Link Prediction with Hypergraphs via Network Embedding
Abstract
1. Introduction
2. Materials and Methods
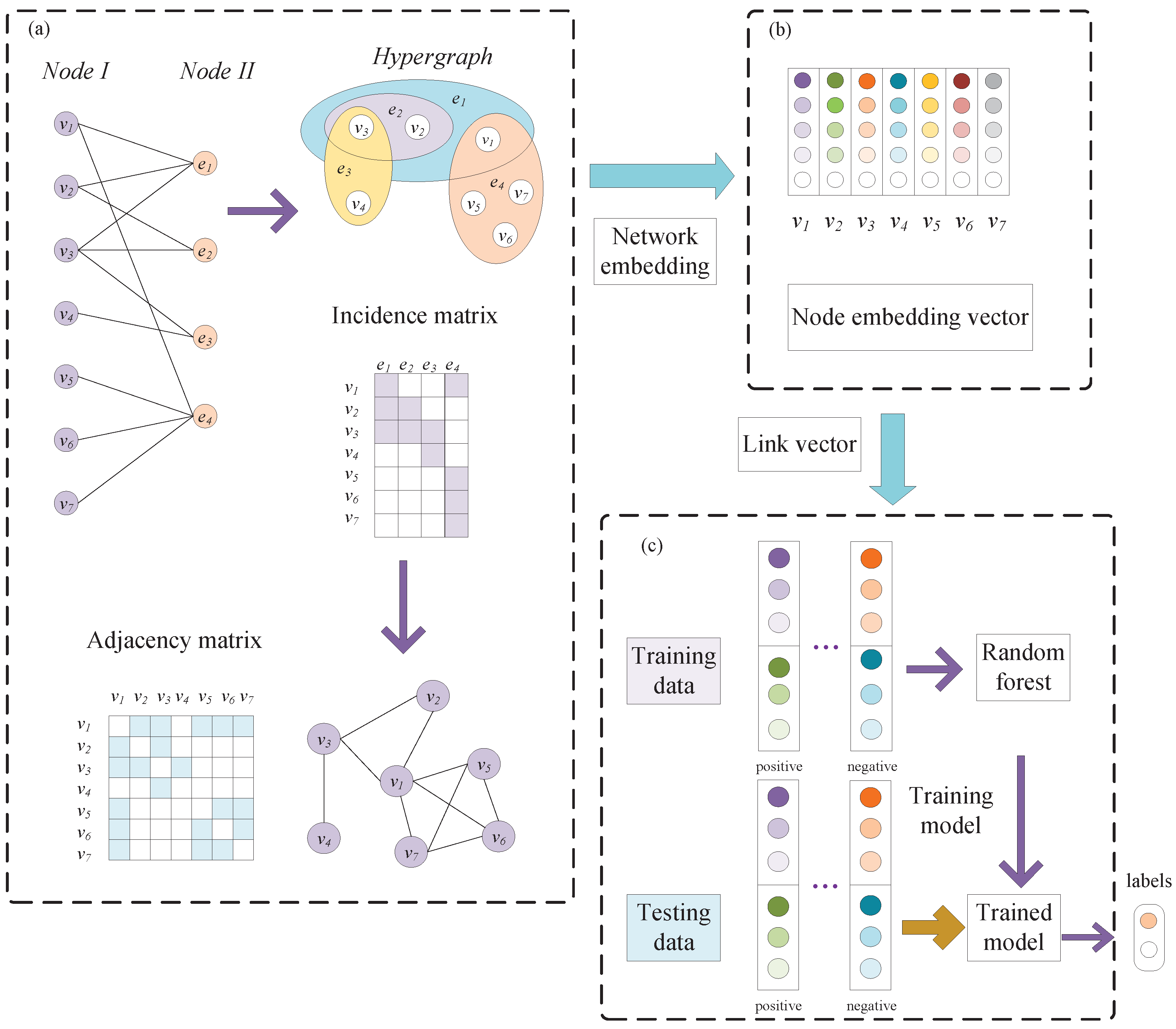
2.1. Hypergraph Construction
2.2. Learning Representations with Network Embedding
2.3. Loss Function
2.4. Datasets
3. Experiments
3.1. Compared Methods
3.2. Results
4. Conclusions and Discussions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Lü, L.Y.; Zhou, T. Link prediction in complex networks: A survey. Phys. A 2011, 6, 1150–1170. [Google Scholar] [CrossRef]
- Zhang, M.H.; Chen, Y.X. Link prediction based on graph neural networks. In Proceedings of the Annual Conference on Neural Information Processing Systems 2018, NeurIPS 2018, Montréal, QC, Canada, 3–8 December 2018; pp. 5171–5181. [Google Scholar]
- Wang, H.W.; Zhang, F.Z.; Hou, M.; Xie, X.; Guo, M.Y.; Liu, Q. Shine: Signed heterogeneous information network embedding for sentiment link prediction. In Proceedings of the Eleventh ACM International Conference on Web Search and Data Mining, Marina del Rey, CA, USA, 5–9 February 2018; pp. 592–600. [Google Scholar]
- Pulipati, S.; Somula, R.; Parvathala, B.R. Nature inspired link prediction and community detection algorithms for social networks: A survey. Int. J. Syst. Assur. Eng. Manag. 2021, 1–18. [Google Scholar] [CrossRef]
- Talasu, N.; Jonnalagadda, A.; Pillai, S.S.A.; Rahul, J. A link prediction based approach for recommendation systems. In Proceedings of the 2017 International Conference on Advances in Computing, Communications and Informatics (ICACCI), Manipal, India, 13–16 September 2017; pp. 2059–2062. [Google Scholar]
- Patel, R.; Guo, Y.H.; Alhudhaif, A.; Alenezi, F.; Althubiti, S.A.; Polat, K. Graph-Based Link Prediction between Human Phenotypes and Genes. Math. Probl. Eng. 2021, 2022, 8. [Google Scholar] [CrossRef]
- Yang, K.; Zhao, X.Z.; Waxman, D.; Zhao, X.M. Predicting drug-disease associations with heterogeneous network embedding. Chaos 2019, 12, 123109. [Google Scholar] [CrossRef]
- Kushwah, A.K.S.; Manjhvar, A.K. A review on link prediction in social network. Int. J. Grid Distrib. Comput. 2016, 2, 43–50. [Google Scholar] [CrossRef]
- Passino, F.S.; Turcotte, M.J.M.; Heard, N.A. Graph link prediction in computer networks using poisson matrix factorisation. Ann. Appl. Stat. 2022, 3, 1313–1332. [Google Scholar]
- Zhou, T.; Kuscsik, Z.; Liu, J.-G.; Medo, M.; Wakeling, J.R.; Zhang, Y.-C. Solving the apparent diversity-accuracy dilemma of recommender systems. Proc. Natl. Acad. Sci. USA 2010, 107, 4511–4515. [Google Scholar] [CrossRef]
- Real, R.; Vargas, J.M. The probabilistic basis of Jaccard’s index of similarity. Syst. Biol. 1996, 3, 380–385. [Google Scholar] [CrossRef]
- Katz, L. A new status index derived from sociometric analysis. Psychometrika 1953, 1, 39–43. [Google Scholar]
- Tong, H.h.; Faloutsos, C.; Pan, J.Y. Fast random walk with restart and its applications. In Proceedings of the Sixth International Conference on Data Mining (ICDM’06), Hong Kong, China, 18–22 December 2006; pp. 613–622. [Google Scholar]
- Gao, Y.; Zhang, Z.Z.; Lin, H.J.; Zhao, X.B.; Du, S.Y.; Zou, C.Q. Hypergraph learning: Methods and practices. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 44, 2548–2566. [Google Scholar] [CrossRef]
- Feng, Y.F.; You, H.X.; Zhang, Z.Z.; Ji, R.R.; Gao, Y. Hypergraph neural networks. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; pp. 3558–3565. [Google Scholar]
- Bai, S.; Zhang, F.h.; Torr, P.H.S. Hypergraph convolution and hypergraph attention. Pattern Recognit. 2021, 110, 107637. [Google Scholar] [CrossRef]
- Xiao, Q. Node importance measure for scientific research collaboration from hypernetwork perspective. Teh. Vjesn. 2016, 2, 397–404. [Google Scholar]
- Gallagher, S.R.; Goldberg, D.S. Clustering coefficients in protein interaction hypernetworks. In Proceedings of the International Conference on Bioinformatics, Computational Biology and Biomedical Informatics, Wshington, DC, USA, 22–25 September 2013; pp. 552–560. [Google Scholar]
- Li, D.; Xu, Z.; Li, S.; Sun, X. Link prediction in social networks based on hypergraph. In Proceedings of the 22nd International Conference on World Wide Web, Rio de Janeiro, Brazil, 13–17 May 2013; pp. 41–42. [Google Scholar]
- Chen, C.; Liu, Y.Y. A survey on hyperlink prediction. arXiv 2022, arXiv:2207.02911. [Google Scholar]
- Sharma, A.; Srivastava, J.; Chandra, A. Predicting multi-actor collaborations using hypergraphs. arXiv 2014, arXiv:1401.6404. [Google Scholar]
- Vaida, M.; Purcell, K. Hypergraph link prediction: Learning drug interaction networks embeddings. In Proceedings of the 2019 18th IEEE International Conference On Machine Learning And Applications (ICMLA), Boca Raton, FL, USA, 16–19 December 2019; pp. 1860–1865. [Google Scholar]
- Liu, J.; Song, L.; Wang, G.; Shang, X.Q. Meta-HGT: Metapath-aware HyperGraph Transformer for heterogeneous information network embedding. Neural Netw. 2023, 157, 65–76. [Google Scholar] [CrossRef]
- Kang, X.; Li, X.; Yao, H.; Li, D.; Jiang, B.; Peng, X.; Wu, T.; Qi, S.H.; Dong, L.J. Dynamic hypergraph neural networks based on key hyperedges. Inf. Sci. 2022, 616, 37–51. [Google Scholar] [CrossRef]
- Fan, H.; Zhang, F.; Wei, Y.; Li, Z.; Zou, C.; Gao, Y.; Dai, Q. Heterogeneous hypergraph variational autoencoder for link prediction. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 4125–4138. [Google Scholar] [CrossRef]
- Cui, P.; Wang, X.; Pei, J.; Zhu, W.W. A survey on network embedding. IEEE Trans. Knowl. Data Eng. 2018, 5, 833–853. [Google Scholar] [CrossRef]
- Chang, S.y.; Han, W.; Tang, J.L.; Qi, G.J.; Aggarwal, C.C.; Huang, T.S. Heterogeneous network embedding via deep architectures. In Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Sydney, Australia, 10–13 August 2015; pp. 119–128. [Google Scholar]
- Arsov, N.; Mirceva, G. Network embedding: An overview. arXiv 2019, arXiv:1911.11726. [Google Scholar]
- Wang, X.; Cui, P.; Wang, J.; Pei, J.; Zhu, W.W.; Yang, S.Q. Community preserving network embedding. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Belkin, M.; Niyogi, P. Laplacian eigenmaps and spectral techniques for embedding and clustering. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 3–8 December 2001; p. 14. [Google Scholar]
- Perozzi, B.; Al-Rfou, R.; Skiena, S. Deepwalk: Online learning of social representations. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 701–710. [Google Scholar]
- Grover, A.; Leskovec, J. node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 855–864. [Google Scholar]
- Donnat, C.; Zitnik, M.; Hallac, D.; Leskovec, J. Learning structural node embeddings via diffusion wavelets. In Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, London, UK, 19–23 August 2018; pp. 1320–1329. [Google Scholar]
- Hu, B.; Fang, Y.; Shi, C. Adversarial learning on heterogeneous information networks. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage AK, USA, 4–8 August 2019; pp. 120–129. [Google Scholar]
- Tsitsulin, A.; Mottin, D.; Karras, P.; Müller, E. Verse: Versatile graph embeddings from similarity measures. In Proceedings of the 2018 World Wide Web Conference, Lyon, France, 23–27 April 2018; pp. 539–548. [Google Scholar]
- Wang, S.H.; Tang, J.L.; Aggarwal, C.R.; Chang, Y.; Liu, H. Signed network embedding in social media. In Proceedings of the 2017 SIAM International Conference on Data Mining, Houston, TX, USA, 27–29 April 2017; pp. 327–335. [Google Scholar]
- Yadati, N.; Nitin, V.; Nimishakavi, M.; Yadav, P.; Louis, A.; Talukdar, P. NHP: Neural hypergraph link prediction. In Proceedings of the 29th ACM International Conference on Information & Knowledge Management, Virtual Event, 19–23 October 2020; pp. 1705–1714. [Google Scholar]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Yadati, N.; Nimishakavi, M.; Yadav, P.; Nitin, V.; Louis, A.; Talukdar, P.P. Hypergcn: A new method for training graph convolutional networks on hypergraphs. In Proceedings of the Annual Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; pp. 1509–1520. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Tu, K.; Cui, P.; Wang, X.; Wang, F.; Zhu, W.w. Structural deep embedding for hyper-networks. In Proceedings of the AAAI Conference on Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018; p. 1. [Google Scholar]
- Ma, T.; Suo, Q. Review of Hypernetwork Based on Hypergraph. Oper. Res. Manag. Sci. 2021, 2, 232. [Google Scholar]
- Service Centre of Huiyuan Sharing Academic Resources. University Library Dataset. Available online: http://hdl.handle.net/20.500.12291/10022 (accessed on 22 July 2021).
- Zhang, C.X.; Song, D.j.; Huang, C.; Swami, A.; Chawla, N.V. Heterogeneous graph neural network. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage AK, USA, 4–8 August 2019; pp. 793–803. [Google Scholar]
- Ding, Y.; Zhang, Z.L.; Zhao, X.F.; Cai, W.; He, F.; Cai, Y.M.; Cai, W.W. Deep hybrid: Multi-graph neural network collaboration for hyperspectral image classification. Def. Technol. 2022; in press. [Google Scholar]
- Wu, C.; Wu, F.; Cao, Y.; Huang, Y.; Xie, X. Fedgnn: Federated graph neural network for privacy-preserving recommendation. arXiv 2021, arXiv:2102.04925. [Google Scholar]
- Huang, C.; Xu, H.C.; Xu, Y.; Dai, P.; Xia, L.H.; Lu, M.Y.; Bo, L.F.; Xing, H.; Lai, X.P.; Ye, Y.F. Knowledge-aware coupled graph neural network for social recommendation. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtually, 2–9 February 2021; pp. 4115–4122. [Google Scholar]

| Datasets | n | m | Density | |||||
|---|---|---|---|---|---|---|---|---|
| SUEP | 906 | 24,362 | 19,530 | 0.0297 | 27 | 29 | 47 | 1.3 |
| SHOU | 2680 | 222,126 | 64,958 | 0.0309 | 81 | 41 | 108 | 1.7 |
| SUFE | 1720 | 148,188 | 35,727 | 0.0501 | 86 | 33 | 62 | 1.6 |
| USST | 2733 | 230,597 | 54,437 | 0.0308 | 84 | 36 | 93 | 1.8 |
| SISU | 3089 | 478,953 | 72,100 | 0.0502 | 155 | 46 | 142 | 2 |
| SHNU | 3557 | 263,305 | 93,996 | 0.0208 | 74 | 43 | 120 | 1.6 |
| TJU | 6150 | 988,516 | 131,199 | 0.0261 | 161 | 42 | 134 | 1.9 |
| SHOU | SUFE | SUEP | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | F1-Score | Precision | Recall | F1-Score | Precision | Recall | F1-Score | |
| CN | 0.6971 | 0.6091 | 0.6499 | 0.8996 | 0.6654 | 0.7650 | 0.7063 | 0.5645 | 0.6275 |
| Jaccard | 0.7569 | 0.6034 | 0.6715 | 0.9034 | 0.6916 | 0.7834 | 0.7412 | 0.5424 | 0.6263 |
| Katz | 0.6705 | 0.8001 | 0.7333 | 0.6722 | 0.8121 | 0.7356 | 0.6663 | 0.8061 | 0.7296 |
| RWR | 0.5446 | 0.5456 | 0.5449 | 0.5929 | 0.6250 | 0.6083 | 0.5328 | 0.5366 | 0.5337 |
| Node2vec | 0.8317 | 0.8037 | 0.8223 | 0.9416 | 0.8566 | 0.898 | 0.8401 | 0.8154 | 0.8275 |
| GCN | 0.7959 | 0.7675 | 0.7814 | 0.9046 | 0.8372 | 0.8696 | 0.7934 | 0.7734 | 0.7832 |
| HNE | 0.8379 | 0.8052 | 0.8212 | 0.9424 | 0.8685 | 0.9040 | 0.8516 | 0.8331 | 0.8422 |
| USST | SISU | SHNU | TJU | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | F1-Score | Precision | Recall | F1-Score | Precision | Recall | F1-Score | Precision | Recall | F1-Score | |
| CN | 0.6902 | 0.6663 | 0.678 | 0.6885 | 0.6161 | 0.8354 | 0.7594 | 0.6821 | 0.7187 | 0.8289 | 0.7467 | 0.7856 |
| Jaccard | 0.7685 | 0.6271 | 0.6906 | 0.7481 | 0.6246 | 0.6808 | 0.8107 | 0.6800 | 0.7396 | 0.8726 | 0.7596 | 0.8122 |
| Katz | 0.6711 | 0.8093 | 0.7337 | 0.6682 | 0.8022 | 0.7291 | 0.6706 | 0.8085 | 0.7331 | 0.6666 | 0.7925 | 0.7241 |
| RWR | 0.5438 | 0.5191 | 0.5306 | 0.5603 | 0.5455 | 0.5526 | 0.5457 | 0.5368 | 0.5411 | 0.5718 | 0.547 | 0.5588 |
| Node2vec | 0.8603 | 0.805 | 0.8317 | 0.866 | 0.8068 | 0.6502 | 0.8706 | 0.8582 | 0.8644 | 0.8668 | 0.8355 | 0.8512 |
| GCN | 0.8185 | 0.7738 | 0.7955 | 0.8328 | 0.7798 | 0.8054 | 0.8358 | 0.8229 | 0.7293 | 0.7848 | 0.7375 | 0.7604 |
| HNE | 0.8632 | 0.8160 | 0.8389 | 0.8657 | 0.8092 | 0.8365 | 0.8706 | 0.8674 | 0.8691 | 0.8365 | 0.7789 | 0.7972 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Yang, K.; Guo, J. Link Prediction with Hypergraphs via Network Embedding. Appl. Sci. 2023, 13, 523. https://doi.org/10.3390/app13010523
Zhao Z, Yang K, Guo J. Link Prediction with Hypergraphs via Network Embedding. Applied Sciences. 2023; 13(1):523. https://doi.org/10.3390/app13010523
Chicago/Turabian StyleZhao, Zijuan, Kai Yang, and Jinli Guo. 2023. "Link Prediction with Hypergraphs via Network Embedding" Applied Sciences 13, no. 1: 523. https://doi.org/10.3390/app13010523
APA StyleZhao, Z., Yang, K., & Guo, J. (2023). Link Prediction with Hypergraphs via Network Embedding. Applied Sciences, 13(1), 523. https://doi.org/10.3390/app13010523






