Traditional Grain-Based vs. Commercial Milk Kefirs, How Different Are They?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Kefir Samples and Commercial Kefirs
2.2. Cultivation-Independent Analysis of Microbial Composition of Kefirs
2.2.1. qPCR
2.2.2. NGS
2.3. Assessment of VOCs by Headspace-Gas Chromatography-Ion Mobility Spectrometry (HS-GC-IMS)
2.4. Targeted Metabolite Analysis
3. Results and Discussion
3.1. Microbial Composition of Kefirs
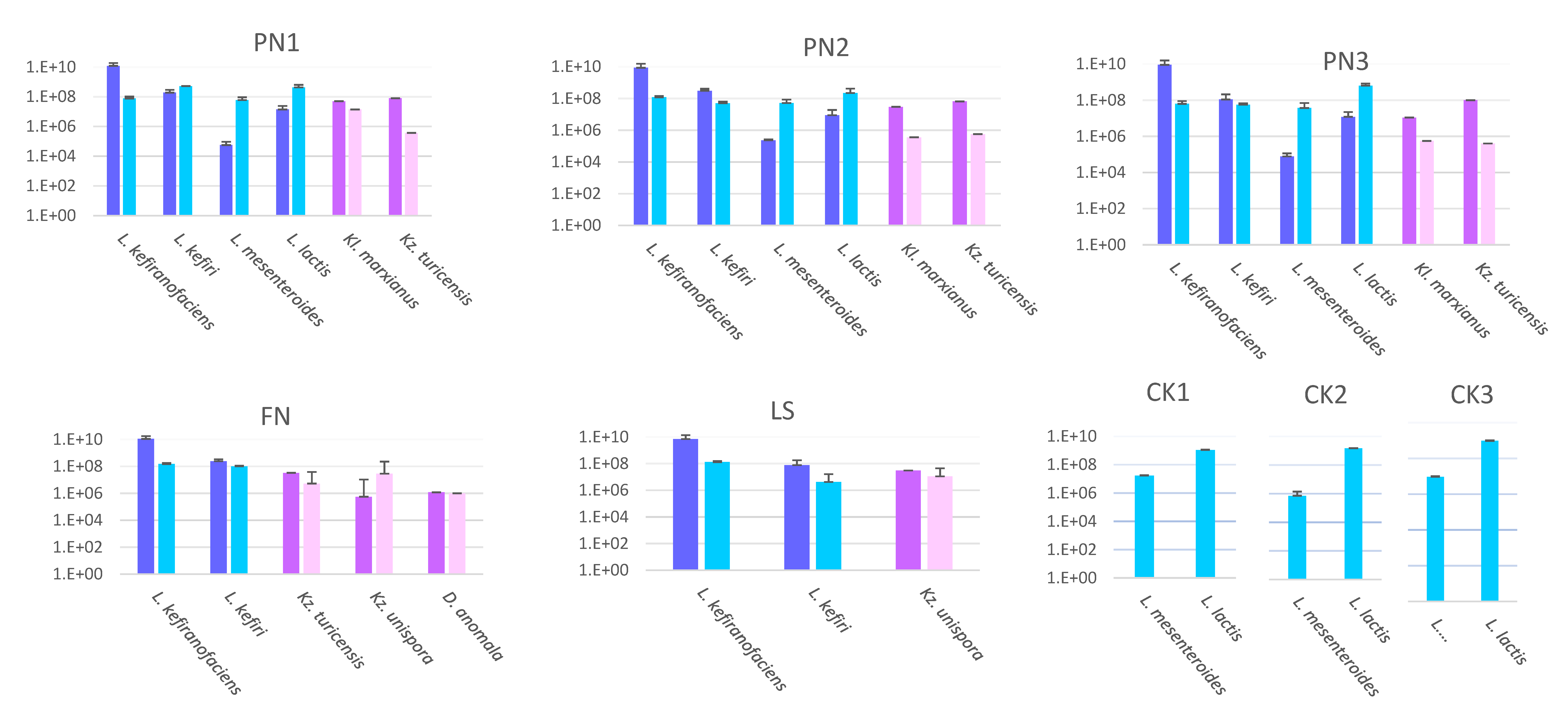
3.1.1. Targeted Quantification Using qPCR
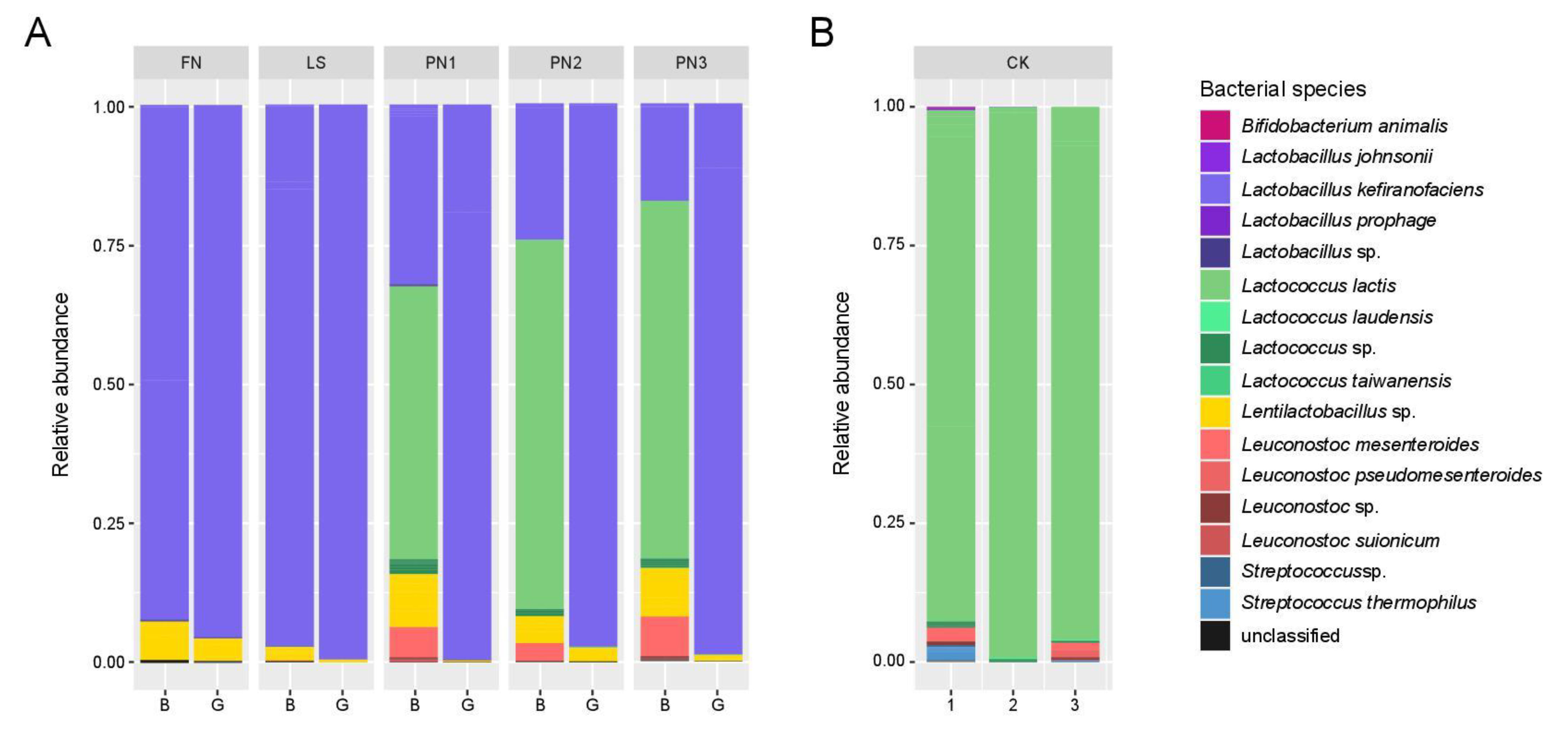
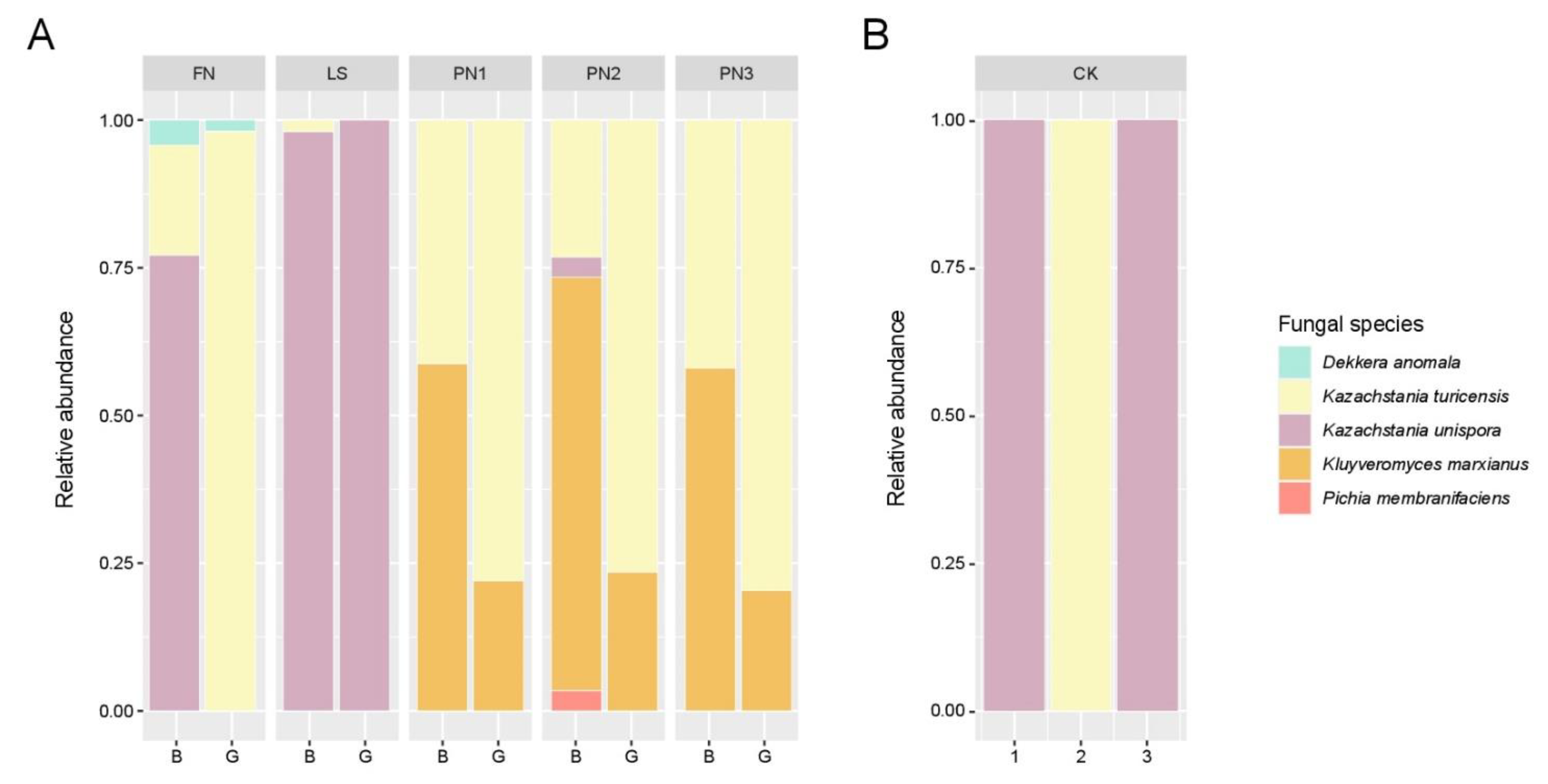
3.1.2. Identification of Microbial Community Using NGS
3.2. Identification of VOCs and Clustering of Yogurt and Kefir Samples
3.3. Targeted Metabolites Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xiang, H.; Sun-Waterhouse, D.; Waterhouse, G.I.; Cui, C.; Ruan, Z. Fermentation-enabled wellness foods: A fresh perspective. Food Sci. Hum. Wellness 2019, 8, 203–243. [Google Scholar] [CrossRef]
- Nejati, F.; Junne, S.; Neubauer, P. A Big World in Small Grain: A Review of Natural Milk Kefir Starters. Microorganisms 2020, 8, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Li, B. Chemical and microbiological characteristics of kefir grains and their fermented dairy products: A review. Cogent Food Agric. 2016, 2, 1272152. [Google Scholar] [CrossRef]
- Kesmen, Z.; Kacmaz, N. Determination of lactic microflora of kefir grains and kefir beverage by using culture-dependent and culture-independent methods. J. Food Sci. 2011, 76, M276–M283. [Google Scholar] [CrossRef]
- Kim, D.-H.; Jeong, D.; Kim, H.; Seo, K.-H. Modern perspectives on the health benefits of kefir in next generation sequencing era: Improvement of the host gut microbiota. Crit. Rev. Food Sci. Nutr. 2019, 59, 1782–1793. [Google Scholar] [CrossRef]
- Ganatsios, V.; Nigam, P.; Plessas, S.; Terpou, A. Kefir as a Functional Beverage Gaining Momentum towards Its Health Promoting Attributes. Beverages 2021, 7, 48. [Google Scholar] [CrossRef]
- Pinto, V.R.A.; Teixeira, C.G.; Lima, T.S.; de Almeida Prata, E.R.B.; Vidigal, M.C.T.R.; Martins, E.; Perrone, Í.T.; de Carvalho, A.F. Health beliefs towards kefir correlate with emotion and attitude: A study using an emoji scale in Brazil. Food Res. Int. 2020, 129, 108833. [Google Scholar] [CrossRef]
- Garofalo, C.; Ferrocino, I.; Reale, A.; Sabbatini, R.; Milanović, V.; Alkić-Subašić, M.; Boscaino, F.; Aquilanti, L.; Pasquini, M.; Trombetta, M.F.; et al. Study of kefir drinks produced by backslopping method using kefir grains from Bosnia and Herzegovina: Microbial dynamics and volatilome profile. Food Res. Int. 2020, 137, 109369. [Google Scholar] [CrossRef]
- Nejati, F.; Junne, S.; Kurreck, J.; Neubauer, P. Quantification of Major Bacteria and Yeast Species in Kefir Consortia by Multiplex TaqMan qPCR. Front. Microbiol. 2020, 11, 1291. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Tedersoo, L.; Lindahl, B. Fungal identification biases in microbiome projects. Environ. Microbiol. Rep. 2016, 8, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker gene amplicon sequences. Microbiome 2018, 6, e3208v2. [Google Scholar] [CrossRef]
- Kõljalg, U.; Larsson, K.-H.; Abarenkov, K.; Nilsson, R.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; et al. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005, 166, 1063–1068. [Google Scholar] [CrossRef]
- Brendel, R.; Schwolow, S.; Rohn, S.; Weller, P. Volatilomic Profiling of Citrus Juices by Dual-Detection HS-GC-MS-IMS and Machine Learning-An Alternative Authentication Approach. J. Agric. Food Chem. 2021, 69, 1727–1738. [Google Scholar] [CrossRef]
- Capitain, C.C.; Nejati, F.; Zischka, M.; Berzak, M.; Junne, S.; Neubauer, P.; Weller, P. Volatilomics-Based Microbiome Evaluation of Fermented Dairy by Prototypic Headspace-Gas Chromatography–High-Temperature Ion Mobility Spectrometry (HS-GC-HTIMS) and Non-Negative Matrix Factorization (NNMF). Metabolites 2022, 12, 299. [Google Scholar] [CrossRef]
- Walsh, A.M.; Crispie, F.; Kilcawley, K.; O’Sullivan, O.; O’Sullivan, M.G.; Claesson, M.J.; Cotter, P.D. Microbial Succession and Flavor Production in the Fermented Dairy Beverage Kefir. mSystems 2016, 1, e00052-16. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Sun, X.; Song, X.; Guo, M. Effects of kefir grains from different origins on proteolysis and volatile profile of goat milk kefir. Food Chem. 2021, 339, 128099. [Google Scholar] [CrossRef]
- Bourrie, B.C.T.; Ju, T.; Fouhse, J.M.; Forgie, A.J.; Sergi, C.; Cotter, P.D.; Willing, B.P. Kefir microbial composition is a deciding factor in the physiological impact of kefir in a mouse model of obesity. Br. J. Nutr. 2021, 125, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Bourrie, B.C.; Cotter, P.D.; Willing, B.P. Traditional kefir reduces weight gain and improves plasma and liver lipid profiles more successfully than a commercial equivalent in a mouse model of obesity. J. Funct. Foods 2018, 46, 29–37. [Google Scholar] [CrossRef]
- de Almeida Brasiel, P.G.; Dutra Medeiros, J.; Barbosa Ferreira Machado, A.; Schuchter Ferreira, M.; Gouveia Peluzio, M.d.C.; Potente Dutra Luquetti, S.C. Microbial community dynamics of fermented kefir beverages changes over time. Int. J. Dairy Technol. 2021, 74, 324–331. [Google Scholar] [CrossRef]
- Kazou, M.; Grafakou, A.; Tsakalidou, E.; Georgalaki, M. Zooming Into the Microbiota of Home-Made and Industrial Kefir Produced in Greece Using Classical Microbiological and Amplicon-Based Metagenomics Analyses. Front. Microbiol. 2021, 12, 621069. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Mclaren, M.R.; Willis, A.D.; Callahan, B.J. Consistent and correctable bias in metagenomic sequencing experiments. eLife 2019, 8, e46923. [Google Scholar] [CrossRef]
- Karstens, L.; Asquith, M.; Davin, S.; Fair, D.; Gregory, W.T.; Wolfe, A.J.; Braun, J.; McWeeney, S. Controlling for Contaminants in Low-Biomass 16S rRNA Gene Sequencing Experiments. mSystems 2019, 4, e00290-19. [Google Scholar] [CrossRef] [Green Version]
- Metras, B.N.; Holle, M.J.; Parker, V.J.; Miller, M.J.; Swanson, K.S. Assessment of commercial companion animal kefir products for label accuracy of microbial composition and quantity. J. Anim. Sci. 2020, 98, skaa301. [Google Scholar] [CrossRef]
- Biçer, Y.; Telli, A.E.; Sönmez, G.; Turkal, G.; Telli, N.; Uçar, G. Comparison of commercial and traditional kefir microbiota using metagenomic analysis. Int. J. Dairy Technol. 2021, 74, 528–534. [Google Scholar] [CrossRef]
- Yegin, Z.; Yurt, M.N.Z.; Tasbasi, B.B.; Acar, E.E.; Altunbas, O.; Ucak, S.; Ozalp, V.C.; Sudagidan, M. Determination of bacterial community structure of Turkish kefir beverages via metagenomic approach. Int. Dairy J. 2022, 129, 105337. [Google Scholar] [CrossRef]
- Jian, C.; Luukkonen, P.; Yki-Järvinen, H.; Salonen, A.; Korpela, K. Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS ONE 2020, 15, e0227285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Llatas, G.; Lagarda, M.J.; Romero, F.; Abellán, P.; Farré, R. A headspace solid-phase microextraction method of use in monitoring hexanal and pentane during storage: Application to liquid infant foods and powdered infant formulas. Food Chem. 2007, 101, 1078–1086. [Google Scholar] [CrossRef] [Green Version]
- Buttery, R.G.; Turnbaugh, J.G.; Ling, L.C. Contribution of volatiles to rice aroma. J. Agric. Food Chem. 1988, 36, 1006–1009. [Google Scholar] [CrossRef]
- Dertli, E.; Çon, A.H. Microbial diversity of traditional kefir grains and their role on kefir aroma. LWT—Food Sci. Technol. 2017, 85, 151–157. [Google Scholar] [CrossRef]
- Beshkova, D.M.; Simova, E.D.; Frengova, G.I.; Simov, Z.I.; Dimitrov, Z. Production of volatile aroma compounds by kefir starter cultures. Int. Dairy J. 2003, 13, 529–535. [Google Scholar] [CrossRef]
- Álvarez-Martín, P.; Flórez, A.B.; Hernández-Barranco, A.; Mayo, B. Interaction between dairy yeasts and lactic acid bacteria strains during milk fermentation. Food Control 2008, 19, 62–70. [Google Scholar] [CrossRef]
- Duran, F.E.; Özdemir, N.; Güneşer, O.; Kök-Taş, T. Prominent strains of kefir grains in the formation of volatile compound profile in milk medium; the role of Lactobacillus kefiranofaciens subsp. kefiranofaciens, Lentilactobacillus kefiri and Lentilactobacillus parakefiri. Eur. Food Res. Technol. 2022, 248, 975–989. [Google Scholar] [CrossRef]
- Beltrán-Barrientos, L.M.; Garcia, H.S.; Reyes-Díaz, R.; Estrada-Montoya, M.C.; Torres-Llanez, M.J.; Hernández-Mendoza, A.; González-Córdova, A.F.; Vallejo-Cordoba, B. Cooperation between Lactococcus lactis NRRL B-50571 and NRRL B-50572 for Aroma Formation in Fermented Milk. Foods 2019, 8, 645. [Google Scholar] [CrossRef] [Green Version]
- Gadaga, T. Growth characteristics of Candida kefyr and two strains of Lactococcus lactis subsp. lactis isolated from Zimbabwean naturally fermented milk. Int. J. Food Microbiol. 2001, 70, 11–19. [Google Scholar]
- Zajšek, K.; Goršek, A.; Kolar, M. Cultivating conditions effects on kefiran production by the mixed culture of lactic acid bacteria imbedded within kefir grains. Food Chem. 2013, 139, 970–977. [Google Scholar] [CrossRef]
- Wu, Q.; Cheung, C.K.; Shah, N.P. Towards galactose accumulation in dairy foods fermented by conventional starter cultures: Challenges and strategies. Trends Food Sci. Technol. 2015, 41, 24–36. [Google Scholar] [CrossRef]
- Grønnevik, H.; Falstad, M.; Narvhus, J.A. Microbiological and chemical properties of Norwegian kefir during storage. Int. Dairy J. 2011, 21, 601–606. [Google Scholar] [CrossRef]
- Delgado-Fernández, P.; Corzo, N.; Lizasoain, S.; Olano, A.; Moreno, F.J. Fermentative properties of starter culture during manufacture of kefir with new prebiotics derived from lactulose. Int. Dairy J. 2019, 93, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Verhue, W.M.; Tjan, F.S.B. Study of the Citrate Metabolism of Lactococcus lactis subsp. lactis Biovar Diacetylactis by Means of 13C Nuclear Magnetic Resonance. Appl. Environ. Microbiol. 1991, 57, 3371–3377. [Google Scholar]
- Comitini, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Vancov, T.; Keen, B. Amplification of soil fungal community DNA using the ITS86F and ITS4 primers. FEMS Microbiol. Lett. 2009, 296, 91–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Cheng, Y.; Wang, Y.; Zhang, G.; Gao, R.; Xiang, C.; Feng, J.; Lou, D.; Liu, Y. Investigation of the fungal community structures of imported wheat using high-throughput sequencing technology. PLoS ONE 2017, 12, e0171894. [Google Scholar] [CrossRef]
- Abarenkov, K.; Henrik Nilsson, R.; Larsson, K.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE database for molecular identification of fungi--recent updates and future perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef]

| Sample | Fermen-Tation Time (h) | Lactose (g L−1) | Glucose (mg L−1) | Galactose (mg L−1) | Lactate (g L−1) | Acetate (mg L−1) | Glycerol (mg L−1) |
|---|---|---|---|---|---|---|---|
| PN1 | 23 ± 1 | 33.6 ± 5.7 ab | 133.3 ± 82.6 ab | 149.3 ± 58.0 bc | 1.7 ± 1.1 c | 328.4 ± 60.5 c | 297.7±58.4 b |
| 47 ± 1 | 9.2 ± 5.4 c | 25.1 ± 3.6 a | 23.2 ± 8.7 d | 3.2 ± 0.1 b | 778.8 ± 68.8 a | 682.2±95.7 c | |
| PN2 | 23 ± 1 | 37.2 ± 2.0 a | 197.2 ± 52.8 b | 266.3 ± 35.5 b | 1.7 ± 0.3 c | 459.5 ± 29.8 b | 232.0±30.0 b |
| 47 ± 1 | 15.3 ± 3.4 c | 29.6 ± 0.5 a | 21.8 ± 10.2 d | 3.9 ± 0.4 b | 944.5 ± 15.2 a | 538.1±76.1 c | |
| PN3 | 23 ± 1 | 37.6 ± 2.8 a | 176.5 ± 74.0 b | 368.8 ± 112.5 b | 1.2 ± 0.2 c | 300.2 ± 21.8 c | 253.9±20.1 b |
| 47 ± 1 | 14.1 ± 1.7 c | 27.0 ± 4.2 a | 12.6 ± 1.7 d | 3.3 ± 0.6 b | 846.9 ± 58.2 a | 639.4±37.2 c | |
| FN | 23 ± 1 | 40.3 ± 2.0 a | 203.5 ± 76.9 b | 228.7 ± 61.4 b | 1.3 ± 0.1 c | 372.8 ± 82.3 bc | 191.0±56.9 b |
| 47 ± 1 | 37.6 ± 5.3 a | 52.2 ± 36.0 a | 33.2 ± 21.2 d | 3.3 ± 0.4 b | 720.7 ± 101.3 a | 186.4±45.6 b | |
| LS | 23 ± 1 | 37.4 ± 2.4 a | 120.0 ± 54.2 ab | 61.0 ± 32.5 cd | 1.7 ± 0.3 c | 326.5 ± 108.6 c | 22.2±6.0 a |
| 47 ± 1 | 30.8 ± 0.5 b | 26.0 ± 1.6 a | 11.7 ± 1.1 d | 4.8 ± 0.1 b | 613.7 ± 68.9 a | 19.7±4.7 a | |
| CK1 | 44.7 ± 7.4 | 33.2 ± 0.8 | 664.1 ± 12.8 | 7.0 ± 0.7 | 745.6 ± 44.0 | 28.0 ± 1.6 | |
| CK2 | 42.0 ± 2.7 | 23.0 ± 0.9 | 352.8 ± 44.2 | 7.1 ± 1.2 | 584.4 ± 105.6 | 23.3 ± 1.2 | |
| CK3 | 39.8 ± 3.0 | 79.5 ± 15.5 | 799.4 ± 19.5 | 7.6 ± 0.8 | 810.1 ± 84.6 | 25.2 ± 5.9 | |
| CK-average | 42.2 ± 4.3 a | 45.1 ± 5.9 a | 605.7 ± 25.5 a | 7.3 ± 0.9 a | 713.3 ± 84.2 a | 25.5 ± 5.9 a | |
| Milk * | 47.2 ± 0.3 | 98.2 ± 2.9 | <TEST RNG # | <TEST RNG | <TEST RNG | <TEST RNG | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nejati, F.; Capitain, C.C.; Krause, J.L.; Kang, G.-U.; Riedel, R.; Chang, H.-D.; Kurreck, J.; Junne, S.; Weller, P.; Neubauer, P. Traditional Grain-Based vs. Commercial Milk Kefirs, How Different Are They? Appl. Sci. 2022, 12, 3838. https://doi.org/10.3390/app12083838
Nejati F, Capitain CC, Krause JL, Kang G-U, Riedel R, Chang H-D, Kurreck J, Junne S, Weller P, Neubauer P. Traditional Grain-Based vs. Commercial Milk Kefirs, How Different Are They? Applied Sciences. 2022; 12(8):3838. https://doi.org/10.3390/app12083838
Chicago/Turabian StyleNejati, Fatemeh, Charlotte C. Capitain, Jannike Lea Krause, Gi-Ung Kang, René Riedel, Hyun-Dong Chang, Jens Kurreck, Stefan Junne, Philipp Weller, and Peter Neubauer. 2022. "Traditional Grain-Based vs. Commercial Milk Kefirs, How Different Are They?" Applied Sciences 12, no. 8: 3838. https://doi.org/10.3390/app12083838
APA StyleNejati, F., Capitain, C. C., Krause, J. L., Kang, G.-U., Riedel, R., Chang, H.-D., Kurreck, J., Junne, S., Weller, P., & Neubauer, P. (2022). Traditional Grain-Based vs. Commercial Milk Kefirs, How Different Are They? Applied Sciences, 12(8), 3838. https://doi.org/10.3390/app12083838








