1. Introduction
Cardiovascular diseases (CVDs) are the leading cause of death worldwide. The World Health Organization (WHO) estimates that 17.9 million people died from cardiovascular disease in 2019, accounting for 32% of deaths worldwide. Of these deaths, 85% were due to coronary heart disease and stroke [
1]. Early diagnosis and appropriate treatment can significantly reduce mortality and improve the quality of life. The diagnostic process commonly consists of two main parts. The first part refers to obtaining images of cardiac structures with the help of imaging devices. Imaging techniques, such as computed tomography (CT) or magnetic resonance imaging (MRI), allow inspection of a human body without surgically cutting into it. CT has become one of the most common imaging techniques for examining the human cardiovascular system. The second part of the diagnostic process is interpreting and quantifying images using advanced image processing methods.
Manual or semiautomatic segmentation of cardiovascular structures from CT images is a time-consuming process, often prone to error. Not only may segmentation drawn by two radiologists differ (inter-observer variability), but there will also not be an agreement between segmentation drawn by the same radiologist at different occasions (intra-observer variability). This is mainly due to the high noise in CT images and their fluctuating contrast. For example, the coronary sinus is almost devoid of contrast in the CT data, making it difficult to distinguish from surrounding tissues. The papillary muscles are complex and small, making them difficult to distinguish from the noise. Segmentation of the right ventricular wall is complex because of the thin myocardium and considerable inter-patient individual variability. Therefore, there is a need to develop automated tools to facilitate cardiovascular images segmentation and interpretation.
Deep learning, particularly convolutional neural networks (CNNs), shows promising results in automatic segmentation for a variety of medical applications [
2,
3]. A popular deep learning architecture in the field of semantic segmentation for biomedical applications is UNet [
4]. UNet’s symmetric, encoder–decoder architecture allows automatic learning of features at different levels. Nevertheless, low-level features learned in the encoder are rich with feature space information but lack semantic information, and high-level features learned from the decoder are the opposite. Thus, the direct concatenation of these features may not produce the most optimal results. Researchers have already offered plenty of improvement schemes based on UNet. For example, addition of residual connections in ResUNet architecture propagates information over layers, allowing building of deeper neural networks and reducing the impact of exploding or vanishing gradients, which ultimately alleviates training performance [
5]. Moreover, contrary to UNet, architectures such as RefineNet [
6], DenseNet [
7], or SegNet [
8] only use the highest layer features while they lack in low-level representation. This was further improved in DeepLabV3 [
9] and ResUNet++ [
10], where extracted features are passed through the Atrous Spatial Pyramid Pooling (ASPP) module to obtain multi-scale information [
9]. Although it is advantageous to have as many extracted features as possible, not all features are equally important. Therefore, distinguishing between feature importance allows the network to focus on the most important features. To solve this problem, SENet architecture [
11] introduces squeeze-and-excitation operations that can capture the importance degree of each feature channel through the feature recalibration strategy. Based on the importance degree, the less useful features are suppressed while more useful features are enhanced. Similarly, the attention mechanism dynamically allocates the input weights of neurons to selectively focus on the most critical part of the information [
12] and are often introduced into UNet-based architectures to improve their performance. Nevertheless, such networks effectively fuse multi-level features but do not fully utilize the contextual information.
The primary motivation behind this work is to introduce a new UNet-based network that will fully explore useful features of the channel and capitalize on the contextual information with an overall aim of increasing segmentation accuracy and training performance. To achieve this, we introduce three modifications to the original UNet. First, we add residual connections to each layer of the encoder and decoder to help network training. Second, we introduce a self-attentive mechanism in skip connections to capture feature dependencies in the channel dimension while ensuring effective fusion of multi-level features. This is achieved by assigning the specific attention weight to channels, which reduces noise and gives more attention to essential regions. Third, we use ASPP blocks as the bottom structure of UNet to effectively increase the receptive field and reduce the impact of learned redundant features, with the overall aim of obtaining higher segmentation accuracy.
1.1. Research Contributions
Therefore, in this paper we present a new AB-ResUNet+ architecture. Our intention is to construct a network that can achieve high segmentation accuracy with a small dataset and use it for segmentation of complex cardiovascular structures.
In summary, the contributions of the paper are as follows:
We present a new AB-ResUNet+ architecture that uses residual learning, squeeze and excitation operations, Atrous Spatial Pyramid Pooling (ASPP), and the attention mechanism.
The channel attention block is inserted into the skip connection to optimize the coding ability of each layer.
The proposed architecture works well with small datasets.
Our proposed architecture significantly improves the segmentation of challenging cardiovascular structures.
We evaluated our model on eleven datasets with different cardiovascular structures. Our experimental evaluations show that the proposed AB-ResUNet+ architecture outperforms the UNet, ResUNet, and ResUNet++ architectures by achieving higher values in terms of Dice coefficient and mean intersection over union (mIoU).
1.2. Paper Overview
The remainder of the paper is structured as follows. In
Section 2, we give an overview of related research.
Section 3 gives details about our proposed method.
Section 4 provides dataset description and implementation details.
Section 5 presents conducted experiments and obtained results for different cardiovascular structures. Finally, the discussion and concluding remarks are given in
Section 6 and
Section 7, respectively.
2. Related Research
In this section, we discuss some related work. First, we briefly review previous methods in cardiovascular segmentation, focusing on CNN-based approaches. Second, we introduce essential deep learning concepts and networks relevant to our research.
2.1. Previous Methods for Cardiovascular Segmentation
Recent advances in medical imaging have been facilitated by the widespread application of deep learning techniques. Two-stage segmentation methods consisting of localization and segmentation steps [
13,
14,
15], FCNs with deep supervision [
16,
17], multi-view CNNs [
18,
19], and residual network variants are most commonly used for various cardiovascular segmentation tasks. Few works use UNet architecture to provide experimental analysis observing the influence of different parameters for final segmentation results. For example, Baumgartner et al. [
20] investigate two-dimensional (2D) UNet and three-dimensional (3D) UNet with various hyperparameters. Patravali et al. [
21] evaluated a 2D and 3D UNet trained with varying Dice and cross-entropy losses. Jang et al. [
22] implemented an M-Net architecture in which the decoding layer’s feature maps are concatenated with those of the previous layer. Luo et al. [
23] propose a method based on UNet combined with image sequence information. They introduce two modules: the contextual extraction module and the segmentation module. The context extraction module can fully extract the context features of the image to be segmented and effectively combine the sequence features. The segmentation module is an encoder–decoder module and the input image can directly predict a segmented image. The module effectively learns the characteristics of the original image and avoids feature loss and gradient dispersion by the design of the skip connection. Isensee et al. [
24] implemented an ensemble of 2D and 3D UNet architectures (with residual connections along with the upsampling layers). Yang et al. [
25] implemented a 3D UNet with residual connections instead of a commonly used concatenation operator. Chen et al. [
26] proposed TransUNet architecture, with inherent global self-attention mechanisms into UNet. In the encoder, the transformer tokenizes image patches from a CNN feature map. At the same time, the decoder upsamples the encoded features before combining them with the high-resolution CNN feature maps to enable exact localization. Transformers overcome UNet’s limited localization ability due to insufficient low-level details. Cao et al. [
27] propose UNet-based architecture named swin-Unet. They use hierarchical swin transformer with shifted windows [
28] as the encoder to extract context features, and symmetric swin transformer-based decoder with patch expanding layer designed to perform the upsampling operation to restore the spatial resolution of the feature maps.
Furthermore, structure-wise, commonly observed are the whole heart and its main substructures, such as left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), pulmonary artery (PA), descending aorta (DA), and coronary arteries [
29,
30]. However, the coronary sinus (CS), right ventricular wall (RVW), left atrial wall (LAW), superior vena cava, and inferior vena cava (SVC, IVC) are somewhat less explored structures. This is mainly due to the lack of annotated datasets. Recently, Baskaran et al. [
31] used the UNet architecture to segment the proximal ascending and descending aorta (PAA, DA), superior and inferior vena cavae (SVC, IVC), pulmonary artery (PA), coronary sinus (CS), right ventricular wall (RVW), and left atrial wall (LAW) and made the dataset publicly available. This dataset is used in our work.
2.2. Residual Learning, Spatial Pyramidal Pooling, and Attention Mechanism
Segmentation accuracy can be improved with increasing network depth. However, this has been shown to hinder the training process and ultimately contribute to accuracy degradation [
32]. He et al. [
33] proposed deep residual learning to facilitate the training process and solve the degradation problem. ResUNet [
5] uses full residual units prior to activation. The residual unit simplifies the training of the deep network, and the skip connection within the network ensures that the information is passed without degradation. The improved version of the ResUNet is the ResUNet++ architecture [
10]. It takes advantage of the residual blocks, the squeeze and excitation block, ASPP, and the attention block. The attention mechanism, placed at the bottom of UNet architecture, determines which parts of the network require more attention [
12]. In this way, it reduces the computational cost of information encoding and enhances the quality of features that boost the results, ultimately enhancing the results.
Convolution is an essential step in both CNN and FCN models, as it allows models to learn increasingly abstract feature representations. However, the pooling operations and the convolution steps between layers in the convolution process reduce the spatial resolution of the feature map, resulting in a loss of spatial detail. To learn the contextual information at multiple scales, dilated or atroid convolutions [
34,
35,
36] are introduced. They are able to increase the receptive field and maintain the resolution of the feature map by injecting holes into the standard convolution. Compared to the original standard convolution, the dilation convolution has a hyperparameter called dilation rate, which refers to the number of intervals of the convolution kernel. The idea of capturing contextual information at different scales subsequently led to the ASPP module [
9,
37]. Here, a large number of parallel atrous convolutions with different rates are fused together in the input feature map to control the field of view and accurately capture information at different scales [
38]. The most prominent network that uses dilated convolution is DeepLabV3 [
39]. This network combines the contextual information at multiple scales by fusing the lower and upper layer features. In addition, PSPNet18 [
40] uses a pyramid pooling module that aggregates contextual information of different grid scales to improve the ability to obtain global information. In DeconvNet [
41], stacked deconvolution layers are used for full resolution recovery. However, this results in a large number of parameters and can lead to difficulties in training.
3. Methodology
This section presents a theoretical overview of the proposed encoder–decoder-based architecture. We give an overall architecture design and introduce the main building blocks and their purpose.
3.1. Architecture Overview
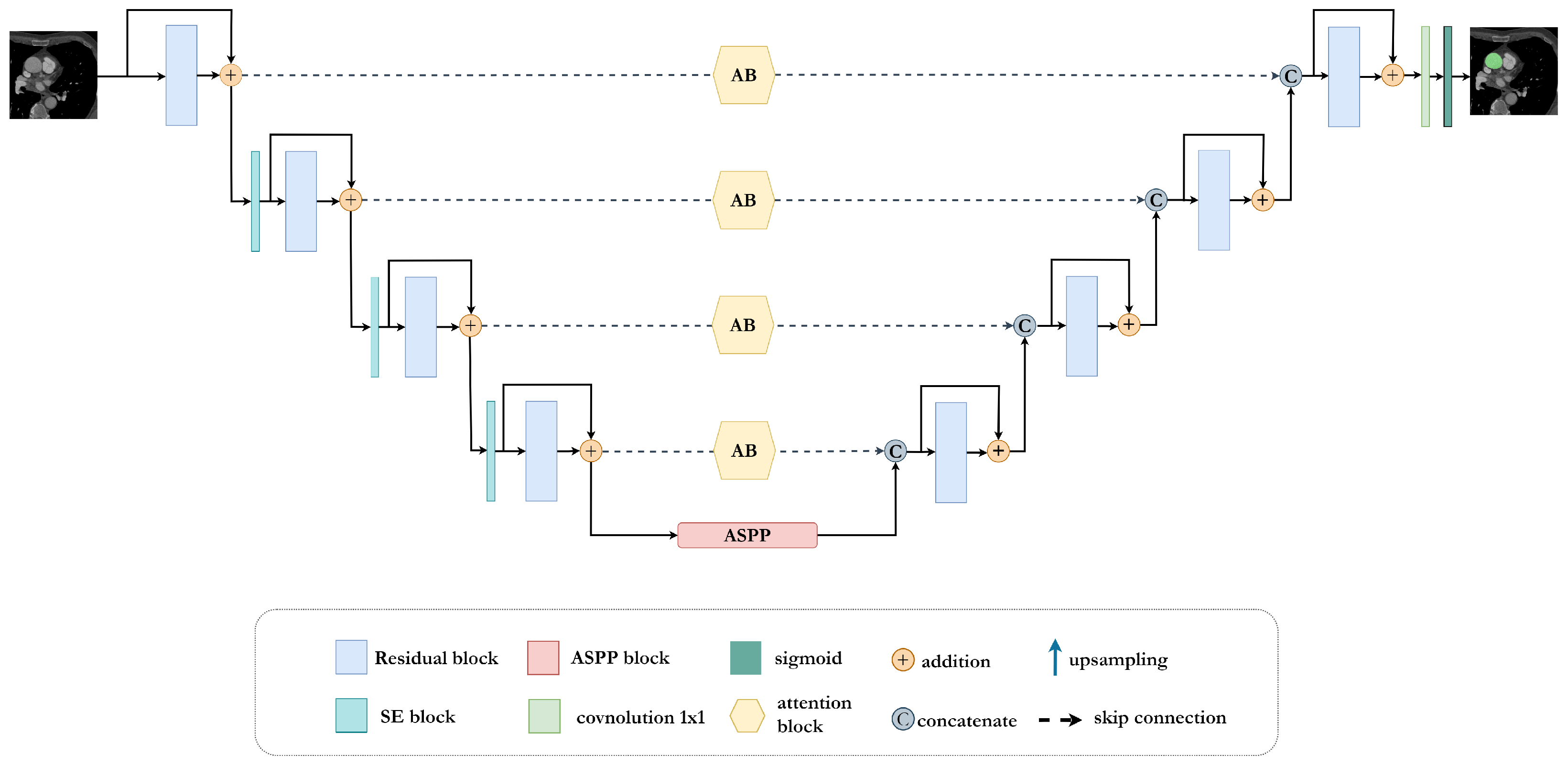
In this work, we present a new UNet-inspired architecture, AB-ResUNet+, that exploits residual learning, squeeze and excitation operations, Atrous Spatial Pyramid Pooling (ASPP), and the attention mechanism for accurate and effective segmentation of complex cardiovascular structures.
The proposed AB-ResUNet+ architecture consists of one stem block and three encoder blocks, ASPP and three decoder blocks. The encoder consists of squeeze-and-excitation and residual blocks, i.e., two successive
convolutional layers and identity mapping. Each convolution layer includes an ReLU activation layer and batch normalization. The identity mapping connects the input and output of the encoder block. The output of the residual blocks in the encoder part is routed through the squeeze-and-excitation block to increase the representational power of the network. The main improvement is mainly achieved by adding the channel attention block into the skip connection. The addition of the channel attention block in each skip connection improves the coding ability in each layer and successfully eliminates irrelevant and redundant information. This improves the network’s ability to distinguish between feature importance and focus on the most important features. The ASPP block is placed at the bottom of the network and acts as a bridge between the encoder and the decoder, increasing the field of view of the filters and allowing them to include a wider context. The decoder consists of residual blocks, a
convolution with sigmoid activation, that provides the final segmentation map. An illustration of the proposed network is shown in
Figure 1.
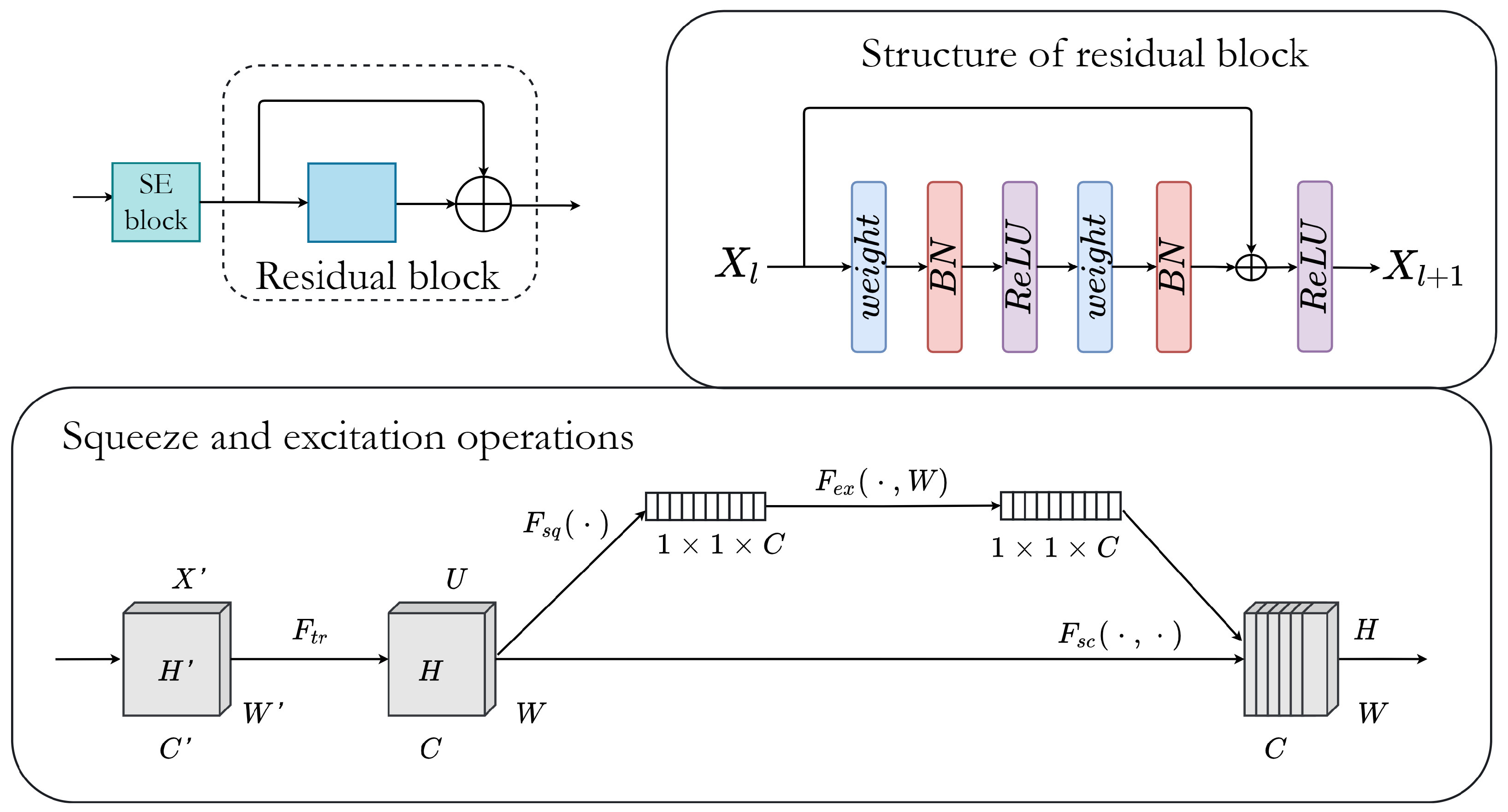
3.2. Residual Block
Deep residual learning is characterized by the addition of shortcut connections between every few stacked layers to build residual blocks. Generally, each residual block can be expressed with the following two formulations:
and
where
is the residual mapping which needs to be learned,
and
are the input and output of the
l-th residual block,
is an activation function, and
is the identity mapping function. The mapping function has a typical form that can be expressed as
In this work, we employ residual blocks in both contracting path and expansive path, as illustrated in
Figure 1. Additionally, the output of residual blocks in the encoder part is passed through the squeeze and excitation block to increase the network’s representational power.
Figure 2 shows the structure of residual and squeeze and excitation blocks.
3.3. Attention Block
An input feature map
can be reshaped in matrices
and
. The channel attention map
is obtained by dividing
K and
Q by the factor
and applying a softmax layer. This can be written as
where
refers to
ith channel’s influence on
jth channel, while function
f calculates their relationship.
The channel statistics of a channel from an original feature map can be acquired with global average pooling (GAP), which can be expressed using
where
,
and
g is GAP function.
GAP is obtained as an attention vector and compresses global information, which allows feature dimensionality reduction and high-level feature extraction. This preserves salient features. Furthermore, by multiplying matrices
A and
V, we obtain the result transformed into
, which can be multiplied by the parameter
. We can then use the original feature map
I to obtain the final output, which can be written as
where
starts from 0 and learns weight during training,
refers to the weight of the
convolution, and
is the bias. Therefore, the final output is a weighted sum between feature maps obtained by convolution and those that came from attention with GAP.
Since each channel corresponds to a specific semantic response, different channels have different contributions for acquiring useful feature information. By modeling the dependencies of each channel and by adjusting features channel by channel, the network can selectively learn to identify which features contain useful information and which contain useless information, and can strengthen or suppress them accordingly. Therefore, addition of attention blocks into skip connections helps in eliminating redundant information and improves networks’ representational power.
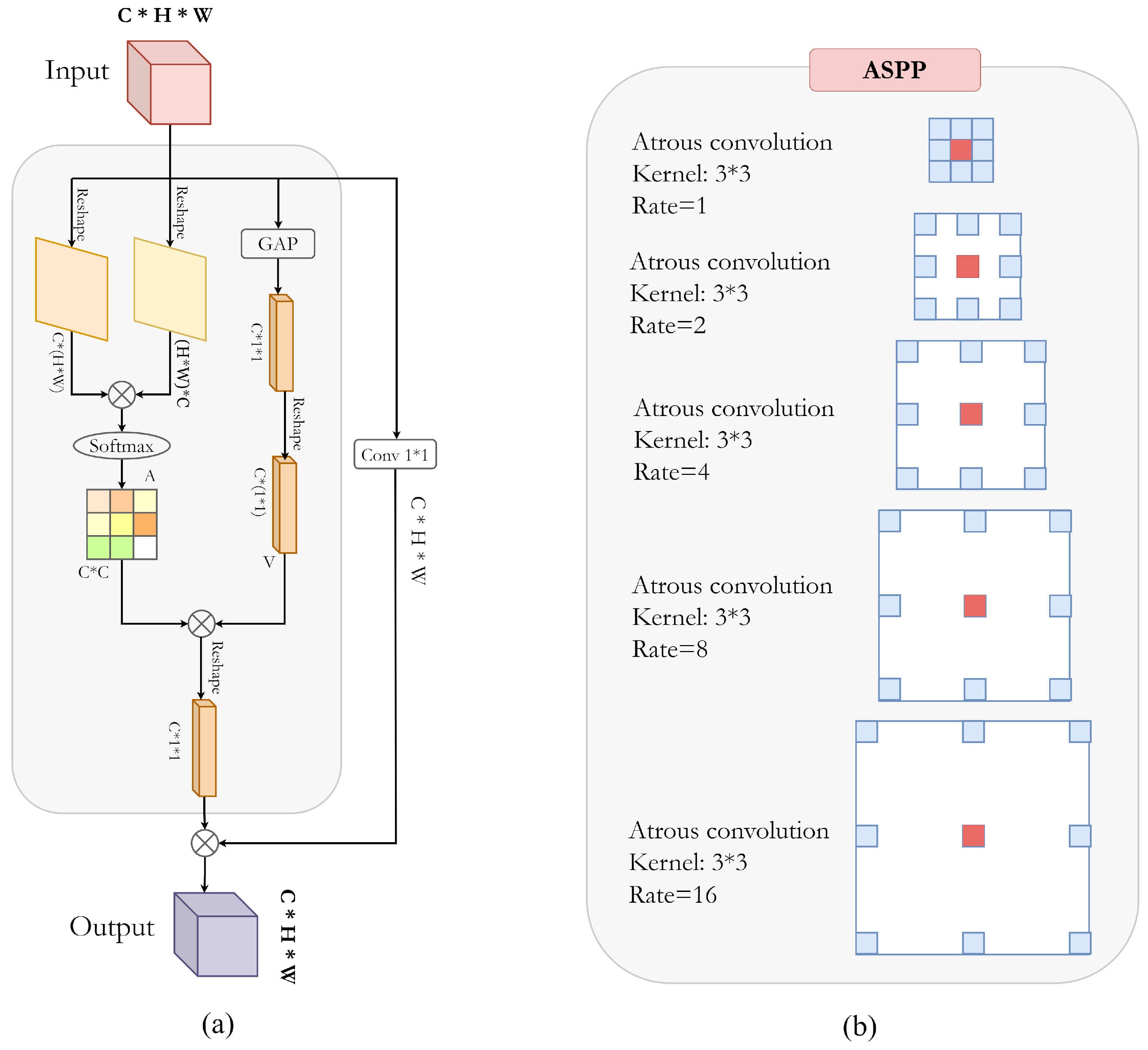
Figure 3a shows the detailed block structure of the described attention block.
3.4. ASPP for Dense Feature Extraction
Atrous convolution allows us to compute the responses of any layer at any desirable resolution. It can be mathematically expressed with the following:
where
refers to an input signal,
is the output of an atrous convolution,
w represents the convolution kernel of length
k, and
r denotes the rate parameter of the stride to which the input signal is sampled.
Figure 3b illustrates the atrous convolution on image with
kernel and target pixel rates of
,
, and
. Therefore, the larger field of view is obtained with higher sampling rates. A standard convolution filter requires more parameters to enlarge the field of view, while an atrous filter can increase the field of view without an increase in parameters. This significantly reduces the computational cost. Similarly, ASPP follows this idea by parallel implementation of multiple atrous convolution layers with different sampling rates. The multi-scale features are then integrated to generate a final feature map [
38,
42].
4. Implementation Details
In this section, we give a dataset description on which we conducted our experiments. After that, we give details about network training and implementation. To provide experimental analysis we train four models: (1) UNet, (2) ResUNet, (3) ResUNet++, and (4) proposed AB-ResUNet+. Finally, we briefly describe used evaluation metrics.
4.1. Dataset Description and Preprocessing
The network presented in the previous section was applied to the task of the multiple cardiovascular structures from the Kaggle Dataset [
31,
43]. This data was collected on patients from the everyday clinical environment. It has various qualities to preserve the robustness of the developed algorithms when it comes to real clinical usage. The cardiac CTA data are obtained using 64-slice CT scanners, and images are reconstructed at 0.50 mm thickness. The datasets include the following eleven cardiovascular structures: coronary sinus (CS), descending aorta (DA), inferior vena cava (IVC), left atrial appendage (LAA), left atrial wall (LAW), papillary muscle (PM), posterior mitral leaflet (PML), proximal ascending aorta (PAA), pulmonary aorta (PA), right ventricular wall (RVW), and superior vena cava (SVC). Dataset details regarding the number of patients and the total number of 2D images are expressed in
Table 1. The PAA begins in the plane corresponding to the nadirs of all three aortic valve cusps, which is also the plane closest to the origin of the brachiocephalic artery. The DA begins distal to the origin of the left subclavian artery and extends to the lowest axial disc. The vena cavae are venous veins that run through the right middle mediastinum adjacent to and to the right of the trachea and PAA and empty into the right atrium. The main left and right pulmonary arteries were all included in the PA. The CS is a venous structure that runs from the great cardiac vein through the atrioventricular groove near the left circumflex coronary artery and empties into the right atrium. The RVW is the volume of myocardial tissue in the right ventricle calculated by delineating the endocardial and epicardial boundaries of the ventricle, omitting the papillary muscles.
4.2. Training Details
In our experiments, we train four encoder–decoder based architectures: (1) original UNet, (2) ResUNet, (3) ResUNet++, and (4) propsoed AB-ResUNet+. All architectures were implemented using the Keras framework with TensorFlow as the backend. We performed our experiment on a single Nvidia GEForce GTX 1070 GPU. We started the training with a batch size of four, and the proposed architecture was optimized by Adam optimizer. The learning rate of the algorithm was set to
. The images of size
pixels were fed to the model. To alleviate the number of training samples, we used the following data augmentation techniques: random crop, horizontal flip, vertical flip, scale, and random rotation. The rotation angle was randomly chosen in between −20 and 20 degrees. Fivefold cross-validation was employed to assess the performance of the proposed model. We trained all models for 200 epochs. We also used the stochastic gradient descent with restart (SGDR) to improve the model’s performance.
Table 2 summarizes used data augmentation methods training parameters.
4.3. Evaluation Metrics
Comparing images to assess the accuracy of segmentation is critical for evaluating progress in this field of research. To evaluate segmentation performance, we use the following four metrics: mean intersection over union (
mIoU), Dice similarity coefficient (
DSC), precision (
P), and recall (
R). These metrics are mathematically defined with the following equations:
where
FP refers to the number of false positives,
TP refers to the number of true positives,
TN refers to the number of true negatives, and
FN refers to the number of false negatives.
The
DSC metric measures the degree of overlap between the ground truth and predicted segmentation and can be written as follows:
where
P is the predicted mask and
G is the ground truth.
5. Experiments and Results
This section gives a quantitative and qualitative analysis of obtained results. To evaluate the AB-ResUNet+ architecture, we train, validate, and test models using eleven datasets of cardiovascular structures. We compare the performance of our proposed AB-ResUNet+ architecture with results obtained using UNet, ResUNet, and ResUNet++. We compare our obtained results with AB-ResUnet+ architecture to other significant architectures in the field of cardiac segmentation. Finally, we provide and discuss qualitative results.
5.1. Quantitative Results
In our experiments, we trained four encoder–decoder-based architectures: (1) the original UNet [
4], (2) ResUNet [
5], (3) ResUNet++ [
10], and (4) the proposed AB-ResUNet+. These experiments provide insight into our proposed architecture’s competitiveness compared to previously published architectures relevant to our work. The detailed qualitative segmentation results are shown in
Table 3,
Table 4,
Table 5 and
Table 6.
According to results, we can observe that addition of attention blocks in the skip connection obtains finer results than original UNet. Generally, after the skip connection, there are two features that are combined—one is from the decoder layer and the others from the matching encoder layer. In the original UNet, these two features are directly combined with a concatenation function. On the other hand, in our proposed AB-ResUNet+ model, we combine these features using attention block. Therefore, after the channel compression, the encoded feature contains more local information from the input sample and global information of the channel, which yields better results. The ASPP block assists the decoded feature to possess more semantic information of the input sample while the interaction between encoding and decoding features may form a group of feature maps with both sample global information and semantic information. Addition of residual connections mostly accelerates networks’ learning process.
Moreover, we may observe that UNet can segment the general outlines of most cardiovascular structures from the results. However, it cannot segment regions with high contrast variability and prominent edges. ResUNet has a better performance than UNet, in which the residual connection enhances the segmentation ability. We can see from the results that the proposed AB-ResUNet+ provides higher segmentation accuracy than the other original UNet, ResUnet, and ResUNet++ regarding DSC and mIoU for all cardiac structures, except for PVW, where ResUNet++ achieves the highest DSC. Furthermore, regarding precision and recall, we observe that the proposed AB-ResUNet+ outperforms in all cardiovascular structures except RVW, LAW, and IVC, respectively. This is probably due to the overfitting that occurs for these structures. Nevertheless, the proposed architecture significantly outperformed the baseline architectures for all other datasets.
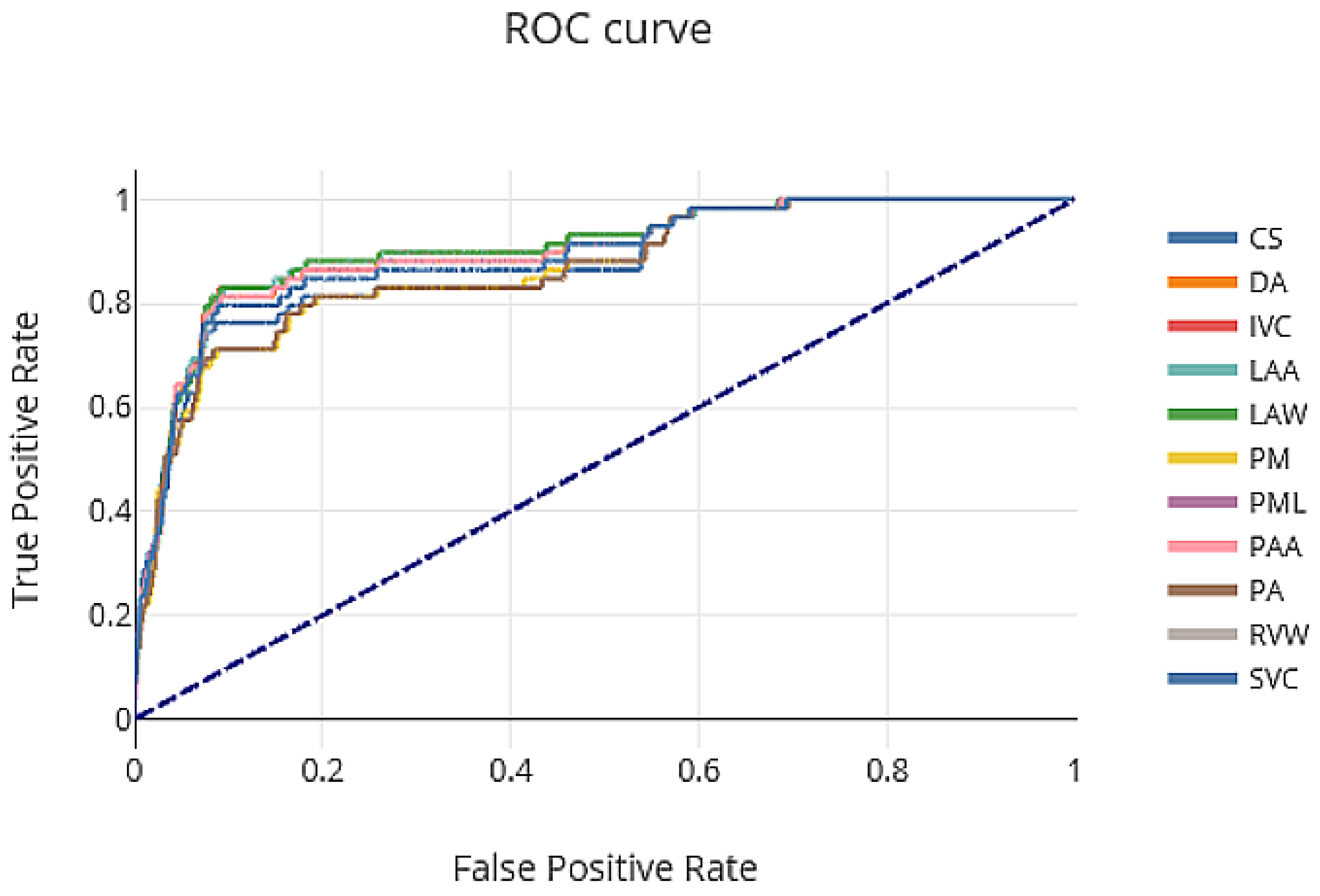
The ROC curve showing the tradeoff between sensitivity and specificity of our proposed AB-ResUNet+ network architecture is shown in
Figure 4. The area under the ROC curve (AUC) for CS, DA, IVC, LAA, LAW, PM, PML, PAA, PA, RVW, and SVC were 0.74, 0.96, 0.91, 0.90, 0.81, 0.75, 0.72, 0.94, 0.93, 0.90, and 0.95, respectively. From the ROC curve, we can see that DA, PA, and SVC obtain a significant altitude, which is reflected in a very robust segmentation performance.
Comparison with State-of-the-Art Methods
The proposed approach was compared with other similar deep learning approaches in terms of image segmentation accuracy, as shown in
Table 7. The most similar work, regarding observed structures, to our work is work by Baskaran et al. [
31]. They used UNet architecture for PAA, DA, SVC, IVC, PA, CS, RVW, and LAW segmentation. Nevertheless, their dataset consisted of 206 patients with 2D images of size
, which contributed to high accuracy. Moreover, they used CAT images. The use of contrast and ECG gating that may have better delineated the border between the vessel wall and lumen may be partly attributable to higher DSC regarding PAA and DA segmentation. Another approach introduced by Jin et al. [
44] uses fully convolutional neural networks (FCNs) with three-dimensional (3D) conditional random fields (CRFs) for LAA segmentation. After manual localization of LAA, they employed the FCNs and fine-tuned them to segment each 2D LAA image slice. Further, they used a dense 3D CRF to refine the segmentations of all slices. Noothout et al. [
45] proposed a method for DA and AA segmentation low-dose chest CT without contrast enhancement. They used a dilated convolutional neural network (CNN) that classifies voxels in axial, coronal, and sagittal image slices. The probabilities of the three planes were averaged per class and voxels were subsequently assigned to the class with the highest class probability to obtain final segmentation. Furthermore, Shi et al. [
46] proposed a probabilistic deep voxelwise dilated residual network named Bayesian VoxDRN that can predict voxelwise class labels with a measure of model uncertainty. By utilizing the dropout process, the model is able to learn weight distributions with a higher degree of data explanation. This considerably reduces the likelihood of overfitting.
Nevertheless, it is important to highlight that compared methods use different datasets, often not publicly available for the research community. Datasets differ in the number of patients, 2D images per patient, and image modality. Moreover, most of the previous studies report the results of the commonly researched cardiovascular structures such as whole heart, left atrium, left ventricle, right atrium, and right ventricle (RV), while cardiac structures investigated in this work are significantly less represented in previous research (due to unavailability of publicly available datasets). This makes it hard to provide representative and quality comparisons with our work.
5.2. Qualitative Results
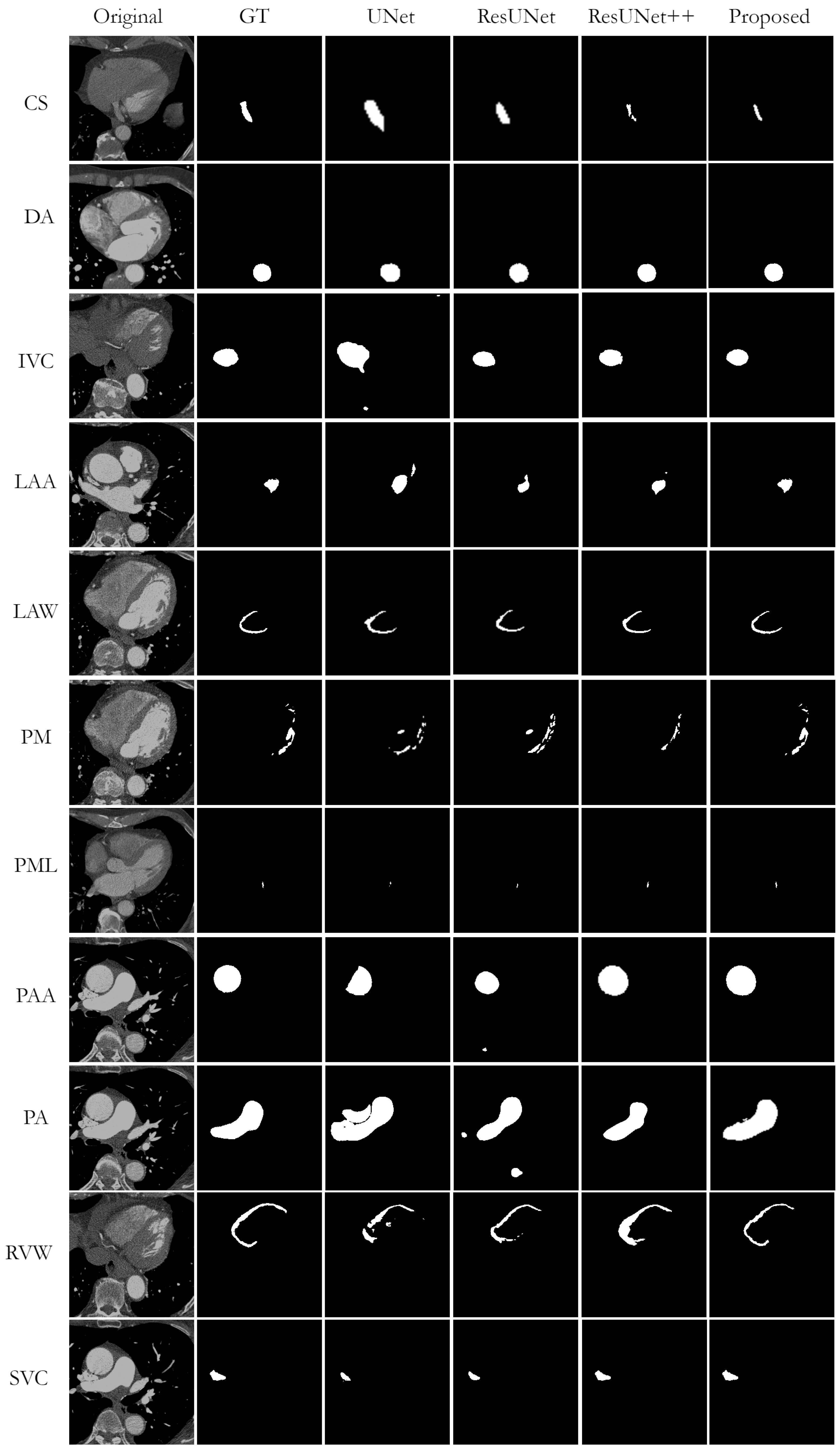
Figure 5 shows a visual comparison of the successful segmentation predictions from the test datasets, generated by UNet, ResUNet, ResUNet++, and the proposed AB-ResUNet+, respectively. The most accurate segmentation results are obtained for DA, IVC, PAA, and PA. This is probably due to their circular structure and the high contrast in the images around these structures.
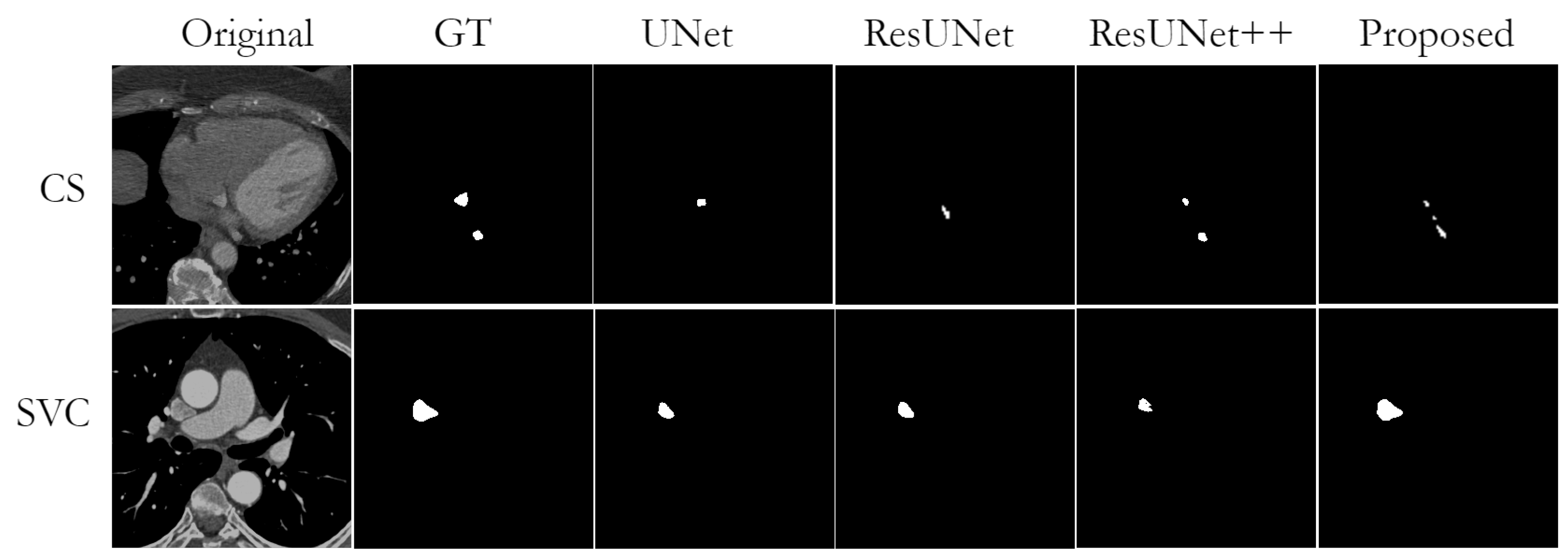
Nevertheless, while observing obtained segmentation predictions, we found some cases of missing or wrong segmentation results. A missing segmentation refers to an incomplete segmentation, i.e., when part of the area to be segmented is missing. This is a common problem in small structures such as CS, PML, and SVC. In particular, since the mask of PML is represented almost as a single point, the model could not segment it completely in case of any segmentation failure. An example of incorrect segmentation of CS and SVC can be found in
Figure 6.
The overfitting issue is successfully overcome in most cases. Nevertheless, we observe some overfitting while segmenting PM and LAA, where the model hardly distinguishes between background and these structures due to low contrast and anatomical complexity. An example of such errors can be found in
Figure 7.
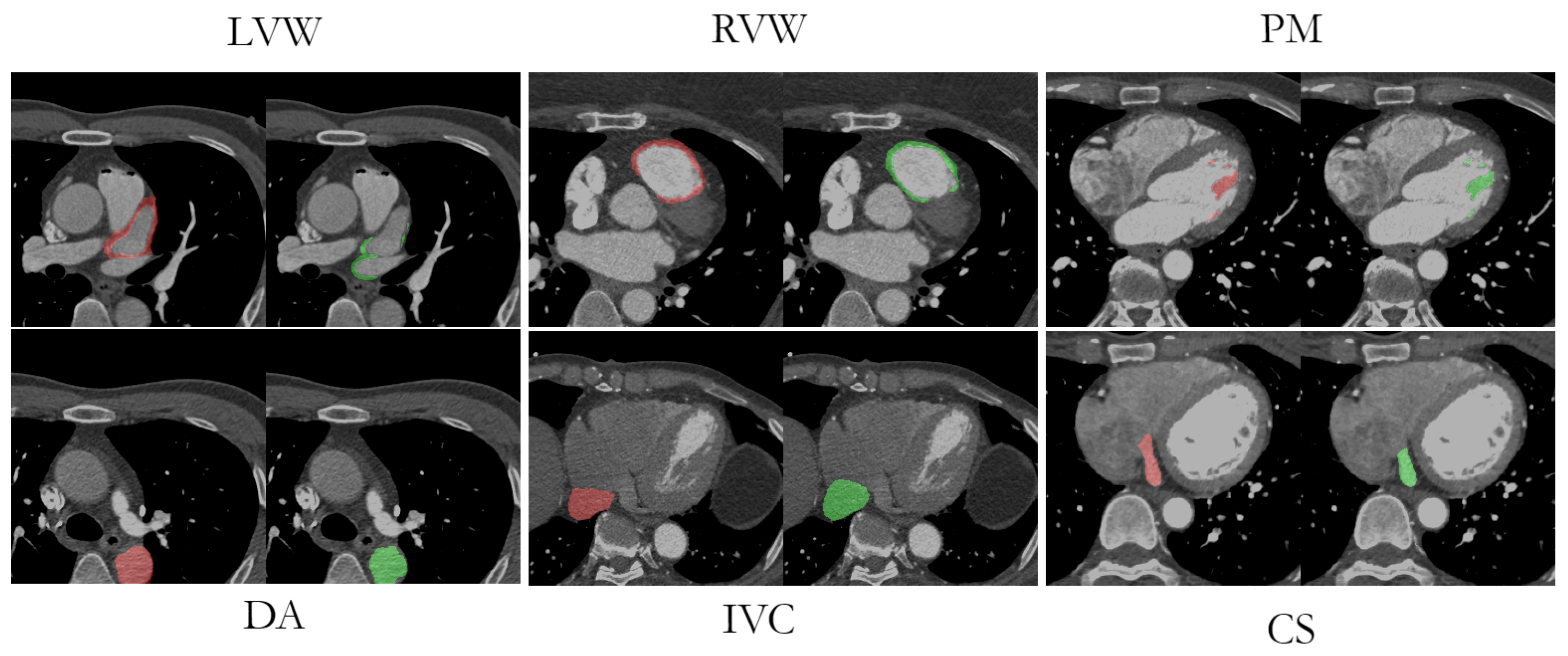
Visual comparisons between the original image, the manual labeling, and the AB-ResUNet+ model prediction are shown in
Figure 8. The segmentation prediction examples and GT overlays over the original image for LVW, RVW, PM, DA, IVC, and CS more accurately illustrate the potential difficulties in segmentation due to low image quality, high and low contrast differences, and the highly anatomical complexity of the structures. The difficulty in identifying the irregularly shaped CS, especially with suboptimal contrast fluoroscopy, may account for the lower accuracy compared to the other vessels. The complex structure of LVWs and RVWs, which varies in different layers and in different patients, makes them a particular challenge for automatic segmentation methods.
6. Discussion
In this work, we aimed to develop a new deep learning model for accurate segmentation of most of the great vessels, left atrial and right ventricular walls, and coronary sinus. So far, we have presented our newly developed AB-ResUNet+ architecture that utilizes residual learning, squeeze and excitation operations, Atrous Spatial Pyramid Pooling (ASPP), and the attention mechanism for accurate and effective segmentation of complex cardiovascular structures. The encoder consists of squeeze-and-excitation and residual blocks. The output of the residual blocks in the encoder part is routed through the squeeze-and-excitation block to increase the representational power of the network. Squeeze and excitation operations capture the importance degree of each feature channel through feature recalibration strategy. Based on the importance degree, the less useful channel features are suppressed while useful features are enhanced. The decoder consists of residual blocks and generates final segmentation predictions. Moreover, the main improvement is mainly achieved by adding the channel attention block into the skip connection. The addition of the channel attention block in each skip connection improves the coding ability in each layer and successfully eliminates irrelevant and redundant information. This improves the network’s ability to distinguish between feature importance and focus on the most important features. The ASPP block is placed at the bottom of the network and acts as a bridge between the encoder and the decoder, increasing the field of view of the filters and allowing them to include a wider context.
To evaluate the quality of our design choice, we implemented and trained a total of four networks, namely, UNet, ResUNet, ResUNet++, and the proposed AB-ResUNet+, and evaluated them on eleven test datasets of complex cardiovascular structures, namely, coronary sinus (CS), descending aorta (DA), inferior vena cava (IVC), left atrial appendage (LAA), left atrial wall (LAW), papillary muscle (PM), posterior mitral leaflet (PML), proximal ascending aorta (PAA), pulmonary aorta (PA), right ventricular wall (RVW), and superior vena cava (SVC). The proposed network achieved more accurate DSC results for most of the datasets used compared to ResUNet++. In particular, our proposed architecture improved the DSC of CS, DA, IVC, LAA, LAW, PM, PAA, PA, and SVC by 4.57%, 2.76%, 2.63%, 3.11%, 0.95%, 3.15%, 1.68%, 2.37%, and 2.68%, respectively. However, for the PML and RVW datasets, we obtained the worst DSC compared to ResUNet++. Moreover, we obtained better results for mIoU compared to ResUNet++, except for the LAA and PAA datasets. Moreover, we observed that the proposed AB -ResUNet+ architecture achieved higher DSC and mIoU metrics, as well as competitive precision and recall, for most datasets compared to the baseline models. Based on the obtained results, it is clear that the inclusion of AB blocks in the proposed AB-ResUNet+ architecture leads to slightly better results than the plain ResUNet++ architecture. The designed AB block in skip connections helped the proposed network to exploit the intra-slice information to a certain extent; thus, the network obtained higher segmentation results.
There are several limitations associated with our current study. First, while obtained results are promising, it is important to determine whether such a model may be applied in clinical practice. Given that clinical practice often involves recordings of live video, a model with a fast inference time is required to process images in real time. Therefore, a proposed method should be improved by using lightweight models. Second, although the proposed method improves performance, the lack of medical data related to observed images limits the segmentation effect. Therefore, for future work, we plan to further improve segmentation accuracy by facilitating the dataset used with more advanced data augmentation methods. For example, generative adversarial networks [
47] have great potential to obtain a larger training dataset by generating synthetic data. In addition, we aim to explore the possibilities of few-shot learning to reduce the impact of the lack of annotated data on segmentation accuracy.
7. Conclusions
In this work, we propose the AB-ResUNet+ for the segmentation of complex cardiovascular structures. Our network follows UNet structure and strengthens its representational power by incorporating residual learning, squeeze and excitation operations, ASPP, and the attention mechanism. The channel attention block is inserted into the skip connection to optimize the coding ability of each layer. The ASPP block is located at the bottom of the network and acts as a bridge between the encoder and decoder to increase the field of view. The proposed AB-ResUNet+ is evaluated on eleven datasets of complex cardiovascular structures. We obtain an average DSC of 71.68%, 93.45%, 91.45%, 88.22%, 85.12%, 77.44%, 58.64%, 94.12%, 94.25%, 87.22%, and 95.44% for CS, DA, IVC, LAA, LAW, PM, PML, PAA, PA, RVW, and SVC, respectively. Moreover, we observe that the proposed AB-ResUNet+ architecture achieves higher DSC and mIoU metrics, as well as competitive precision and recall, for most datasets compared to the baseline models.