Abstract
Data science techniques have increasing importance in medical data analysis, including detecting and predicting the probability of contracting a disease. A large amount of medical data is generated close to the patients in the form of a stream, such as data from sensors and medical devices. The distribution of these kinds of data may change from time to time; adaptive Machine Learning (ML) consists of a continuous training process responding to the distribution’s change. Adaptive ML models require high computational resources, which can be provided by cloud computing. In this work, a classification model is proposed to utilize the advantages of cloud computing, edge computing, and adaptive ML. It aims to precisely and efficiently classify EEG signal data, thereby detecting the seizures of epileptic patients using Adaptive Random Forest (ARF). It includes a global adaptive classifier in the cloud master node and a local light classifier in each edge node. In this model, the delayed labels consider missing values, and the Model-based imputation method is used to handle them in the global classifier. Implementing the proposed model on a real huge dataset (CHB-MIT) showed an accurate performance. It has a 0.998 True Negative Rate, a 0.785 True Positive Rate, and a 0.0017 False Positive Rate, which overcomes much of the research in the state-of-the-art.
1. Introduction
Epilepsy is one of the prevalent chronic diseases that affect the human central nervous system of all ages, and it is widespread around the world [1]. In 2016, 45.9 million people had epilepsy, and there were more active cases with increasing age [2]. The number of deaths and disabilities caused by this disease is still significant; the epilepsy-related death rate in the U.S increased from 0.3 per 100,000 population in 1999 to 0.5 in 2019. In India, there were 658 deaths related to epilepsy per week in 2016, while there were 44 in Germany in the same year [2].
Nowadays, data-mining techniques are becoming increasingly important in medical data analysis. Large amounts of these data are generated in the form of a stream, such as data from sensors and medical devices. Furthermore, many personal devices of daily use, such as the smartphone and smartwatch, contain sensors for the acceleration, location, and biometric data of their bearers, and these sensors generate large and continuous amounts of data [3]. Data-mining techniques are commonly used to classify patient data, thereby detecting or predicting values often associated with diseases. The performance of the classification model based on a single classifier is insufficient in dealing with sensitive tasks such as monitoring the elderly with chronic diseases and classifying critical medical conditions. Ensemble models that build multiple classifiers often produce more reliable performance but require high computational resources.
Adaptive Machine Learning (ML) consists of a continuous training process for the classification model to respond to the change in data distribution. The delay in obtaining the actual values of the class label after the classification decision restricts the adaptation process. For this reason, the classification should have the ability to overcome this delay by considering them as missing values, for example, after waiting for a specified period of time. Imputation refers to predicting the most reasonable values instead of the missing values. The prediction is performed using the available information, which has known values [4]. The model-based imputation utilizes statistical methods for building the prediction model for the missing values based on the data distribution. The task of building an ensemble and adaptive classifier required centric and high computational resources to handle the ongoing patient data stream, and these resources can be provided by cloud computing services [5].
The cloud provides storage and computing capabilities that are highly expandable. Despite that, it is not practical to store and process all the data there, especially in applications that require a real-time or near-real-time response. Edge computing models have been developed to handle this type of application by adding a new layer of storage and computing resources that are closer to the source of data (end-user device) [6]. In addition to minimizing the computation latency, utilizing edge computing can provide more reliability against failures of the network, minimizing the network traffic and increasing the capability of the cloud by shifting the computing to the edge infrastructure. On the other hand, edge devices have a limitation to utilize the adaptive ensemble classifiers due to the limited resources.
In this work, we focus on processing and analyzing Electroencephalography (EEG) data to classify the brain activity signals of epileptic patients in the cloud environment. This goal can be achieved by building an adaptive classification model that accurately detects an epileptic seizure, thereby warning the patient’s relatives and the medical staff. It deals with EEG data in their original form as a stream (not a batch file) when generating it without the need to store and retrieve it.
The proposed model is characterized by utilizing the adaptive mechanism of online bagging for handling the concept drift and the fast response of VFDT as a base learner in the ensemble model. It utilizes Edge computing by combining a light, fast, and local classifier that will be used only for the inference process with an adaptive and global classifier in the master cloud node that performs the training process and sends the latest version of the classifier to all edge nodes. The model reduces the effect of delayed labels by considering them as missing values, and the model-based imputation method is used to handle them in the global classifier.
2. Literature Review
Most of the current research on stream data mining focus only on improving the accuracy of adaptive classification regardless of its efficiency. In addition, the research in Federate learning focuses on applying the training process on both local edge devices and the central cloud server. In this section, the theoretical background for the methods used in this work will be explained in addition to many related works.
2.1. EEG Signals
An electroencephalogram (EEG) is a graphical record of the electrical activity of the brain recorded from the scalp. This method can be performed in two different ways: (1) Record invasive EEG directly from the brain through surgically implanted electrodes; (2) Record non-invasive EEG from the surface of the human brain [7,8]. A sufficient amount of information about brain activity can be extracted from the EEG signal. Feature extraction involves transforming the original signal into a related data structure by removing noise and highlighting important data, called a feature vector [9].
The features extracted from biological signals can be divided into three types [10]: (1) Time-domain features (TDF) are calculated based on the signal amplitude, and the result value gives a measure of the waveform amplitude, frequency, and duration within some limited parameters; (2) Frequency domain feature (FDF), based on the estimated power spectral density (PSD) of the signal; and (3) Time-frequency domain characteristics (T-FDF), using time-frequency representation to explain the energy that can locate the signal in time and frequency [11,12].
The information extracted from EEG signals can be used for the diagnosis of brain diseases, such as disorders, coma, and epilepsy [8,13]. The Fourier transform (FT) is an effective technique for signal processing used in feature extraction. The Fourier transform (FT) of the function is [14,15]:
Discrete Fourier Transform (DFT) maps a sequence into the frequency domain. It is used in the case of discrete and periodic signals [15]. Let the signal be q for and q = q for all m and j. The discrete Fourier transform of q is [14]:
2.2. Data Mining Classification Techniques
Data mining is defined as “extracting or mining knowledge from large amounts of data” [16]. In data analysis, working with data commonly includes descriptive and predictive tasks. A descriptive analysis could visualize relationships in data in an understandable form for decision makers. In the prediction task, the model is used to predict the target attribute for the given data [16,17,18]. The ensemble methods have been developed to enhance the performance of the single classifier [19]. The main idea of an ensemble model is to combine a set of single models, and each one tries to solve the same original task [18,20,21]. Random forest is one of the most popular ensemble techniques that are widely used for medical data mining [18].
2.3. Stream Data Classification
There are many constraints that must be taken into account when mining streaming data: (1) the impossibility of storing data samples due to their infinite arrival; (2) real-time processing is required due to the rapid arrival of data samples; and (3) the possibility of changing items’ distributions, which may require altering the current structure of the classification model [22,23].
Concept drift means that the data being collected may change from time to time, each time based on a certain minimum persistence. There are two important issues related to change: the cause of the change and the speed of the change [24]. ADWIN (Adaptive Sliding Window) is an estimation technique designed to detect changes in a data stream based on a sliding window with an adaptive size. In this technique, as long as the average value in the window does not change, the length of the window will not be updated [23,24].
2.4. Very Fast Decision Tree
A Very Fast Decision Tree (VFDT) is a variation of the typical decision tree designed for streaming data. The learning of these techniques depends on replacing the leaves of the tree with decision nodes. Each terminal node (leaf) in the tree stores enough information statistics about the feature value, and the heuristic function uses this information to perform the split test. After reaching the new data instance, it starts to transfer from the root until it reaches a specific leaf node. At this point, the statistical information is evaluated, and a new decision node can be created based on the evaluation [23,24]. VFDT relies on the concept of the Hoeffding bound [24,25,26], in which r is a random variable and its real value range is R, where the range R is in information gain, and is the number of classes. Consider there are n independent observations from the random variable r and the computed mean for n is . The Hoeffding bound points out that, with probability , the actual mean for r is at least , where:
where is a value that is specified by the user.
2.5. Adaptive Random Forest
Ensemble modeling aims to build a strong cumulative classifier from many weak classifiers. Adaptive random forest (ARF) is a variant of the typical random forest algorithm used for data stream mining tasks. The main idea is to use VFDT with the ability to adapt to changes in distribution as the base classifier of the bagging integration method [22,23,27]. In order to detect changes in the data stream, ADWIN is used in this technique, and it utilizes the Online Bagging as a resampling method [22,23].
Table 1 shows the equivalent of four popular batch classification techniques for classifying the data stream.

Table 1.
Equivalent of four popular classification techniques for classifying the data stream.
2.6. Cloud Computing
Due to its many features, such as mobility, scalability, and on-demand resources, cloud computing has become one of the important topics in information technology [35]. According to [36], cloud computing has a significant role in applications of the Internet of Things for healthcare systems. It can provide the required resources such as servers, networks, operating systems, and software applications. From the service coverage point of view, cloud computing can be categorized into three types: public, private, and hybrid. According to the kind of service, cloud computing can be divided into three categories [37]:
- Infrastructure as a Service (IaaS), which focuses on proving the required computing resources such as the processor, storage, and the network;
- Platform as a Service (PaaS), which provides the deployment in the cloud infrastructure using provided tools, software libraries, and programming languages;
- Software as a Service (SaaS), which runs the applications of the clients within the cloud infrastructure.
2.7. Related Works
In [38], Chiang utilized the bivariate pattern classification method for developing an online retraining technique that can gradually scale the training set. Their proposed model reduced false alarms using a post-processing scheme. Three datasets were used to evaluate the model, including the ECoG Freiburg database, the CHB-MIT EEG database, and the national Taiwan university hospital database. The results showed an improvement in sensitivity by 29.0 in the ECoG database and 17.4 in the CHB-MIT EEG database compared with the offline training method.
Khan [39] developed an algorithm for recognizing epileptic seizures automatically. They utilized discrete wavelet transform (DWT) for feature extraction and decomposing EEG signals. They compared the performance of normalized coefficient of variation (NCOV), which was computed on the coefficients extracted from signals range of 0–32 Hz, with a typical coefficient of variation (COV), and the results show the advantage of NCOV according to the evaluation on five subjects from CHB-MIT EEG dataset using Linear discriminant analysis (LDA).
In [40], Ahammad utilized a linear classifier with EEG-extracted features using wavelets and some other statistical features (mean, minimum, maximum, entropy, and standard deviation). EEG datasets from Bonn University and CHB-MIT are used in their implementation. The accuracy of seizure onset detection was 84.2 on the CHB-MIT dataset. In [41], Zhang reduced the hardware complexity required for predicting epileptic seizures from EEG signals. Overall, 44 extracted features were ranked and selected, then processed using a second-order Kalman filter. A Support Vector Machine (SVM) classifier was used in their model, and the implementation included two datasets: the Freiburg dataset and the CHB-MIT dataset. The results showed a sensitivity of 100 for the Freiburg dataset and of 98.68 for the CHB-MIT dataset.
A prediction method for patient-specific seizures was presented by [42], which applied Convolutional Neural Networks (CNNs) to the EEG dataset. The features were extracted by using Short-Time Fourier Transform (STFT); then, a standardization process was used to prevent any possible influence between high and low frequencies. CNN was used in their model in both features selection and classification steps. The best result of their proposed model was an 82.3 sensitivity.
Zhou [43] compared the performances of frequency and time domain for the detection of epilepsy in two datasets: the Freiburg dataset and the CHB-MIT EEG dataset. The comparison included two binary classification tasks and one three-class task. The best result of the frequency domain experiments was an accuracy of 96.7% with the Freiburg dataset and of 97.5% with the CHB-MIT EEG dataset. It was 91.1% accurate in the Freiburg dataset and 62.3% accurate in the CHB-MIT EEG dataset with time-domain signals experiments. A cloud-based healthcare system was proposed by [44]; it utilized Parallel Particle Swarm Optimization (PPSO) to improve the selection of Virtual Machines (VMs). The implementation of their model included using linear regression and a neural network model for chronic kidney disease (CKD) diagnosis and prediction. Their results showed an improvement in the time of data retrieval in addition to a reduction in the total execution time.
Common Spatial Patterns (CSP) and Convolutional Neural Networks (CNN) are utilized by [45] for seizure prediction. Wavelet packet decomposition and CSP are used to extract the number of selected features from generated artificial EEG signals. Their implantation was evaluated using EEG data from 23 patients in CHB-MIT dataset. It achieved a sensitivity of 92.2% with the cross-validation method. Seo [46] developed a classification model for recognizing the emotions of Alzheimer’s disease (AD) patients. EEG data from 30 Korean patients were used to compare the performance of three machine learning techniques: multilayer perceptron, support vector machine, and recurrent neural network. The best result was gained by using the multilayer perceptron, in which its average accuracy was 70.97%.
Ayodele [47] combined data from two datasets using the domain generalization method. Transfer component analysis was utilized for extracted data from CHB-MIT and TUSZ EEG datasets based on reproducing the kernel Hilbert space method. The transformation of 3D electrode coordinates into a 2D space is applied, then Azimuthal Equidistant Projection is used, followed by the Clough–Tocher technique. A recurrent CNN was trained using the generated vectors with 128 units, and the testing result was a sensitivity of 74.5%.
Ref. [48] proposed a fall detection model for the elderly by utilizing the accelerometer and gyroscope in personal smart devices. They used machine learning in the local edge device and in the cloud to classify fall status from the collected sensors data. Their model reduced the amount of transmitted data and thereby the network traffic. A selective model for image classification using Deep Neural Network DNN is proposed by [49]. They presented a federate aggregation approach by evaluating the local DNN models and sent the best to the central cloud server. The implantation of their method showed an improvement in terms of efficiency and accuracy compared with the federated averaging method.
The applications of graph theory in neuroeconomics using EEG data were reviewed systematically by [50]. They proposed a method for building an EEG data-based brain network by utilizing sensor and source methods. Their results showed that the majority of 57 research articles aimed at cognitive tasks. In [51], a review of 38 articles that utilized the two-photon imaging (2PI) technique for epilepsy research was introduced. They concluded that 2PI is a suitable imaging technique for the research of epilepsy, and it can extend the incoming research on human brains. In [52], a hybrid classification model was proposed for predicting epileptic seizures. Their model used DWT to obtain time-frequency domain data from the original EEG signals. The training process in their model is performed using both Long Short-Term Memory and a Dense Convolutional Network, and the evaluation used the CHB-MIT dataset.
3. Methodology
Generally, the adaptive classifier has two states: classifying a new arrival data stream according to the current model and adapting the model to the change in data distribution. The first state needs low computational resources, while the second state may require the model’s reconstruction and therefore more resources. Considering that there is no control over the time and order of the arrival of the stream data and the change in its distribution, the classification process requires the provision of high computational resources all the time. The utilization of the cloud as a computational platform represents an efficient solution for adapting the classifier as follows:
- Scalability of cloud resources can provide a fast training process even when an unexpectedly large amount of data arrives from patients. The resource scheduling in the cloud environment provides a reduction in the execution time by choosing the optimal virtual machines.
- In the cloud, an on-demand service with auto-scale resources can reduce the cost of the classification task by utilizing the high computational resources only in the classifier adapting process.
The goal of the proposed model is to precisely classify the EEG signal for detecting epileptic seizures. The proposed model, entitled Adaptive Random Forest in the cloud (ARFC), can be used to protect the patient from sudden epileptic seizures, which can cause injury or more critical situations. It also provides more time for the medical staff to take the appropriate medical procedure. The model contains two major stages: (1) the stream data preprocessing stage, which is performed on the patient’s side, and (2) the building of an adaptive classifier stage in the cloud environment. Figure 1 illustrates all steps of the proposed model. The paradigm can utilize the advantages of both Edge computing and adaptive ML. It includes two classifiers: (1) a global classifier in the cloud master node, which receives the data stream from all edge nodes for adaptive training processes, and (2) a local classifier in each edge node that contains the latest version from the global classifier. The local classifier is used to classify the local node’s stream data rows then send them to the master node. The global classifier applies machine learning imputation for retrieving the missing label values. It adapts itself with the change in the data distribution from all nodes, and finally, updating the local classifier with the newest version will be performed.
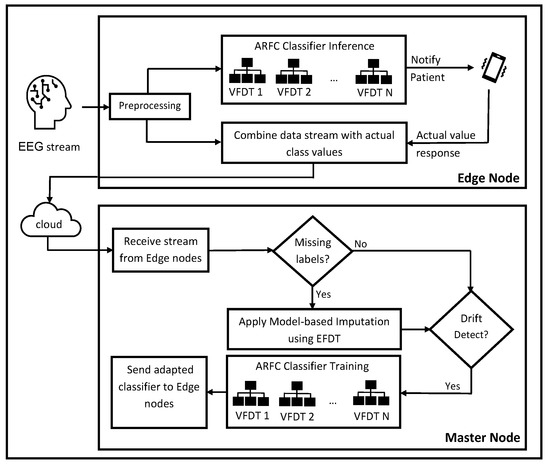
Figure 1.
The diagram of ARFC for classifying EEG data stream in the Cloud.
3.1. Stream Data Preprocessing
The signals from the patient’s brain are read using EEG sensors, which are placed in particular places in the cortex, as explained in Section 2.1. These sensors produce a large amount of data because there are multiple channels, so the preprocessing stage aims to prepare the EEG data in an appropriate form for building the classification model. In the first step, the important features are extracted from the EEG signal using Fast Fourier Transform (FFT). FFT is an implementation of Discrete Fourier Transform, which is explained in Section 2.1. The importance of this step is based on the fact that building a classification model depends on choosing the best feature for the data splitting process during building decision trees. For this reason, the quality of extracted features and their correlation to the target of the classification task has significant importance in preprocessing operations.
While the previous step is trying to reduce the complexity of the data steam horizontally by extracting the most valuable features, the aggregation aims to make a vertical reduction in the data stream by minimizing the number of rows. This step includes the aggregation of a set of patient rows that are generated during the same time moment and with the same target class value. The preprocessing ends with sending the preprocessed EEG data stream to the cloud where the adaptive classifier is built. Algorithm 1 summarizes the steps of preprocessing stage.
| Algorithm 1 EEG Stream data preprocessing—Edge Node. |
|
The classification in this part of the proposed model can be considered static, whose primary goal is a fast response due to the rapid generation and large quantity of EEG signals. If the signal is classified as an epileptic seizure, the patient, the contact person, and the medical authority responsible for his care are notified. The patient’s actual status is acquired after the notification, and then it combines with the feature data of that instance before sending them to the master node across the cloud. The patient or the medical staff may dismiss the notification, thinking it is a false warning or due to the lack of information to confirm this at present. The feedback for the actual value of the patient’s status will wait for a specific period. If there is no response after the time expires, the class label is considered a missing value, and only the features values are sent to the master node.
3.2. Building of Adaptive Classification Model
The master node is the centerpiece of the proposed model, receiving instances sent from all the edge nodes for adapting the classifier with these instances. First, the received data are tested for missing values, as these reduce the accuracy of the classification process. The fact is that the data received for different patients make it impractical to use arithmetic methods to impute the value, such as the average. In addition, the EEG signals often have unbalanced distribution in the target values, in which the number of normal signal values greatly exceeds the number of the seizure values, and this makes choosing the most frequent value to missing value imputation also impractical. Therefore, the process of imputation in the proposed model depends on the employment of machine learning to build a quick and simple classifier to infer the missing value through training on the rest of the known values.
The imputation is performed by using an extreme fast decision tree classifier, which was proposed by [53]. It is an efficient modified version of VFDT in which the data will be split when there is a good splitting; then, if a better split is detected, the old one is replaced. The result will be tested for concept drift; if there is a drift in the distribution of the data, the new training operation starts. The ensemble classifier is built using an Adaptive Random forest.
At this stage, the process begins with reading a specified number of data rows sent from all patients’ devices, and this number represents the default size of the ADWIN window. The default window size must be carefully chosen because if the number is too small, the changes in the model will be frequent. In addition, if the window size is too large, the response of the model to the change in data distribution will be limited. The next step is using the online Bagging [32] of K-based classifiers, and each base classifier is constructed using VFDT, as proposed in [25]. In the beginning, this stage sets the size of the ARF model and then the number of subset ensembles generated by re-sampling the data samples from the previous stage. The number of subsets is equal to the number of base classifiers. Each subset is used to train a single VFDT classifier. Finally, each data element is classified by voting among all base classifiers.
3.2.1. Initializing the Size of Ensemble Model
Although Online Bagging is used for ensemble model construction, the size of the model, that is, the number of base classifiers is a user-defined parameter whose value is required before the model generation operation starts. The importance of this parameter comes from the fact that it represents the stopping condition of each learning step, which is related to the complexity of the required computing resources. It is recommended that the value of this parameter in the model tends to be in the low range value (about ten base classifiers). With the continuous arrival of stream elements, mining stream classifiers are expected to have a fast response in the learning and classification process.
In online bagging, any new data sample is selected according to the Poisson(1) distribution. The classification decision of the ensemble bagging model is based on voting for all K base classifiers with the same weight. It assigns an initial weight w = 1 to each new data example and then passes it to the first weak learner. If this data example is misclassified, its weight will increase before passing it to the next weak learner. The base learner in the proposed model is a VFDT classifier, and the ensemble size used is 10 learners. The ARF classifier is presented in [54]; it uses the resampling method of Online Bagging with a different adaptive method. The parameters of ARF can be categorized into two groups, the first one related to the bagging model and the second related to the base learner. The first group includes:
- The ensemble size, which is the total number of base learners in the model;
- Maximum number of features that can be tested in each split;
- Warning threshold, which detects when the background tree will be built as a response for distribution change;
- Drift threshold, which detects when the current tree will be replaced by the background tree.
The second group includes the parameters for building VFDT, which are:
- The tree size that can be controlled by its depth or/and the maximum number of terminal nodes;
- Minimum number of data instances in a terminal node to make the splitting;
- The type of criterion that should be used for splitting.
All the seven parameters in both groups are user-defined, and they should be chosen carefully. The reason for that is because their values have a direct effect on the model performance in terms of efficiency and accuracy.
3.2.2. VFDT Classifier Building
It includes two types of nodes: decision nodes and terminal nodes; each terminal node in the tree stores enough statistical information about the characteristic value. The heuristic function uses this information to perform split testing. After reaching the new data instance, it starts to transfer from the root node (root) until it reaches a specific leaf node. At this point, if the class value of the new instance is not available (i.e., unknown value) or has not been seen before (i.e., a new class in multi-class classification tasks), the instance is classified according to the majority of the current terminal nodes. Otherwise, the statistical information is evaluated, and the evaluation of a new decision node can be created according to the following conditions.
The evaluation includes computing the gain for all features in all possible split points. For each split point, the impurity of class distribution of the current node and the possible child nodes will be computed using Entropy, according to the following equation, for node a:
where c refers to the number of classes. The difference between the entropy of the current node and the average value of the entropy of its possible child nodes after splitting represents the gain of the splitting operation. A split that produces a more uniform class distribution, that is, a higher gain, is desirable. If the difference between the highest two features is greater than the Hoeffding boundary value, which is computed according to Equation (3), the current terminal node will be replaced with a decision node based on the highest feature. At the same time, for each split branch of this new node, a new empty terminal node will be added.
The final decision of classifying the data instance will be made using majority voting among all the decision trees in the forest. If the instance is classified as a seizure, an immediate alert will be sent to the patient’s device and also to the responsible medical staff. Algorithm 2 summarizes the steps of the building classifier stage.
| Algorithm 2 Adaptive Classification Model-Master Node. |
|
4. Implementation and Experimental Results
The implementation included using CHB-MIT Scalp EEG database [55,56], which contains EEG recordings with intractable seizures gathered at Children’s Hospital Boston. After the cessation of anti-seizure medication, subjects were observed for up to several days to define their seizures and determine their suitability for surgical intervention. These recordings were made using the International 10–20 method of EEG electrode physical locations and names, as shown in Figure 2. In this method, the electrode’s name points to its placement on the brain; C refers to the center of the head, F: to the front, T is temporary, P is a rear part, and O means occipital. In addition, the name also contains a number that refers to the side of the cerebral; for the right side, even numbers are used, and odd numbers are used on the left [57,58].
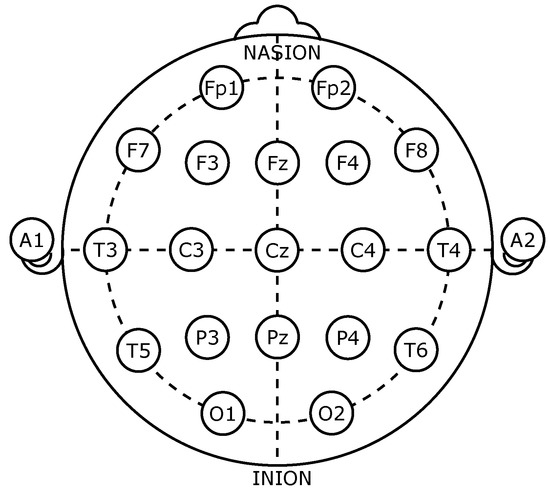
Figure 2.
The placement of EEG electrode based on the 10–20 International system.
EEG signals of the CHB-MIT dataset were gathered from 24 cases (23 human subjects). Each case contains 9–42 continuous data files from a single subject. The sampling rate for the signals was 256 samples per second with 16-bit resolution. In this work, 20 files from 10 patients were chosen randomly to generate a stream containing 110,644 s and included normal and seizures signals. The total number of extracted rows was 28,324,864 rows with 23 columns and the class label.
4.1. The Platform of the Data Stream Classification
In this work, four main tools were used: scikit-multiflow, Sklearn, Weka, and RapidMiner. Scikit-multiflow is an open source package that is used in Python programming language for data stream mining tasks, including classification and clustering. It was developed with ECOSYSTEM project by the University of Waikato, which includes a Massive Online Analysis (MOA) platform for stream mining and the Waikato Environment for Knowledge Analysis (Weka) for batch mining. Scikit-multiflow provides many popular mining techniques, flow generators, and concept drift detection techniques. In our implementation, it has been used to perform the data streaming and classification techniques.
Sklearn is a free Python library for machine learning tasks. It contains many classification techniques, such as random forest and boosting. In this work, Sklearn was used for feature extraction from EEG data and also for data row aggregation. RapidMiner is another software that is used in this implementation for calculating the correlation among the features of the CHB-MIT dataset. It is developed for data science, including deep learning, machine learning, and text mining. Amazon AWS cloud service has been utilized in this implementation as an on-demand computational resource. The EC2 virtual machine VM was used as a master node which has 8 CPU cores, 32 GB memory, and a 300 GB SSD hard drive. Ubuntu Linux operating system, Python, scikit-multiflow, and Sklearn have been installed in the VM to perform the stream classification. On the other hand, a Raspberry Pi3 Model A+ unit is used as an edge node with a 1.4 GHz quad-core processor, 512 MB memory, and Raspberry Pi operating system with Python and the same packages in the master node. Pickle python object has been used for transmitting the classifier object from the master node to the edge node byte by byte across the network.
4.2. Dataset Analysis
The CHB-MIT EEG dataset has many interesting points that represent challenges to the typical data mining techniques as follows:
- The frequency of classes values is an imbalance, and the dataset contains only two values for the target class, zero for the normal EEG signal and one for the EEG signal. The total number for the normal class instances in the extracted rows was 109,141, and that represents 99.64% of the whole extracted data. This majority leads most classification techniques to incorrect high accuracy; for example, if the classifier predicts all extracted rows as a normal EEG signal, the accuracy will be 99.64%.
- Very weak correlation between the features on the one hand and the target class on the other hand. In Table 2 and Figure 3, we can notice that all correlation values (in columns correlation B) are very close to zero, which makes the classification task more difficult.
 Table 2. The Correlation between features and target class of CHB-MIT dataset.
Table 2. The Correlation between features and target class of CHB-MIT dataset.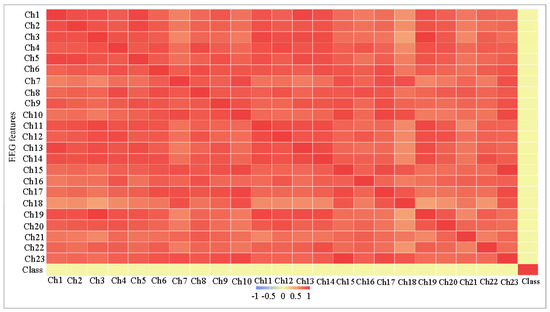 Figure 3. The Correlation matrix among all features of CHB-MIT dataset.
Figure 3. The Correlation matrix among all features of CHB-MIT dataset. - High correlation between most of the features, as can be seen in Table 2 and Figure 3, the average correlation between each feature and other features is shown in Table 2 in columns (correlation A), and most values are more than 0.8. The features are not completely matched (i.e., the correlation is not 1) to be considered as a frequent feature and will be removed.
4.3. Stream Mining Configuration
The configuration of the EEG stream included the aggregation of each 256 rows to one single row because all rows related to a single second lead to either the normal or seizure class. This will reduce the required computational resources. Then, the 10 files were merged, in which the model will receive 10 rows from 10 patients every second. The parameters in scikit-multiflow were set as follows: sample size-frequency, which refers to the default window size, was set to 100; ensemble size was 11 base learners; info gain was chosen as the split criterion for building the random tree classifier; and split confidence was set to 0.01.
4.4. The Experimental Results
The model performance evaluation depended on the Test then Train method because the data are a stream, not a batch file. In this method, each instance that arrives in the current sliding window will be considered a test instance and used to evaluate the model. Then, the instance will be used to train the model. The mean accuracy will be calculated for every 100 instances depending on testing performance. So, in this method, all data elements will be used for testing and training to avoid bias such as the cross-validation in batch data mining. Applying the proposed model using the CHB-MIT dataset led to some significant results that confirm its advantage. Considering the normal signal is a negative class and the seizure is a positive class, Table 3 shows that the proposed model had the best True Positive Rate TPR compared to Random Forest, J48 Decision Tree, Reduced Error Pruning Tree (REPTree), AdaBoost, and Bayes Network classifier. In addition to the high value of True Negative Rate TNR, ARFC also has a low False Positive Rate, which is most important.

Table 3.
Comparison of the accuracy between ARFC and five batch classifiers.
The implementation detected 37 changes in the extracted EEG data stream. The size of the ADWIN window has been changed accordingly; as we can see in Figure 4, the average window size was 1251, while the maximum size was 10,240. The large size of ADWIN points to more stability in the distribution of data stream and vice versa. Table 4 showed that the proposed model has the best performance in comparison to five stream classifiers: Rule-based, VFDT, Random VFDT, OzaBagging, and Adaptive-NN. The comparison is included in Table 4 using five metrics: accuracy, Kappa, precision, Recall, and F1 score.
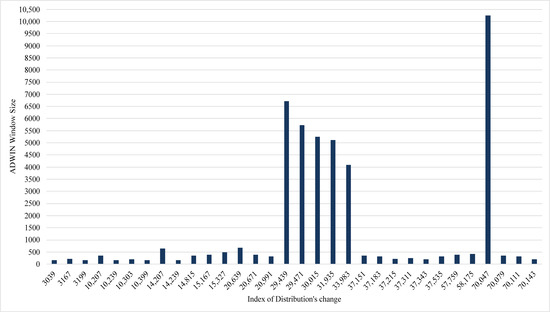
Figure 4.
ADWIN window size of ARFC when a change in data distribution occurs.

Table 4.
Comparison of the performance between ARFC and three stream classifiers using five metrics.
Tracking of the proposed model accuracy during the classification of the EEG data stream is illustrated in Figure 5. The stability of its accuracy can be noticed; the accuracy of seizures also tends to increase. Figure 5 illustrates that the obvious change in ADWIN’s size had an effect on the True Positive Rate (i.e., seizures class), as we can see in the periods (20,991–37,151) and (58,175–70,079). In both Figure 4 and Figure 5, the index of the distribution’s change refers to the position of the instance where the change in distribution was declared by ADWIN.
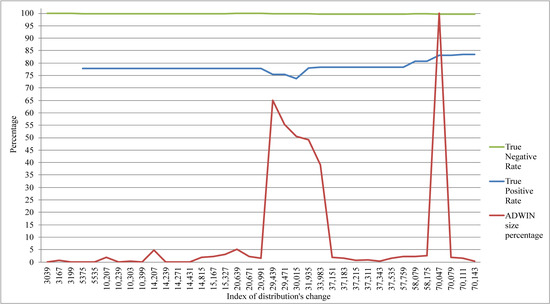
Figure 5.
Tracking the performance of ARFC and the change in ADWIN window size.
Another observation that has been noticed from the performance of the proposed model is its robustness of it against the missing values in the class label, which is one of the most difficult challenges for data stream classification tasks. Table 5 shows a comparison with the other five stream classifiers using two versions of the CHB-MIT EEG dataset. In the first one, we removed 1% from the values of the class label, while in the second one, we removed 3% from these values. The results in Table 5 confirm the advantage of ARFC in both cases before applying the model-based imputation.

Table 5.
Comparison of ARFC accuracy with four stream classifiers in presence of missing class values without Model-besed imputation.
Figure 6 and Figure 7 illustrates the ARFC performance with utilizing EFTD for imputation. They show the ability of EFDT to recover the original values for missing class labels and increase the accuracy of ARFC from 0.958 to 0.986 with a 3% missing ratio.
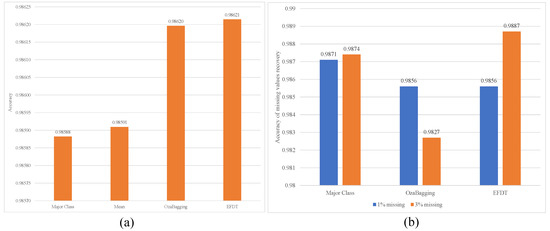
Figure 6.
Comparison of Model performance with Model-besed imputation: (a) ARFC accuracy with 3% missing values; (b) Accuracy of missing value recovery with 1% and 3% ratios.
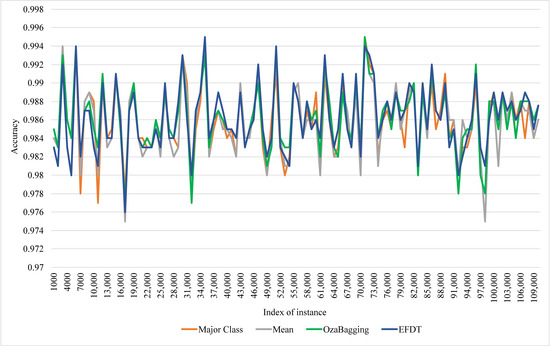
Figure 7.
Comparison of ARFC performance with using EFDT as Model-based imputation technique against the other three techniques.
An evaluation of the computational costs of the proposed model has been performed in terms of training time, testing time, and memory space. The evaluation using the CHB-MIT dataset included a comparison with the performance of Oza bagging, which is a popular bagging classifier, and it also has accuracy results close to the proposed model. The comparison in Figure 8 showed that ARFC has an average (0.8 s) training time and (0.2 s) testing time, while the average of Oza bagging was (4 s) training time and (3.6 s) testing time. In terms of memory space cost, ARFC consumed 1269.084 kB on average during the training processes compared with 10,966.367 kB memory consumed by Oza bagging, as shown in Figure 9. In Table 6, a significant reduction can be noticed in the response time of ARFC in the edge node without training reach to 90%; OzaBagging also has a reasonable reduction, and that confirms the benefit of applying the proposed method with other ensemble classifiers.
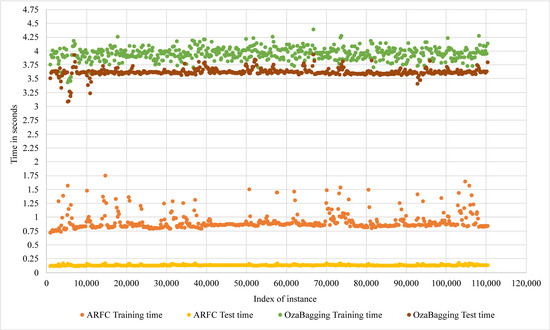
Figure 8.
Comparison of Computational time costs of ARFC and OzaBagging in Master node.
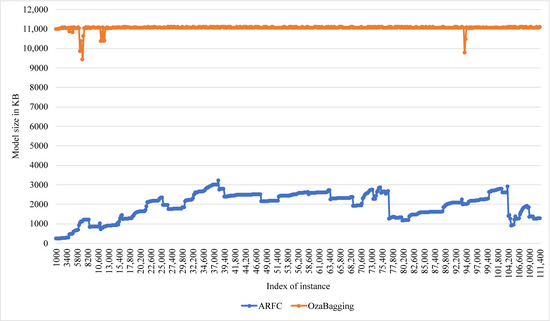
Figure 9.
Comparison of Model size in Memory of ARFC and OzaBagging in Master node.

Table 6.
Comparison of the response time’s reduction in seconds between ARFC and OzaBagging in edge node.
The CHB-MIT dataset has been used in many research articles, and the used techniques included deep learning, support vector machines, and linear classifiers. Table 7 shows a comparison between ARFC and eight randomly selected research articles. The comparisons confirm that ARFC overcomes the results of 15 articles.

Table 7.
The performance of ARFC compared to 15 related works.
5. Discussion
The results above showed an effective performance of the adaptive classification with EEG data stream regarding accuracy and response time in the cloud environment. TPR (or sensitivity) is a popular metric for evaluating the performance of seizure detectors, as in this work. However, depending only on the high value of the True Negative Rate (or specificity) may lead to an unreliable evaluation. Epilepsy patients often have normal EEGs; consequently, if the classifier mostly receives normal EEG signals without seizures, then TNR tends to be very high. This metric does not necessarily reflect the actual performance in actual clinical use. The False Positive Rate is a more critical metric because false alarms about seizures waste the neurologists’ time. However, the results in Table 3 show that ARFC has a low FPR equal to 0.0017. This means, on average, from 1000 detections, only 1.7 can be detected falsely as a seizure.
Most of the previous work dealt with data as a batch file (i.e., not a stream) and focused on improving its classification accuracy regardless of its efficiency. Some of these works suggested a distributed classification environment, but the training process was carried out at both the edge and the master nodes. In our work, the results confirm the possibility of centralizing the training process of the adaptive model in a node with high computational resources and using the resulting model each time to perform the inference in the edge nodes close to the patient. This facility produces a lightweight and constantly adaptive classifier that can be implemented in low-resources devices such as smartphones, tablets, and even smartwatches.
The fast response of the proposed classifier can provide efficient seizure onset detection. This kind of detection focuses on starting the warning procedures, diagnosis, and the required therapy. The instant notification for an epilepsy patient would save his life by preventing severe injuries caused by falling or car accidents through driving. The delayed alarm may cause worsening of seizure symptoms, thereby restricting the patient’s ability to respond. The immediate seizure onset detection can also help the medical staff instantly give the patient the necessary anticonvulsant. Another essential characteristic of the proposed model is providing a resistant classification to the problem of connection loss, which is common on the user’s side. The inference process continues in the edge node using the newest classifier regardless of a connection with the master node; the new data are sent after the connection returns.
Regarding reliability, handling the delayed labeling as missing values can make the proposed detector more applicable for patients and medical staff because we do not expect high feedback, especially during seizure time. Despite that, the experiments were limited to a limited percentage of the values that did not exceed 3%. The missing rate could be more extensive in practical applications due to the lack of user response. It should be noted that the challenges of EEG signal classification are not limited to missing data. Non-invasive EEG depends on the signals from the outer brain surface; thereby, its effectiveness may be reduced with epilepsy that begins deep in the brain. The detection of this kind of seizure can be affected by the signals of daily physical activities such as eye flutter.
Accessible and affordable wearable sensor devices such as EMOTIV can facilitate the preprocessing of EEG signals in the edge node. Despite the reduction in the received data during preprocessing steps, the expected amount of uploading data can still be significant because of the rapid signals received from EEG sensors. Regarding the implementation, the CHB-MIT dataset used to evaluate the model’s performance has been collected from a young sample of 23 cases whose ages do not exceed 22 years for males and 19 years for females. Since epilepsy is common in all age groups, exploring the efficiency of the proposed model in middle-aged and older people is necessary. Generally, large medical datasets are difficult to obtain and utilize due to patients’ privacy rights. Finally, the benefits of our model go beyond the home use of the patient, as health institutions can use it to monitor inpatients within their private cloud.
6. Conclusions
EEG signal data are an effective source for exploring the activities of the human brain. In this work, stream data mining techniques were utilized to build an adaptive classification model in the cloud environment that can classify EEG signals for detecting epileptic seizures. The proposed model included extracting the important features using Fast Fourier Transform, data row aggregation, and utilizing ARF as an adaptive classifier and EFDT for recovering missing values of the class label. The results of applying the proposed model to a real big dataset showed its preference over many typical batch classifiers, online classifiers, and many related research articles. It has a 0.998 True Negative Rate, a 0.785 True Positive Rate, and a 0.001 False Positive Rate. Applying EFDT to recover the original values of missing class labels improved the accuracy of ARFC from 0.958 to 0.986, with a 3% missing ratio. In terms of efficiency, a significant reduction was obtained in the response time of ARFC by using it for inference in the edge node, reaching 90%. The medical staff can utilize this fast response for applying the necessary therapy procedures as immediately after seizure onset to prevent complications in patients with epilepsy. The low computational requirements in the edge node facilitate the monitoring of patients outside health centers and prevent serious accidents while performing their daily activities. The proposed classifier can be developed in the future to handle missing EEG data itself and utilize Active Learning techniques to improve the classification process.
Author Contributions
Conceptualization, H.K.F. and A.K.; methodology, H.K.F. and A.K.; software, H.K.F.; validation, H.K.F. and A.K.; investigation, H.K.F. and A.K.; writing—original draft preparation, H.K.F. and A.K.; writing—review and editing, H.K.F. and A.K.; supervision, A.K.; project administration, A.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants of “Application Domain Specific Highly Reliable IT Solutions” project that has been implemented with the support provided from the National Research, Development and Innovation Fund of Hungary, financed under the Thematic Excellence Programme TKP2020-NKA-06 (National Challenges Subprogramme) funding scheme. This research was also supported by the ÚNKP-21-3 New National Excellence Program of the Ministry for Innovation and Technology from the source of the National Research, Development and Innovation Fund.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets and the source code of ARFC model were analyzed in this study. The files can be found at https://github.com/HayderFatlawi/ARFC, accessed on 29 November 2021.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| EEG | Electroencephalography |
| CHB-MIT | This database, collected at the Children’s Hospital Boston, consists of EEG recordings |
| TDF | Time-domain features |
| FDF | Frequency domain feature |
| PSD | Power spectral density |
| T-FDF | Time-frequency domain feature |
| FT | Fourier transform |
| DFT | Discrete Fourier Transform |
| ADWIN | ADaptive WINdowing |
| VFDT | Very Fast Decision Tree |
| IaaS | Infrastructure as a Service |
| PaaS | Platform as a Service |
| SaaS | Software as a Service |
| ARF | Adaptive random forest |
| ECoG | Electrocorticography |
| DWT | Discrete wavelet transform |
| COV | Coefficient of variation |
| NCOV | Normalized coefficient of variation |
| LDA | Linear discriminant analysis |
| SVM | Support vector machine |
| CNN | Convolutional Neural Network |
| STFT | Short-Time Fourier Transform |
| PPSO | Parallel Particle Swarm Optimization |
| VM | Virtual Machine |
| CKD | Chronic Kidney Disease |
| CSP | Common Spatial Pattern |
| AD | Alzheimer’s disease |
| TUSZ | Temple University EEG Seizure Corpus |
| ARFC | Adaptive Random Forest in the Cloud |
| REPTree | Reduced Error Pruning Tree |
| EFDT | Extremely Fast Decision Tree |
| LSTM | Long Short-Term Memory |
| LGCN | Linear graph convolution network |
| TNR | True Negative Rate |
| TPR | True Positive Rate |
| FPR | False Positive Rate |
References
- Neligan, A.; Hauser, W.A.; Sander, J.W. The epidemiology of the epilepsies. In Handbook of Clinical Neurology; Stefan, H., Theodore, W.H., Eds.; Elsevier: Amsterdam, The Netherlands, 2012; Volume 107, pp. 113–133. [Google Scholar]
- Beghi, E.; Giussani, G.; Nichols, E.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; Abraha, H.N.; Adib, M.G.; Agrawal, S.; Alahdab, F.; et al. Global, regional, and national burden of epilepsy, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 357–375. [Google Scholar] [CrossRef] [Green Version]
- Greengard, S. The Internet of Things; MIT Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Zainuri, N.A.; Jemain, A.A.; Muda, N. A comparison of various imputation methods for missing values in air quality data. Sains Malays. 2015, 44, 449–456. [Google Scholar] [CrossRef]
- Rajabion, L.; Shaltooki, A.A.; Taghikhah, M.; Ghasemi, A.; Badfar, A. Healthcare big data processing mechanisms: The role of cloud computing. Int. J. Inf. Manag. 2019, 49, 271–289. [Google Scholar] [CrossRef]
- Iqbal, M.A.; Hussain, S.; Xing, H.; Imran, M.A. Enabling the Internet of Things: Fundamentals, Design and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Abdulkader, S.N.; Atia, A.; Mostafa, M.S.M. Brain computer interfacing: Applications and challenges. Egypt. Inform. J. 2015, 16, 213–230. [Google Scholar] [CrossRef] [Green Version]
- Alyasseri, Z.A.A.; Khader, A.T.; Al-Betar, M.A.; Papa, J.P.; Alomari, O.A. EEG feature extraction for person identification using wavelet decomposition and multi-objective flower pollination algorithm. IEEE Access 2018, 6, 76007–76024. [Google Scholar] [CrossRef]
- Rechy-Ramirez, E.J.; Hu, H. Stages for Developing Control Systems Using EMG and EEG Signals: A Survey; TECHNICAL REPORT: CES-513; University of Essex: Essex, UK, 2011. [Google Scholar]
- Phinyomark, A.; Limsakul, C.; Phukpattaranont, P. A novel feature extraction for robust EMG pattern recognition. J. Comput. 2009, 1, 71–80. [Google Scholar]
- Oskoei, M.A.; Hu, H. GA-based feature subset selection for myoelectric classification. In Proceedings of the 2006 IEEE International Conference on Robotics and Biomimetics, Kunming, China, 17–20 December 2006; pp. 1465–1470. [Google Scholar]
- Englehart, K. Signal Representation for Classification of the Transient Myoelectric Signal. Ph.D. Thesis, University of New Brunswick, Fredericton, NB, Canada, 1998. [Google Scholar]
- Ramadan, R.A.; Vasilakos, A.V. Brain computer interface: Control signals review. Neurocomputing 2017, 223, 26–44. [Google Scholar] [CrossRef]
- Heckbert, P. Fourier transforms and the fast Fourier transform (FFT) algorithm. Comput. Graph. 1995, 2, 15–463. [Google Scholar]
- Rao, K.R.; Kim, D.N.; Hwang, J.J. Fast Fourier Transform: Algorithms and Applications; Springer: Cham, Switzerland, 2010; pp. 5–41. [Google Scholar]
- Han, J.; Pei, J.; Kamber, M. Data Mining: Concepts and Techniques, 3rd ed.; Elsevier: Waltham, MA, USA, 2011; pp. 330–348. [Google Scholar]
- Tan, P.N.; Steinbach, M.; Kumar, V. Introduction to Data Mining, 2nd ed.; Pearson Education: London, UK, 2016; pp. 113–137. [Google Scholar]
- Al-Fatlawi, A.H.; Fatlawi, H.K.; Ling, S.H. Recognition physical activities with optimal number of wearable sensors using data mining algorithms and deep belief network. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Korea, 11–15 July 2017; pp. 2871–2874. [Google Scholar]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013; Volume 26, pp. 369–413. [Google Scholar]
- Rokach, L.; Maimon, O.Z. Data Mining with Decision Trees: Theory and Applications, 2nd ed.; World Scientific: London, UK, 2014; pp. 53–59. [Google Scholar]
- Seni, G.; Elder, J.F. Ensemble methods in data mining: Improving accuracy through combining predictions. Synth. Lect. Data Min. Knowl. Discov. 2010, 2, 1–126. [Google Scholar] [CrossRef]
- Babenko, B.; Yang, M.H.; Belongie, S. A family of online boosting algorithms. In Proceedings of the 2009 IEEE 12th International Conference on Computer Vision Workshops, ICCV Workshops, Kyoto, Japan, 27 September–4 October 2009; pp. 1346–1353. [Google Scholar]
- Fatlawi, H.K.; Attila, K. Differential privacy based classification model for mining medical data stream using adaptive random forest. Acta Univ. Sapientiae Inform. 2021, 13, 1–20. [Google Scholar] [CrossRef]
- Gama, J. Knowledge Discovery From Data Streams; Chapman and Hall: Boca Raton, FL, USA; CRC Press: Boca Raton, FL, USA, 2010; pp. 33–47. [Google Scholar]
- Domingos, P.; Hulten, G. Mining high-speed data streams. In Proceedings of the Sixth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Boston, MA, USA, 20–12 August 2000; pp. 71–80. [Google Scholar]
- Rutkowski, L.; Jaworski, M.; Duda, P. Stream Data Mining: Algorithms and Their Probabilistic Properties; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Fatlawi, H.K.; Kiss, A. On robustness of adaptive random forest classifier on biomedical data stream. In Proceedings of the 13th Asian Conference on Intelligent Information and Database Systems, Phuket, Thailand, 23–26 March 2020; pp. 332–344. [Google Scholar]
- Quinlan, J.R. C4.5: Programs for Machine Learning. Morgan Kaufmann Publishers: San Mateo, CA, USA, 2014; pp. 17–25. [Google Scholar]
- Hulten, G.; Spencer, L.; Domingos, P. Mining time-changing data streams. In Proceedings of the Seventh ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 26–29 August 2001; pp. 97–106. [Google Scholar]
- Law, Y.N.; Zaniolo, C. An adaptive nearest neighbor classification algorithm for data streams. In Proceedings of the European Conference on Principles of Data Mining and Knowledge Discovery, Prague, Czech Republic, 3–7 October 2005; pp. 108–120. [Google Scholar]
- Freund, Y.; Schapire, R.E. Experiments with a new boosting algorithm. In Proceedings of the Thirteenth International Conference on International Conference on Machine Learning, Bari, Italy, 3–6 July 1996; pp. 148–156. [Google Scholar]
- Oza, N. Online bagging and boosting. In Proceedings of the 2005 IEEE International Conference on Systems, Man and Cybernetics, Waikoloa, HI, USA, 10–12 October 2005; Volume 3, pp. 2340–2345. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Schwing, A.G.; Zach, C.; Zheng, Y.; Pollefeys, M. Adaptive random forest—How many “experts” to ask before making a decision? In Proceedings of the Conference on Computer Vision and Pattern Recognition (CVPR 2011), Colorado Springs, CO, USA, 10–15 June 2011; pp. 1377–1384. [Google Scholar]
- Dang, L.M.; Piran, M.; Han, D.; Min, K.; Moon, H. A survey on internet of things and cloud computing for healthcare. Electronics 2019, 8, 768. [Google Scholar] [CrossRef] [Green Version]
- Sultan, N. Making use of cloud computing for healthcare provision: Opportunities and challenges. Int. J. Inf. Manag. 2014, 34, 177–184. [Google Scholar] [CrossRef]
- Manvi, S.; Shyam, G.K. Cloud Computing: Concepts and Technologies; CRC Press: Boca Raton, FL, USA, 2021; pp. 9–16. [Google Scholar]
- Chiang, C.Y.; Chang, N.F.; Chen, T.C.; Chen, H.H.; Chen, L.G. Seizure prediction based on classification of EEG synchronization patterns with on-line retraining and post-processing scheme. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston MA, USA, 30 August–3 September 2011; pp. 7564–7569. [Google Scholar]
- Khan, Y.U.; Rafiuddin, N.; Farooq, O. Automated seizure detection in scalp EEG using multiple wavelet scales. In Proceedings of the 2012 IEEE International Conference on Signal Processing, Computing and Control, Solan, India, 15–17 March 2012; pp. 1–5. [Google Scholar]
- Ahammad, N.; Fathima, T.; Joseph, P. Detection of epileptic seizure event and onset using EEG. BioMed Res. Int. 2014, 2014, 450573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Parhi, K.K. Low-complexity seizure prediction from iEEG/sEEG using spectral power and ratios of spectral power. IEEE Trans. Biomed. Circuits Syst. 2015, 10, 693–706. [Google Scholar] [CrossRef]
- Truong, N.D.; Nguyen, A.D.; Kuhlmann, L.; Bonyadi, M.R.; Yang, J.; Kavehei, O. A generalised seizure prediction with convolutional neural networks for intracranial and scalp electroencephalogram data analysis. arXiv 2017, arXiv:1707.01976. [Google Scholar]
- Zhou, M.; Tian, C.; Cao, R.; Wang, B.; Niu, Y.; Hu, T.; Guo, H.; Xiang, J. Epileptic seizure detection based on EEG signals and CNN. Front. Neuroinform. 2018, 12, 95. [Google Scholar] [CrossRef] [Green Version]
- Abdelaziz, A.; Elhoseny, M.; Salama, A.S.; Riad, A. A machine learning model for improving healthcare services on cloud computing environment. Measurement 2018, 119, 117–128. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, Y.; Yang, P.; Chen, W.; Lo, B. Epilepsy seizure prediction on EEG using common spatial pattern and convolutional neural network. IEEE J. Biomed. Health Inform. 2019, 24, 465–474. [Google Scholar] [CrossRef]
- Seo, J.; Laine, T.H.; Oh, G.; Sohn, K.A. EEG-Based Emotion Classification for Alzheimer’s Disease Patients Using Conventional Machine Learning and Recurrent Neural Network Models. Sensors 2020, 20, 7212. [Google Scholar] [CrossRef]
- Ayodele, K.P.; Ikezogwo, W.O.; Komolafe, M.A.; Ogunbona, P. Supervised domain generalization for integration of disparate scalp EEG datasets for automatic epileptic seizure detection. Comput. Biol. Med. 2020, 120, 103757. [Google Scholar] [CrossRef] [PubMed]
- Mrozek, D.; Koczur, A.; Małysiak-Mrozek, B. Fall detection in older adults with mobile IoT devices and machine learning in the cloud and on the edge. Inf. Sci. 2020, 537, 132–147. [Google Scholar] [CrossRef]
- Ye, D.; Yu, R.; Pan, M.; Han, Z. Federated learning in vehicular edge computing: A selective model aggregation approach. IEEE Access 2020, 8, 23920–23935. [Google Scholar] [CrossRef]
- Ismail, L.E.; Karwowski, W. A graph theory-based modeling of functional brain connectivity based on eeg: A systematic review in the context of neuroergonomics. IEEE Access 2020, 8, 155103–155135. [Google Scholar] [CrossRef]
- Khan, L.; van Lanen, R.; Hoogland, G.; Schijns, O.; Rijkers, K.; Kapsokalyvas, D.; van Zandvoort, M.; Haeren, R. Two-photon imaging to unravel the pathomechanisms associated with epileptic seizures: A review. Appl. Sci. 2021, 11, 2404. [Google Scholar] [CrossRef]
- Ryu, S.; Joe, I. A Hybrid DenseNet-LSTM model for epileptic seizure prediction. Appl. Sci. 2021, 11, 7661. [Google Scholar] [CrossRef]
- Manapragada, C.; Webb, G.I.; Salehi, M. Extremely fast decision tree. In Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, London, UK, 19–23 August 2018; pp. 1953–1962. [Google Scholar]
- Gomes, H.M.; Bifet, A.; Read, J.; Barddal, J.P.; Enembreck, F.; Pfharinger, B.; Holmes, G.; Abdessalem, T. Adaptive random forests for evolving data stream classification. Mach. Learn. 2017, 106, 1469–1495. [Google Scholar] [CrossRef]
- Shoeb, A.H. Application of Machine Learning to Epileptic Seizure Onset Detection and Treatment. Ph.D. Thesis, Massachusetts Institute of Technology, Cambridge, MA, USA, 2009. [Google Scholar]
- CHB-MIT Scalp EEG Database. Available online: https://www.prb.org/resources/2016-world-population-data-sheet (accessed on 12 July 2021).
- Rojas, G.M.; Alvarez, C.; Montoya, C.E.; De la Iglesia-Vaya, M.; Cisternas, J.E.; Gálvez, M. Study of resting-state functional connectivity networks using EEG electrodes position as seed. Front. Neurosci. 2018, 12, 235. [Google Scholar] [CrossRef]
- Rich, T.L.; Gillick, B.T. Electrode placement in transcranial direct current stimulation—How reliable is the determination of C3/C4? Brain Sci. 2019, 9, 69. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Chen, W.; Li, M. Symplectic geometry decomposition-based features for automatic epileptic seizure detection. Comput. Biol. Med. 2020, 116, 103549. [Google Scholar] [CrossRef]
- Gómez, C.; Arbeláez, P.; Navarrete, M.; Alvarado-Rojas, C.; Le Van Quyen, M.; Valderrama, M. Automatic seizure detection based on imaged-EEG signals through fully convolutional networks. Sci. Rep. 2020, 10, 21833. [Google Scholar] [CrossRef] [PubMed]
- Kaziha, O.; Bonny, T. A convolutional neural network for seizure detection. In Proceedings of the 2020 Advances in Science and Engineering Technology International Conferences (ASET), Dubai, United Arab Emirates, 4 February–9 April 2020; pp. 1–5. [Google Scholar]
- Chen, Z.; Lu, G.; Xie, Z.; Shang, W. A unified framework and method for EEG-based early epileptic seizure detection and epilepsy diagnosis. IEEE Access 2020, 8, 20080–20092. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.; Liu, W.; Chang, Z.; Kärkkäinen, T.; Cong, F. One dimensional convolutional neural networks for seizure onset detection using long-term scalp and intracranial EEG. Neurocomputing 2021, 459, 212–222. [Google Scholar] [CrossRef]
- Liu, G.; Tian, L.; Zhou, W. Patient-Independent Seizure Detection Based on Channel-Perturbation Convolutional Neural Network and Bidirectional Long Short-Term Memory. Int. J. Neural Syst. 2021, 2150051. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Dong, C.; Zhang, G.; Wang, Y.; Chen, X.; Jia, W.; Yuan, Q.; Xu, F.; Zheng, Y. EEG-Based Seizure detection using linear graph convolution network with focal loss. Comput. Methods Programs Biomed. 2021, 208, 106277. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).