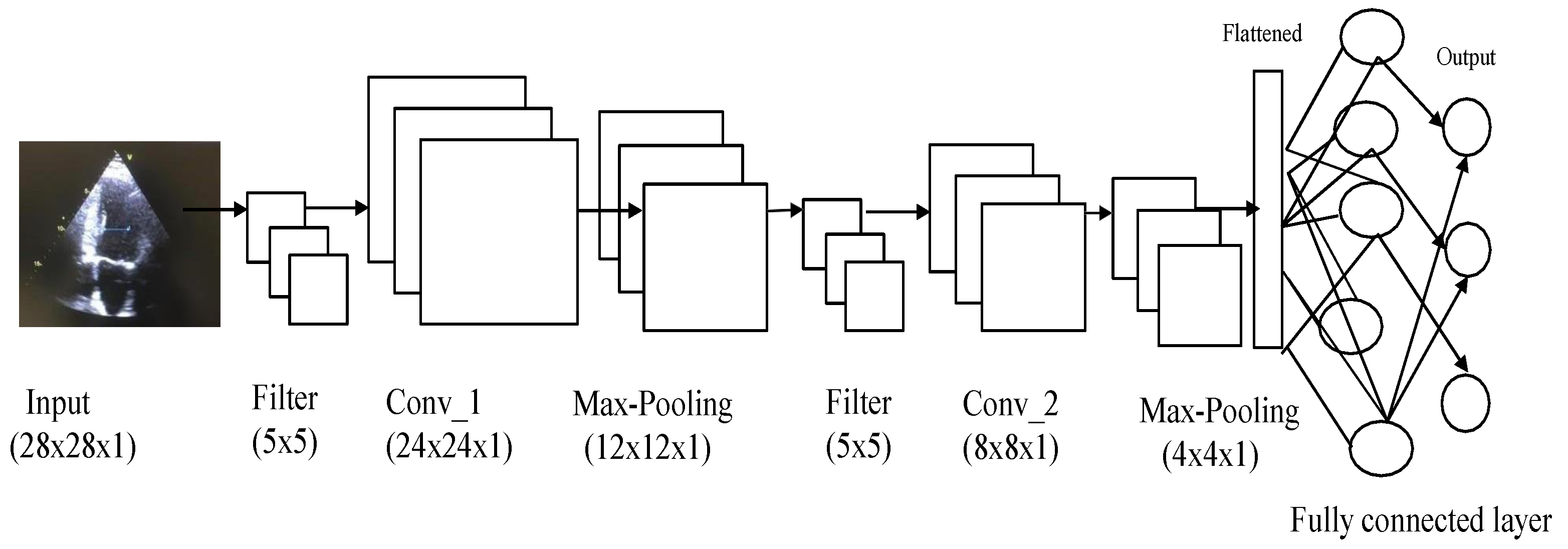
CNNs are of prime importance in the context of deep-learning-based video analysis. CNNs consist of three types of layers, namely convolution, pooling, and fully connected, as shown in
Figure 3 [
17]. The CNN architecture shown in
Figure 3 is just an example of how CNNs are applied to extract features from data and then classify them. The CNN architecture can be modified based on many parameters, such as data and computational speed.
4.2. Fully Convolutional Network
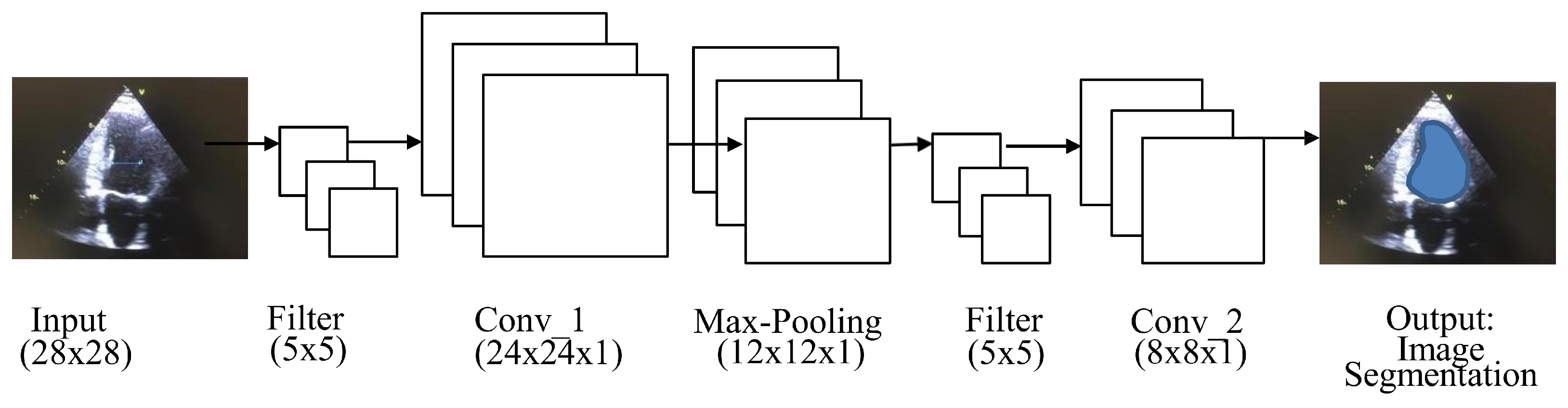
A fully convolutional network (FCN) is used for semantic segmentation and consists of convolution, pooling, and upsampling layers [
18], as shown in
Figure 4. FCN performs in-network upsampling and pixel-to-pixel inference by using a fully convolutional approach, such that it can store the pixelwise spatial correspondence by transforming all fully connected layers to convolutions and enable per-pixel segmentation. Arbitrary-sized video clips can be given input to FCN for the segmentation of ROI. The output provided by the FCN would be in the form of corresponding probability maps. The probability map’s values represent the network output of one subwindow in the input video clips. The FCN probability map approach is much more efficient than the sliding window method, which repeatedly crops overlapping samples [
19].
4.3. Generative Adversarial Networks
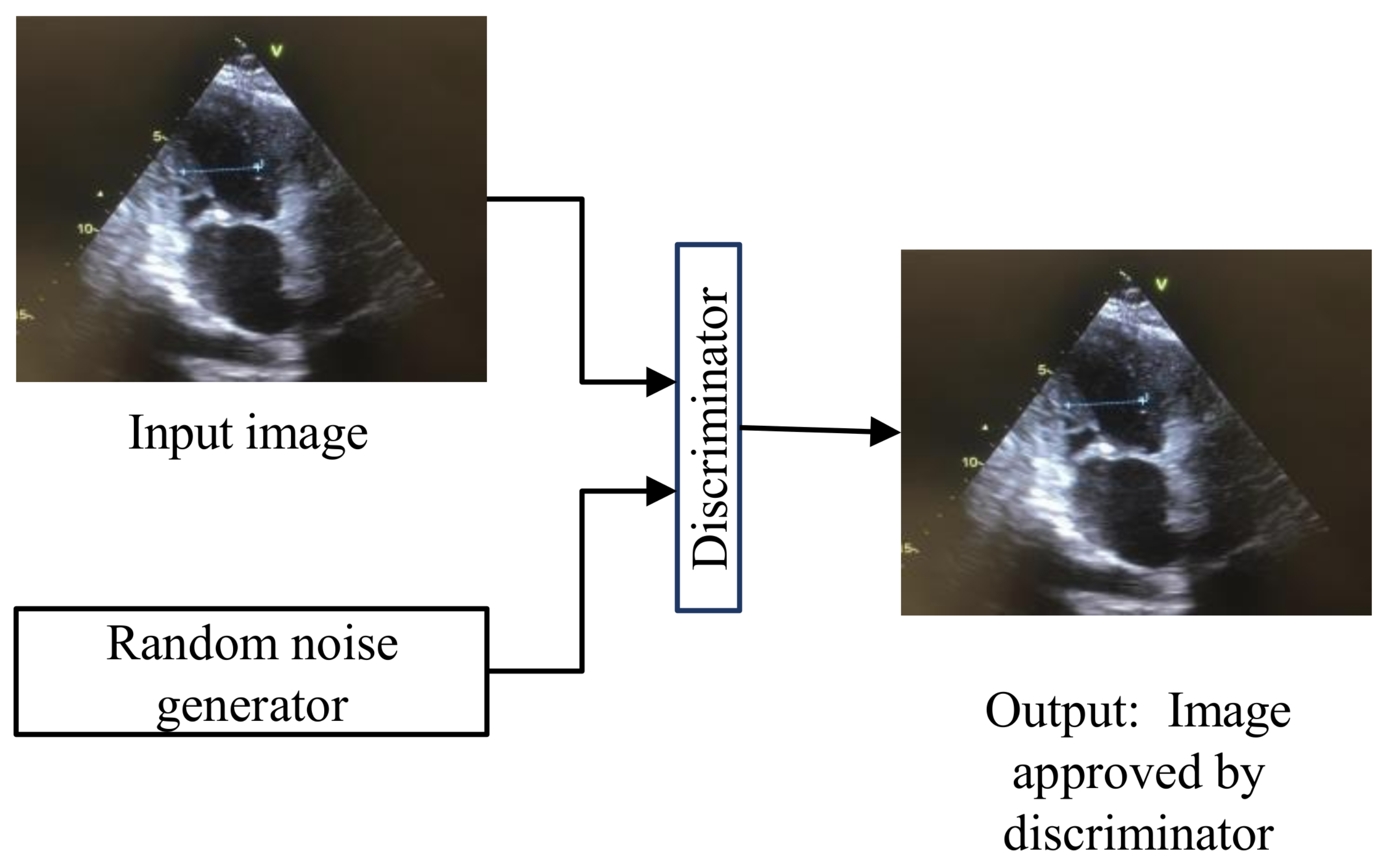
Generative adversarial networks (GANs) [
20] are used for training generative models for image synthesis. The architecture of GANs is given in
Figure 5, where the generator is trying to copy the input data distribution. When the original and generated images are passed through the discriminator, it will decide whether the generated image is accepted or not. GANs can be trained to separate the scene’s foreground from the background. GANs comprise such robust architecture that they can create tiny videos for up to a second at full frame rate. This ability of GANs can predict the next plausible futures of static images. GANs’s ability to internally learn useful features is useful for recognizing actions with minimal supervision in medical videos.
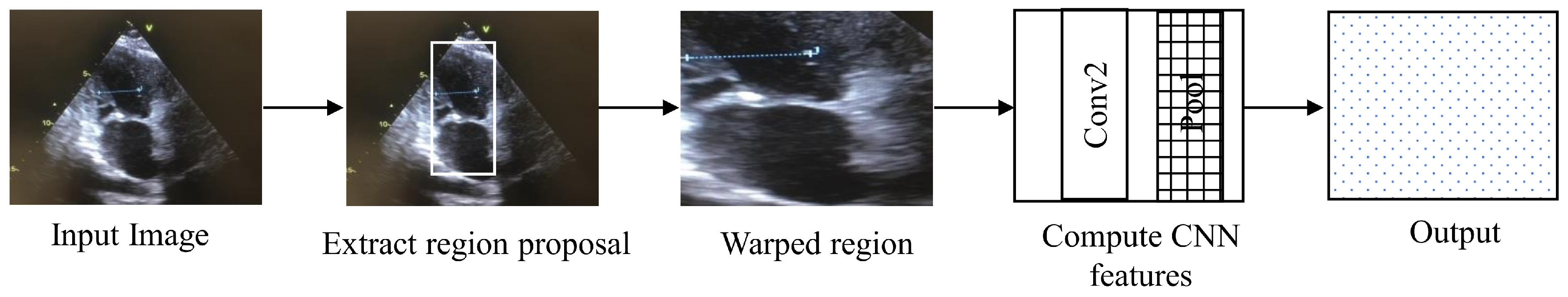
4.4. Regions with CNN Features
Regions with CNN features (RCNN) is an architecture used for segmentation and object detection. RCNN is called region-based because the input is divided into several regions to detect the object of interest, as shown in the
Figure 6 [
21]. RCNN takes around 45 s for processing per video frame, making it unsuitable for real-time application.
4.5. Single-Shot Multibox Detector
The single-shot multibox detector (SSD) is an object detection technique. The architecture of an SSD is based on the VGG-16 architecture but discards the fully connected layers [
22], as shown in
Figure 7. Generally, an SSD is faster than RCNN for video analysis because it eliminates the need for the region proposal network.
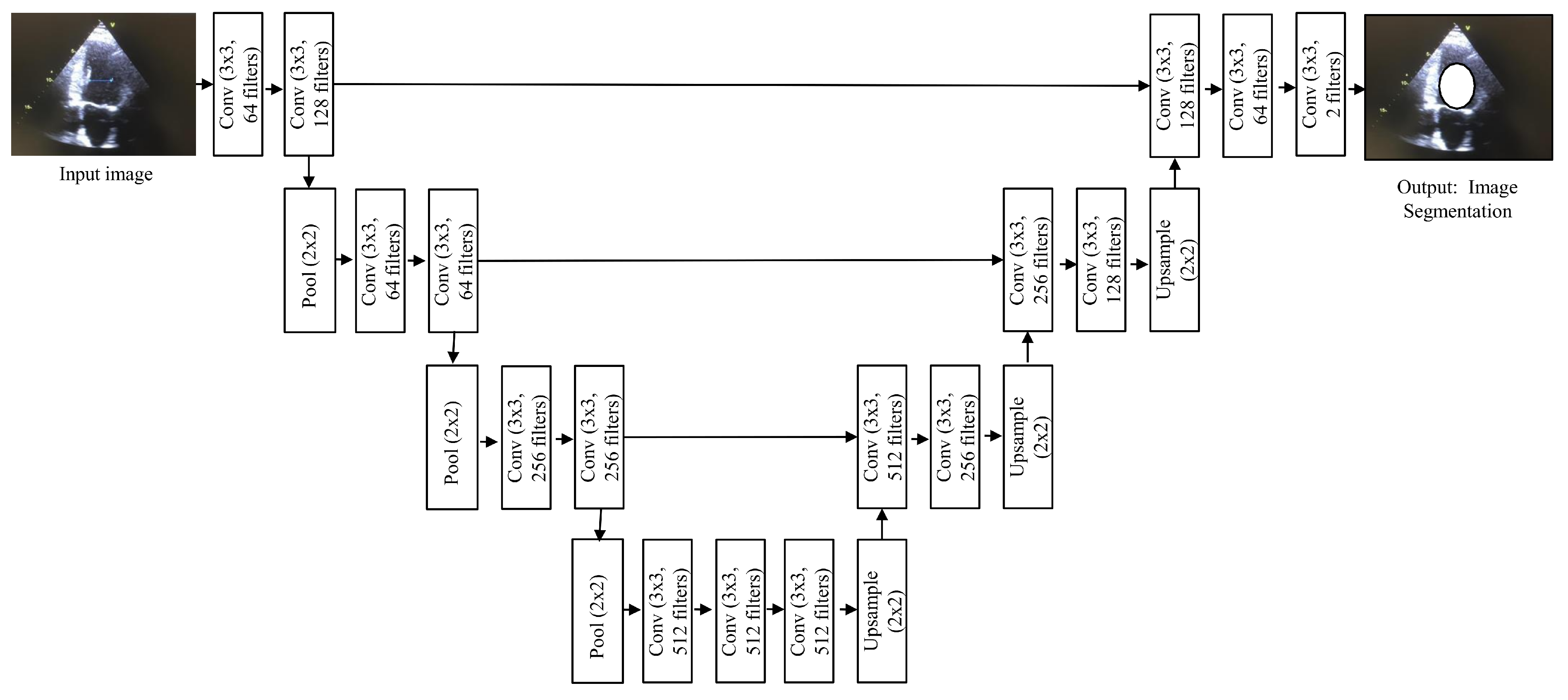
4.6. UNet
The name UNet is derived from the symmetric shape of the architecture [
23], as shown
Figure 8, and it is one of the evolved forms of CNNs. UNet has been widely used for medical video segmentation to detect polyps, blood clots, etc. Two characteristics of UNet make it a suitable choice for video data. Firstly, it performs well even when limited training data are available. Secondly, since it does not have any fully connected layers, there is no limitation on the size of the input. As medical videos are rich in content, having no input size restrictions assures effective ROI segmentation.
4.7. Recurrent Neural Networks
Recurrent neural networks (RNNs) are designed to model sequences of data types, such as text, videos, and images. An RNN uses its internal state to maintain a ‘memory’ of the sequence.
Figure 9 represents a typical RNN.
The most popular type of RNN is a long short-term memory (LSTM) network [
24]. LSTM is effectively used for dealing with long sequences. The fundamental architecture of LSTM networks is similar to an RNN; however, their hidden states are computed differently. Informally, a hidden state takes in the previous state and the input at a given time stamp and decides what to remember and delete from its memory. The current, previous, and memory states are combined for the next time stamp.
4.8. Models Used for Extracting Temporal Data
CNNs can be applied to video data in the same way as image data, where every frame is treated as a separate image. On the other hand, if the temporal information needs to be extracted from the video, other approaches can be used. For example, CNNs can be combined with an RNN model [
25], where the CNN model is used for extracting the image features, and the RNN model is utilized for capturing temporal dependencies. This method is known as the fusion approach, where the model architecture consists of convolutional layers, LSTM layers, and finally fully connected and softmax layers [
26]. The CNN and LSTM models can be fused in three ways. A feature-level fusion can be implemented by combining the outputs after the features are extracted by the sequence of convolution and LSTM layers from the input data. After the fusion, a classifier can be used to develop a feature-level fusion model. Score-level fusion is another type of fusion that can take place between or after the softmax and fully connected layers. In this type of fusion, the input data are passed through the CNN, LSTM layers, and a classifier, resulting in a probability score. The fusion of multiple scores from different models is a score-level fusion. The decision-level fusion is similar to a score-level fusion, except that the fusion is performed after the network’s prediction. This type of fusion is entirely associated with the network’s predicted output and is not related to the score/probability used for the decision.
Another deep learning architecture suitable for extracting temporal features is a 3D CNN [
27]. The 3D CNN comprises several consecutive layers of 3D convolutions and operates by convolving a four-dimensional kernel over a four-dimensional data input in both space and time. These four dimensions for the input data and kernel come from the temporal dimension (i.e., the number of video frames), two spatial dimensions, and the channel dimension (e.g., an RGB image has three channels).
5. Publicly Available Datasets for Medical Video Analysis
This section discusses publicly available datasets, because they play a crucial role in the development and evaluation of deep learning models in medical video analysis. By providing an overview of these datasets, researchers and practitioners can choose the most appropriate dataset for their specific application and compare their results with those of other studies in the field. Additionally, the availability of standardized datasets helps to promote reproducibility and transparency in research.
5.1. Kent Integrated Dataset
The Kent Integrated Dataset (KID) [
28] is an open-source and nonprofit dataset that contains 2500 endoscopy images and three videos. The dataset includes various types of diseased and normal organs, e.g., colon, stomach, and esophagus, and the disease conditions include bleeding, polyps, ulcer, and stenosis. Most of these data were captured using a microcam wireless capsule endoscope.
5.2. Cardiac Acquisitions for Multistructure Ultrasound Segmentation Dataset
The Cardiac Acquisitions for Multistructure Ultrasound Segmentation (CAMUS) dataset was introduced by Leclerc et al. in 2019 [
29]. This is the largest available echocardiogram dataset and contains images and video sequences of 500 patients. The reports were acquired from Saint-Etienne hospital (France), and the dataset reflects daily clinical practice data. The CAMUS dataset contains images and video sequences of good, poor, and medium quality, and labeling was performed by three cardiologists. This dataset provides end-systolic (ES) and end-diastolic (ED) frames in four-chamber and two-chamber views of the heart for each patient. Each video has a different resolution, and all of them are larger than 1024 × 512. All video sequences are stored in .mhd format.
5.3. EchoNet Dynamic Dataset
Ouyang et al. [
30] created the EchoNet-Dynamic dataset, containing 10,030 echocardiography videos and covering a variety of typical lab imaging acquisition conditions. All images have labeled measurements, including LV volume at end-systole and end-diastole, ejection fraction, and expert tracings of the left ventricle. The dataset contains apical-4-chamber echocardiography videos from patients who underwent echocardiography tests between 2016 and 2018 at Stanford University Hospital. Each video was cropped to delete the text and information outside of the scanning sector, and the resulting images were downsampled into standardized 112 × 112 pixel videos. The videos are in .avi format, containing an average of 50 frames per second and 176 in total.
5.4. ASU-Mayo Clinic Colonoscopy Video Dataset
The ASU-Mayo clinic is the largest colonoscopy video dataset [
11]. The training set comprises 20 colonoscopy videos, of which ten videos show polyps, and the other 10 show normal physiology. Furthermore, the test set comprises 18 unlabeled colonoscopy videos to assess the performance of the applied algorithm. The resolution of the videos is 688 × 550 and contains varying amounts of frames ranging from 324 to 1925. The videos are stored in .wmv format.
5.5. GastroLab Dataset
The GastroLab dataset [
31] includes various endoscopy videos for research and educational purposes. The captured videos are of different organs, e.g., the colon, duodenum, oesophagus, and stomach, and they show several medical conditions such as cancer, ulcer, Crohn’s disease, and adenoma. All the videos are of different sizes and resolutions.
5.6. HyperKvasir Dataset
The HyperKvasir dataset is the largest and most diverse endoscopy video dataset [
32]. This dataset contains 374 videos, corresponding to 9.78 h of videos and 889,372 video frames. The videos are divided into two categories: upper GI tract and lower GI tract. These categories are divided into 30 classes, such as ulcers, polyps, and cancer. All the videos are of different sizes and resolutions and stored in .avi format.
5.7. Nerthus Dataset
The Nerthus dataset [
33] consists of 21 videos, showing a gastrointestinal tract with different degrees of bowel cleansing. The dataset has four classes showing bowel cleansing quality. The number of videos per class varies from 1 to 10 with different durations. The dataset consists of videos with a resolution 720 × 576.
5.8. CVC-ClinicVideoDB
This publicly available dataset contains 18 colonoscopy videos with 768 × 576 resolution [
34]. All videos are of different duration and contain polyp and nonpolyp frames.
6. Echocardiography
In this section, the research articles which explore deep learning models applied to echocardiography videos are discussed. All the analysis tasks are performed on echocardiography videos in two ways: (1) by extracting spatial features from individual frames of the videos and (2) by extracting spatial and temporal features from the videos. We mention this information in the respective tables. The section is divided into four subsections: Classification, Segmentation, Detection, and Miscellaneous, in accordance with our taxonomy discussed in
Section 3.
6.1. Classification
Madani et al. [
35] applied a deep learning approach to the classification of echocardiographic views and segmentation of LV hypertrophy. Their data-efficient, semisupervised model was a combination of a CNN, GANs, and UNet. This research work can provide support to doctors and cannot be used as a replacement for the clinician. Moreover, a limited dataset was used due to patients’ privacy and consent issues. Ejection fraction (EF) is a measurement which determines the amount of blood the LV pumps out with each heartbeat, and it can be calculated through echo. Silva et al. [
36] proposed a pipeline in which the echocardiography video sequences and metadata of a patient were given as input to a 3D CNN. They built a custom 3D CNN with 3D convolutions, asymmetric kernels, and residual learning blocks integrated into the 3D convolutional layers. The deep learning model consists of three main blocks, and its input consists of thirty sequential frames of echocardiography sequence. The initial layers are used for obtaining a smaller representation of the data, which is used as input in the middle layers. The middle layers, which use small asymmetric filters, are responsible for most of the computation. Finally, the top layers converge the activations into a vector that is fed to a softmax classifier. Their research aimed to classify a person’s health based on EF. Their accuracy is 78%, which may not be sufficiently high for practical use. Deep learning techniques are also used for viewpoint classification from echocardiography videos [
37,
38]. A fused approach was introduced, wherein two CNNs were trained; spatial and temporal information was combined at the end for the final classification score. The spatial CNN network processes the original echo video images to automatically extract the spatial features, while for the temporal CNN network, the optical flow approach is applied to obtain acceleration and velocity images. The method gave 96.3% accuracy, which was superior to other state-of-the-art approaches. The A5C had the worst classification score due to a limited dataset for that view. Another research work was carried out by Feng et al. [
39] for the classification of normal and abnormal echocardiography videos. They proposed a two-stream deep network which extracted the optical flow and spatial context for the classification of echocardiography videos. A CNN extracts the spatial features, and each LSTM with attention learns the temporal features. For the output, the features from these two streams are fused. They achieved an accuracy of 91% for the dataset of 170 videos.
For classification tasks in echocardiography videos, CNN and LSTM seem popular among researchers. The evident limitation is difficulty in classifying look-alike cardiac views. The reason behind this is the lack of training data for some classes and the noisy nature of the echocardiography modality.
6.2. Segmentation
Deep learning provides new possibilities for echocardiography procedures to produce an accurate and automated interpretation of echos, thus potentially decreasing the risk of human error. Leclerc et al. [
29] presented a large publicly available dataset, namely CAMUS, consisting of echocardiography images and video sequences from 500 patients. They applied several deep learning and non-deep-learning techniques for the segmentation of the LV endocardium and myocardium. The techniques include UNet, UNet++, anatomically constrained NN, encoder–decoder, stacked hourglasses, B-Spline explicit, the active surface model (BEASM) framework, and structured RF. They implemented two UNet architectures and referred to them as UNet-1 and UNet-2. Both had different feature maps, resolutions, upsampling schemes, etc. According to their research, the best accuracy was achieved by the Unit-2 architecture. Moradi et al. [
40] also applied UNet and feature pyramid network (FPN) [
41] on the CAMUS [
29] dataset for LV segmentation. FPN was applied for feature extraction and used a pyramid concept with improved accuracy and speed. The FPN generated multiple feature maps with certain quality information than other feature pyramids for object detection. In the UNet architecture, two more downsampling layers were added for extracting the global information of the image. Due to the low resolution of images, a few encoding–decoding layers were used, which had an impact on overall accuracy. Zeng et al. [
42] proposed an encoder–decoder approach along with FPN for the left ventricle segmentation. They achieved a Dice similarity coefficient of 93.10.
One of the important aspects of training a segmentation model is having a relatively rich initial training set. This implies that the model outperforms when it uses 100 labeled videos from different patients rather than one hundred videos from the same patient. Due to the nature of the echocardiographic modality, augmentation methods can generate a limited number of training data. Therefore, increasing the training data is achievable by using more videos from more instances, which is sometimes difficult due to privacy laws. Moreover, the number of decoding and encoding levels in the segmentation network depends on the resolution of the input frame. More powerful hardware is required to perform segmentation on input frames with higher resolutions, which will increase the number of semantic strengths.
6.3. Detection
Dezaki et al. [
43] proposed an architecture for the detection of ES and ED phases from echocardiography videos. The architecture included a CNN model, an RNN, and a regression module. For the CNN module, residual network (ResNet) performance was better than DenseNet. For the RNN module, long short-term memory (LSTM) and gated recurrent units yielded comparable performance. Only high-quality data were considered in this study. The measurement of the aortic valve area helps in the diagnosis of certain conditions, such as aortic stenosis. Nizar et al. [
44] proposed a detection system for automatic segmentation of aorta valves using an SSD and faster RCNN. The dataset consisted of 30 videos, out of which 23 were used for training, 2 were used for testing, and 5 for validation. MobileNet and Inception were used as feature extractors for this model. A faster RCNN gave the best accuracy of 94%, while an SSD with mobileNet achieved the highest mean frame rate of just 34.21%. The former’s accuracy was promising, but minimal data were used for testing and validation purposes. Jafari et al. [
45] proposed a Bayesian framework for detecting the keyframe in echocardiography videos. They achieved an
score of 66%.
For the detection task, the limitations are the same as segmentation, such as the quality and size of the dataset.
6.4. Miscellaneous
The state-of-the art deep learning techniques used for echocardiography are discussed in [
46]. The main challenges identified were the lack of uniform performance evaluation across different algorithms and annotated data. The paper also mentioned that physicians and medical staff should actively indicate areas where automation is required and provide guidance. All of the research works discussed in this section are summarized in
Table 1,
Table 2 and
Table 3.
7. Endoscopy
This section discusses the research articles that explore deep learning models applied to endoscopy videos. All the analysis tasks were performed on endoscopy videos in two ways: (1) by extracting spatial features from individual frames of the videos and (2) by extracting spatial and temporal features from the videos, as mentioned in the respective tables. The section is divided into four subsections: Classification, Segmentation, Detection, and Miscellaneous, following our taxonomy discussed in
Section 3.
7.1. Classification
Different researchers combined deep learning techniques such as CNN and LSTM to extract spatial and temporal features from endoscopy videos. The proposed model consists of ResNet to extract temporal features, LSTM to extract spatial features, and a stack of fully connected and softmax layers to classify the image into one of the 37 classes. The proposed classification model is also used to design a retrieval system for similar frames. To classify ulcer images from endoscopy videos, a pretrained CNN-based architecture is proposed, which uses principal component analysis for dimension reduction and Xception for classification. Another architecture is developed to classify frames from colonoscopy videos into informative and noninformative categories using handcrafted features and a pretrained Inception V3. Additionally, a novel method utilizing a CNN, channel attention mechanism, and a classifier achieves the best accuracy by using ResNet with a proposed blockwise channel squeeze and excitation attention module.
The availability of large and fully annotated databases, such as ImageNet, is crucial for facilitating the development of deep classification models for endoscopy imaging. This poses a primary challenge in terms of training and testing the network. The active participation of endoscopists is also critical for establishing a large medical image dataset for training and thorough clinical validation.
7.2. Segmentation
Remarkable research is performed using GANs and the LIRE framework as the basis for polyp segmentation. Another study explores an encoder–decoder approach for the real-time segmentation of polyps from colonoscopy, achieving high recall, precision, F2, and accuracy scores on the KvasirCapsule-SEG dataset.
7.3. Detection
Researchers explore 3D CNNs to extract spatial–temporal features and employ a 3D FCN for detecting polyps in colonoscopy videos. Deep CNN architectures are used to classify gastrointestinal endoscopy video frames as abnormal or normal, and deep saliency detection algorithms are applied to detect salient points in abnormal images. Various models based on GoogleNet, AlexNet, VGG 16, and reinforcement learning are proposed for detecting lesions from wireless capsule endoscopy frames. A framework for ureteral orifice detection from ureteroscopy videos achieved high accuracy, although the number of false positive and false negative classifications is notable. Polyp detection from colonoscopy videos is addressed using a bootstrapping method, combining a base detector and temporal consistency verification.
Endoscopy segmentation and detection tasks present several challenges. Separate training data are required for every kind of ulcer or pathology, and further confirmation is often necessary for accurate diagnoses, especially when lesions are histologically indistinguishable from other conditions.
7.4. Miscellaneous
In the research work of Mohammed et al. [
47], they propose an automated capsule endoscopy video summarizing framework by combining deep and handcrafted features. For feature extraction, a pretrained GoogleNet is used. Hence, the deep features do not overfit the endoscopy images. Five hundred sample images are chosen from the KID [
28] by a gastroenterologist and GivenImaging capsule videos from different parts of the colon. Guerre et al. [
48] investigated using the state-of-the-art FlowNet algorithm for motion estimation in ocular endoscopy videos. Because FlowNet is strongly supervised, an artificial dataset of consecutive images paired with ground-truth optical flow was generated using eye fundus photographs from Kaggle’s Diabetic Retinopathy Detection dataset [
49]. A review paper discussed deep learning and non-deep-learning techniques applied to endoscopy videos to detect and classify polyps [
50]. According to the authors, the main challenges were the presence of noninformative frames in wireless endoscopy videos and difficulty in real-time detection due to time complexity. Another study surveyed research articles for cancer detection using deep learning techniques in WCE videos [
51]. The authors discussed several challenges in this field: a lack of annotated data and standard evaluation methods for the model’s performance. The authors also suggested that for future research, domain knowledge and real-time diagnosis should be taken into consideration. The research articles reviewed for endoscopy video analysis are summarized in
Table 4,
Table 5 and
Table 6.
8. Ultrasound
This section discusses the research articles that have explored deep learning models applied to ultrasound videos. All the analysis tasks are performed on ultrasound in two ways: (1) by extracting spatial features from individual frames of the videos and (2) by extracting spatial and temporal features from the videos. We mention this information in the respective tables. The section is divided into four subsections: Classification, Segmentation, Detection, and Miscellaneous, according to our taxonomy discussed in
Section 3.
8.1. Classification
To classify the quality and prevent calculation errors in fetal ultrasound videos, a scheme was proposed by Wu et al. [
68]. The proposed approach was realized with two CNN models, which were denoted as C-CNN and L-CNN. The purpose of the L-CNN was to find the fetal abdominal region in the ultrasound frame. The C-CNN evaluated the frame quality based on the output of L-CNN. Perception ultrasound by learning sonographic experience (PULSE) [
69] was a research project at the University of Oxford that combined state-of-the-art machine learning techniques with probe and eye movement data during an ultrasound. While working on this project, Patra et al. [
70] proposed a model for efficient ultrasound analysis consisting of a teacher and student module. The teacher module was trained on ultrasound images and sonographer eye movement data during ultrasound video, while the student model was trained just over the ultrasound images. The purpose of the model was to classify fetal abdomen, head, and femur frames. VGG-16 was used as a feature extractor in the model, and MobileNet V2 was explored for the student module. Chen et al. [
71] combined a radiologist’s domain expertise to propose a contrast-enhanced ultrasound (CEUS) video classification model for breast cancer. The researchers explored 3D CNN with a domain-knowledge-guided temporal attention module and a channel attention module. Due to these modules, the proposed model is able to focus on critical time slots of CEUS videos and learn features more efficiently, which helps to improve the classification performance of the model. The model achieved an accuracy of 86.3.% and a sensitivity of 97.2% on a dataset containing 221 cases. Zhou et al. [
72] proposed a fused model with hand-crafted and deep features to classify atypical hepatocellular carcinoma. The deep features are extracted by the 3-D-CNN, and the hand-crafted features are extracted by state-of-the-art methods. Then they are combined and given to a classifier such as Support Vector Machine (SVM) for classification.
The limitations for the classification task in ultrasound videos include data from the same source, such as videos captured by the same machine or by the same technician. Ideally, the training dataset should be larger, heterogeneous, and more balanced in terms of classes for learning accurate deep models.
8.2. Segmentation
Chi et al. [
73] used a combination of random forest with GoogleNet for the segmentation of thyroid nodules in ultrasound videos. Their model comprised two main steps. The first was image preprocessing, where they normalized and removed the artifacts from images. The second was data augmentation to prevent the model from overfitting. The model could only classify thyroid nodules into malignant and benign classes. It depended on a doctor’s expertise to mark the region of interest (ROI), as the system did not automatically detect it. Roy et al. [
74] collected and annotated a dataset for COVID-19 containing lung ultrasound videos from 35 patients. They proposed a model based on a CNN, spatial transformers network, and soft ordinal regression to help medical professionals estimate the severity of COVID-19 by assigning pathological scores to the ultrasound video frames. They also explored UNet for pixel-level segmentation of COVID-19 pathological artifacts.
8.3. Detection
Gao et al. [
75] compared a directly learned CNN with a transfer-learned CNN (TCNN) for object detection in ultrasound videos, such as the skull, abdomen, and heart. The accuracy of the TCNN was proven to be better than the directly learned CNN. The detection of fetal standard planes provides fetal development information during pregnancy. Chen et al. [
76] proposed using a multitask CNN to extract features from videos. In this case, the CNN classifier was explored to extract a region of interest (ROI), which is fetal planes. LSTM was applied for scoring frames based on between-planes knowledge. A classified frame would be labeled as a fetal plane if the inferred score was more significant than the predefined threshold score. The proposed system was unsuitable for real-time feedback due to the long processing time, which is around one minute for a video containing 40 frames. Another limitation of the model was that all ultrasound images were taken from healthy mothers and babies, so the researchers could not anticipate how the system would respond when applied to a fetus with abnormalities. Another work for fetal plane detection is proposed by [
77]. The proposed model is used to identify four fetal planes and is based on a CNN and an RNN. The model extracts the spatial and temporal features from US videos.
The challenges faced by detection and segmentation tasks are low accuracy, dependency on radiologist decisions, and high processing time. One of the reasons behind these shortcomings is, firstly, that when an ultrasound test is performed, the technician tries to capture as many details as possible. This causes the ultrasound video to have many uninformative frames. Secondly, the ultrasound videos are noisy in nature, contain speckles, and are usually grayscale. Lastly, ultrasounds may generate blurry results if the patient is obese, resulting in difficulty in analysis.
8.4. Miscellaneous
Jarosik et al. [
78] suggested a real-time processing architecture for reconstructing ultrasound images augmented with deep learning methods. To achieve this, they implemented WaveFlow, a collection of ultrasound data acquisition and processing tools integrated with TensorFlow. WaveFlow includes ultrasound environments and signal processing operator libraries. However, the system was only tested on five ultrasound videos. To assess the state of skeletal muscle from ultrasound videos in general conditions, Cunningham and Loram [
79] proposed a CNN-based architecture. The two muscles selected for the study were gastrocnemius medialis and soleus, and the performance varied across all 32 participants, with accuracy ranging from 45% to 56.9%. The survey of current imaging and deep learning techniques applied to ultrasound technology was conducted by different researchers [
80,
81]. Transfer learning, 3D ultrasound data, speckle noise removal, and publicly available standard datasets were identified as the areas that need attention to improve accuracy. The research papers discussed in this section are summarized in
Table 7,
Table 8 and
Table 9.
9. Discussion
There are many deep learning methods which are applied to the analysis of medical videos. For echocardiography videos, deep learning is mainly used for LV segmentation, because it provides prognostic information and diagnostic clues. LV size helps cardiologists in diagnosing heart failure and LV hypertrophy. UNet is considered the top choice for segmentation in the medical field because CNNs impose some limitations. These limitations include the unavailability of a large number of samples and other problems, such as gradient exploding or gradient vanishing. UNet was introduced to cater to these drawbacks. UNet can be trained on a limited number of samples. Furthermore, UNet realizes image features with multiscale recognition and fusion. However, UNet does not consider the contribution of all semantic strengths during the segmentation process. To tackle this, combining UNet with FPN can improve segmentation accuracy. Deep learning is also applied to echocardiography videos to classify different views, as every standard view of the heart during echocardiography is crucial for other measurements; for example, the parasternal long-axis view is considered the most appropriate view to measure LV size. Most researchers have explored CNNs for view classification and reported greater than 90% accuracy overall. However, the performance of CNN models was not uniform for all personal views. To avoid this issue, segmentation can be used as a preprocessing step, although this additional step can prove to be resource- and time-consuming.
For endoscopy videos, deep learning is usually applied to detect or classify polyps and lesions in different body organs. Some researchers have interpreted this as classifying informative (with polyps) and noninformative (without polyps) frames from videos. Researchers have widely explored transfer and machine learning models for endoscopy videos [
83]. GoogleNet has achieved up to 100% accuracy in detecting ulcers from endoscopy videos. However, the runtime cost for GoogleNet can be as high as 37 min, which is very high compared with a CNN trained from scratch. Another approach that can improve the accuracy of the pretrained model is the fusion of handcrafted and CNN-extracted features. This approach is beneficial when data are limited and domain knowledge is available. However, this approach is not suitable for real-time analysis of the videos.
For ultrasound videos, deep learning is usually applied for classifying or detecting vital fetal organs. To capture the temporal features, LSTM is widely used for all kinds of medical diagnostic videos, including ultrasound [
59]. LSTM can be fused with a pretrained CNN model or a CNN model which is trained from scratch. The combination of ResNet and LSTM has produced promising results for detection and classification tasks. Researchers also explore 3D CNNs to capture the temporal features of all three kinds of videos. However, 3D CNNs require more extensive training time than 2D CNN models.
For newcomers venturing into the field of medical diagnostic videos using deep learning, it is imperative to follow a systematic approach to effectively utilize the existing models for their research. Firstly, it is highly recommended for newcomers to go through various literature reviews of medical videos and images to gain a comprehensive understanding of the current state-of-the-art methods and their practical applications in the medical domain. As discussed earlier, LSTM, 3D CNNs, and transfer learning models are some of the promising approaches for beginners to start with.
In addition, it is highly advisable to stay updated with newly emerging concepts and technologies in the field of deep learning. For instance, explainable AI [
84] is an increasingly important topic that enables clinicians to better interpret and explain the predictions made by deep learning models. Similarly, Federated Learning [
85] is another recent advancement in the field of deep learning that allows multiple institutions to collaboratively train models on their respective datasets without sharing sensitive patient data. By being aware of these emerging concepts and technologies, newcomers can broaden their horizons and enhance their understanding of the latest trends and developments in the field of medical diagnostic using deep learning.
10. Advancing Medical Diagnostic Videos: Limitations and Future Work
This section discusses the limitations and future directions for deep-learning-based methods for medical video analysis. The lack of available data and class imbalance in medical videos are some of the significant challenges. Another issue is the sparse labeling of videos, which results in biased and incomplete datasets. Furthermore, the inability to know the reason behind decisions made by the model and time complexity are major concerns. Medical data impose unique challenges that need to be handled during preprocessing, including video format, resolution, and computation time. To address these challenges, downsampling can be performed while attempting to avoid the loss of features useful in differentiating dissimilar classes. However, the ideal resolution size can vary depending on the complexity of visual structures and data type. This section also discusses different techniques and approaches to improve medical video analysis using deep learning. Transfer learning is explored as a solution to the limited availability of labeled data in medical cases. Multitask learning helps learn different parameters from limited available video data. Data augmentation techniques are widely used for imbalanced or small datasets. Incorporating expert knowledge and telemedicine can help improve the quality of medical videos. Semisupervised learning is helpful for medical videos because labeling a massive amount of videos for supervised learning is time-consuming and expensive. N-shot learning, including few-shot, zero-shot, and one-shot learning, is a data-efficient approach that can be used efficiently in the medical field. Further details about these limitations and future work are given below.
10.1. Data Limitation
The most important aspect of deep learning is its capability to model highly complex mathematical calculations, which makes it different from the rest of artificial intelligence algorithms. Generally, more layers are introduced to learn more complex functions; however, a deeper network must also be trained over a large number of parameters. Such a model can only learn well if we correspondingly use a large amount of data to infer the parameter values. A complex model trained using a small dataset normally overfits and becomes data-dependent. Such modeling gives a false impression of performance and is not reliable. The lack of availability of the required data has been a limitation for various research works (Feng, Year; Yao, Year; Gao, Year). This is the reason that makes the medical field challenging for deep learning. Crowd-sourcing can be used to label the data of daily life objects, but for medical videos, experts in the field are required. Privacy and consent of patients are the other two aspects contributing to the lack of publicly available medical data.
10.2. Class Imbalance
Another challenge is the class or data imbalance in medical videos, where only a limited number of the frames show ROI, and most frames are normal. Class imbalance can also be a matter of concern when one of the classes is rare or underrepresented, as observed in the research work by Gao et al. [
75] and Shahin et al. [
38]. This data imbalance can cause a model to be biased towards one class, resulting in unexpected errors. Balancing data for rare conditions, such as cystic fibrosis, is as difficult as developing large-scale datasets. To cater to the problem of data unavailability, Leclerc et al. (Year) introduced the largest echo dataset of 500 patients. However, the dataset is still not balanced regarding the image quality and size of LV.
10.3. Labeling of Medical Videos
During the labeling of medical videos, physicians or technicians usually only label the landmark markers at the selected informative frames. As a result, the video datasets suffer from two constraints: (1) the labeled frames are hugely biased towards specific points in time, i.e., only informative frames in each video are labeled, and (2) videos are sparsely labeled, i.e., a limited portion of frames in each video have ground-truth marker labels. Moreover, there is a lack of quantitative criteria for image annotation. Some frames show the ROI and uninformative features simultaneously, making the annotation process difficult. Adding uncertainty grading during the annotation process and integrating the uncertainty in the training process may improve model performance.
10.4. Lack of Confidence Interval
Deep learning is said to be a black-box approach, where we cannot observe how the model is taking the decision. The inability to know the reason behind decisions made by the model makes it unreliable to be used in the medical field. Doctors and patients find it difficult to trust something working with a phenomenon they are unaware of. There is a need to explain the working of deep learning methods to experts and nonexperts in the form of graphs or interactive media.
10.5. Time Complexity
Processing videos is a time-consuming process, and when the medical field is involved, it is even more complex, because medical videos have lots of uninformative frames, noise, and background details. Some of the work above [
76,
86] has proved to give acceptable accuracy, but the time taken by the model to process the video is very high. Due to time complexity, these models can be used offline rather than in real time.
One more problem observed during the survey is a lack of standard performance metrics [
87]. Time complexity is a very important factor but is completely ignored by some of the research works. The lack of standard metrics makes it difficult to compare the performance of different approaches.
10.6. Preprocessing of Medical Videos
For the medical domain, important considerations must be taken into account in developing deep learning models. Medical data impose a few challenges that should be handled at the preprocessing step. The first challenge is regarding the format of the medical videos and their resolution. Medical videos often come in a DICOM format with varying resolutions per acquisition. Secondly, processing all pixels in a medical video can lead to excessive computation time and weights to learn. Lastly, medical videos show a slight difference in dissimilar classes; hence, there is a risk of losing vital information if the resolution is reduced.
To tackle these challenges, the primary prerequisite step is to preprocess the data to standardize the resolution before developing the deep learning models with nontrivial implications. Downsampling can be performed on videos while attempting to avoid the loss of features that are useful in differentiating dissimilar classes. The ideal resolution is one which has a minimal amount of visual information necessary for accurate classification or segmentation. Ideal resolution size can highly depend on the complexity of visual structures and data type. However, in general, there is a rapid increase in computation time with increasing input resolution as the network architecture expands in depth.
10.7. Transfer Learning
Some models, such as VGG-16 and GoogleNet, are already trained on one million images and can be used for video data processing. Our discussion shows that such heavily labeled data is not readily available for medical cases, so researchers are exploring transfer learning to resolve this problem. Several studies, such as [
70,
73,
75], have exploited transfer learning to solve the problem of limited data and have achieved good results. The pretrained models are further fine-tuned to be applied to medical problems. Another form of transfer learning is yet to be explored in the medical field. As we know, deeper models need a large number of parameters to be trained. However, there is a possibility to divide the data into low-level and high-level features and then freeze the features that are similar in all videos. This way, the number of learnable parameters can be reduced; thus, fewer data will be required. The low-level features can also be transferred by another pretrained model which is already trained on similar data.
10.8. Multitask Learning
Multitask learning, also known as joint learning, refers to the process where multiple deep models are trained and then combined to compute the result. Each model has its complexity, architecture, and loss function. This approach helps to learn different parameters from limited available video data. Multitask learning is being successfully used in several fields, such as natural language processing, computer vision, and pattern recognition [
88].
10.9. Data Augmentation
Data augmentation is a widely used technique for imbalanced or small datasets. It involves flipping, rotating, and cropping already available video frames to increase the size of the training dataset. Several researchers have used this technique [
52,
70] to generate endoscopy and ultrasound frames. Patra et al. [
70] augmented video frames by a rotation of 20 degrees and flipping both ultrasound and gaze map frames horizontally, while Owais et al. [
52] applied plain rotation and random translation to augment the data. GANs are another approach successfully used for generating artificial data, as previously explained in the paper. Scientists are now using GANs to predict the next few frames in a video, and this technique can also be helpful for medical videos. This approach can help predict the subsequent frames if the medical video is captured by an undertrained medical staff and ROI is not fully captured. It is recommended that precaution should be taken while applying GANs to medical videos, because GANs do not learn the distribution of original videos; instead, they only copy them. Hence, the original videos are different from GAN-generated synthetic ones.
10.10. Incorporating Expert Knowledge
Knowledge from stakeholders, such as doctors, sonographers, patients, and technicians, can be incorporated into the process of automation in many ways. Firstly, medical personnel can help researchers understand the most important features of data critical for the diagnosis. This way, more efficient and effective models can be trained. Secondly, medical terminologies are usually very complex to be understood by laymen, which makes it difficult for people to take an interest in or explore it. Researchers and programmers can only excel in a subject if they understand it completely. Doctors and experts can make an effort to explain medical terminologies in simple language on platforms, such as Kaggle, where competitions are held.
10.11. Semisupervised Learning
In semisupervised learning, the model is trained upon a collection of labeled and unlabeled data. Typically, the dataset will have a minimal amount of labeled data and a huge amount of unlabeled data. This type of learning is helpful for medical videos, because labeling a massive amount of videos for supervised learning is time-consuming and expensive.
10.12. N-Shot Learning
N-shot learning is a broad concept, as it includes few-shot, zero-shot, and one-shot learning. Few-shot learning is a classification task where a very small number of training examples are given for each class that is used to prepare a model. In zero-shot learning, the model observes images from categories that were not monitored while training and predicting the class they belong to. Similarly, one-shot learning is a classification task where one example is given for each class. One of the networks used for N-shot learning is the Siamese neural network. A Siamese neural network consists of twin networks that accept two images but are joined by an energy function at the output. In Siamese networks, feature vectors are learned by using convolutional neural networks, which are obtained from labeled nonmatching and matching image pairs. The parameters between the twin networks are identical. Similar initial weights assure that two similar images could not possibly be mapped by their respective networks to very different locations in feature space, because each network computes the same function. Then, the similarity between these feature vectors is measured by Euclidean distance. The image is classified based on the nearest-neighbor approach’s similarity score. N-shot learning is a data-efficient approach which can be used efficiently in the medical field. One-shot learning specifically is helpful for the classification of rare diseases.
10.13. Live Feedback from Experts
The collection process of ultrasound videos is different from other imaging technologies. The technicians need to not only be experts in ultrasound imaging and related diagnosis but also be masters in capturing videos from standardized views and planes. Therefore, a diagnosis from ultrasound videos is very challenging for physicians at secondary care hospitals. Telemedicine is an approach that combines electronic technology, computer network, modern communication, and medical diagnostic data [
89]. If experts can integrate telemedicine with deep learning models for remote consultation of patients, then real-time feedback from experts in the field can help not only the diagnosis [
90] but also improve the efficiency of the deep learning models. Training data can be refined based on the medical experts’ analysis of the model’s performance.
10.14. Incorporating Medical History
Deep learning models should not only rely on one type of data, such as text or images. The models can be fed a combination of text, videos, and images, for example, the current state of the particular organ in the form of a video or image, the previous medical history of the patient, and other vital data such as age, gender, and weight. This will improve performance and help the models make informed decisions about the patient.
10.15. Rapid Reading Software
Recently, EndoCapsule [
91] introduced a rapid reading software [
92] to reduce the number of uninformative frames from endoscopy videos. The rapid reading software decreases the time taken to process the video and increases the efficiency of the diagnosis process. A study [
93] has shown a reduction of 64% in reading times associated with a 0.93 sensitivity in the diagnosis of lesions in preselected cases. The application of such software for other diagnostic videos is still an open challenge for researchers.
11. Conclusions
Medical diagnostic video processing using deep learning is a thriving and challenging area of research that combines the fields of medicine and information technology. In this manuscript, we provided a comprehensive review of this field, covering several key areas of focus. First, we outlined the methodology used to conduct our review. Next, we provided an overview of medical videos and deep learning techniques, giving readers a foundation for the subsequent sections. We then summarized different medical video datasets that have been used for classification, detection, and segmentation. Moving on, we reviewed many research articles focusing on deep learning applications in medical diagnostic videos. This section provided readers with a broad understanding of the current state of the art in this field. Lastly, we discussed the limitations and future work perspectives for deep learning in medical videos. We believe that this perspective is distinct from other related reviews and provides a deeper level of detail on the topic. In conclusion, we suggest that medical diagnostic video analysis could significantly benefit from deep learning techniques through collaboration with domain experts and patients. By continuing to explore this area, we can improve the accuracy and efficiency of medical diagnoses, ultimately leading to better patient outcomes.