Generative Adversarial CT Volume Extrapolation for Robust Small-to-Large Field of View Registration
Abstract
:1. Introduction
2. Materials and Methods
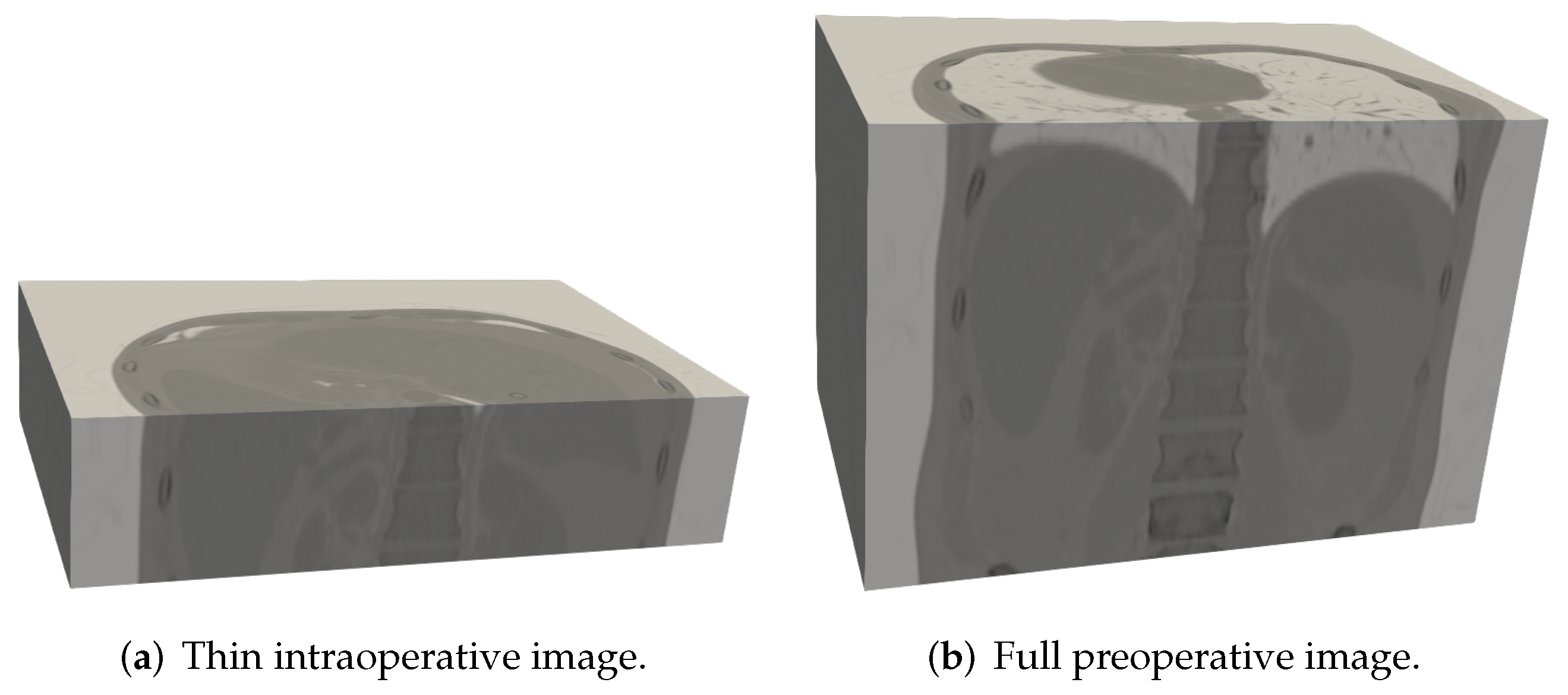
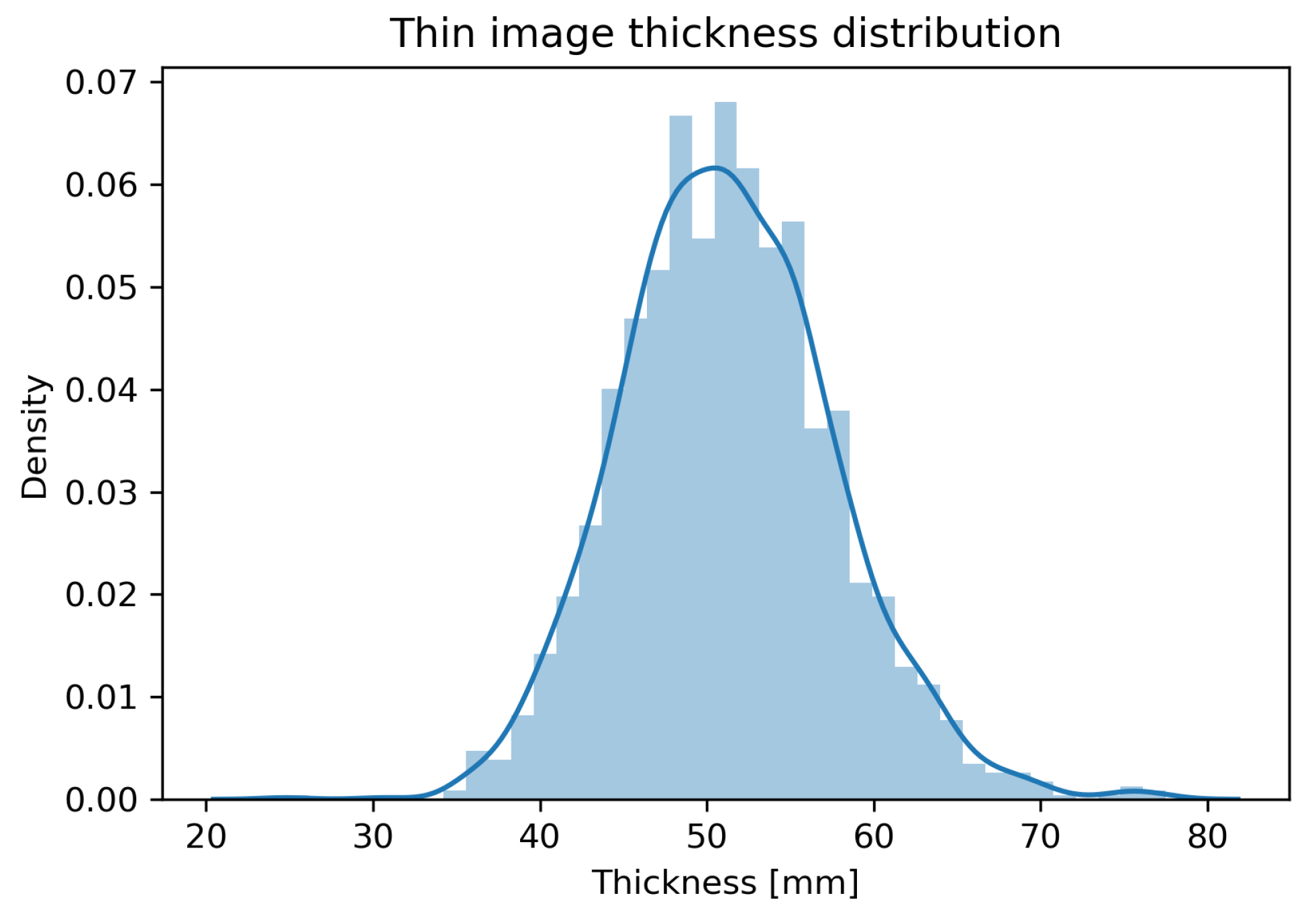
2.1. Dataset
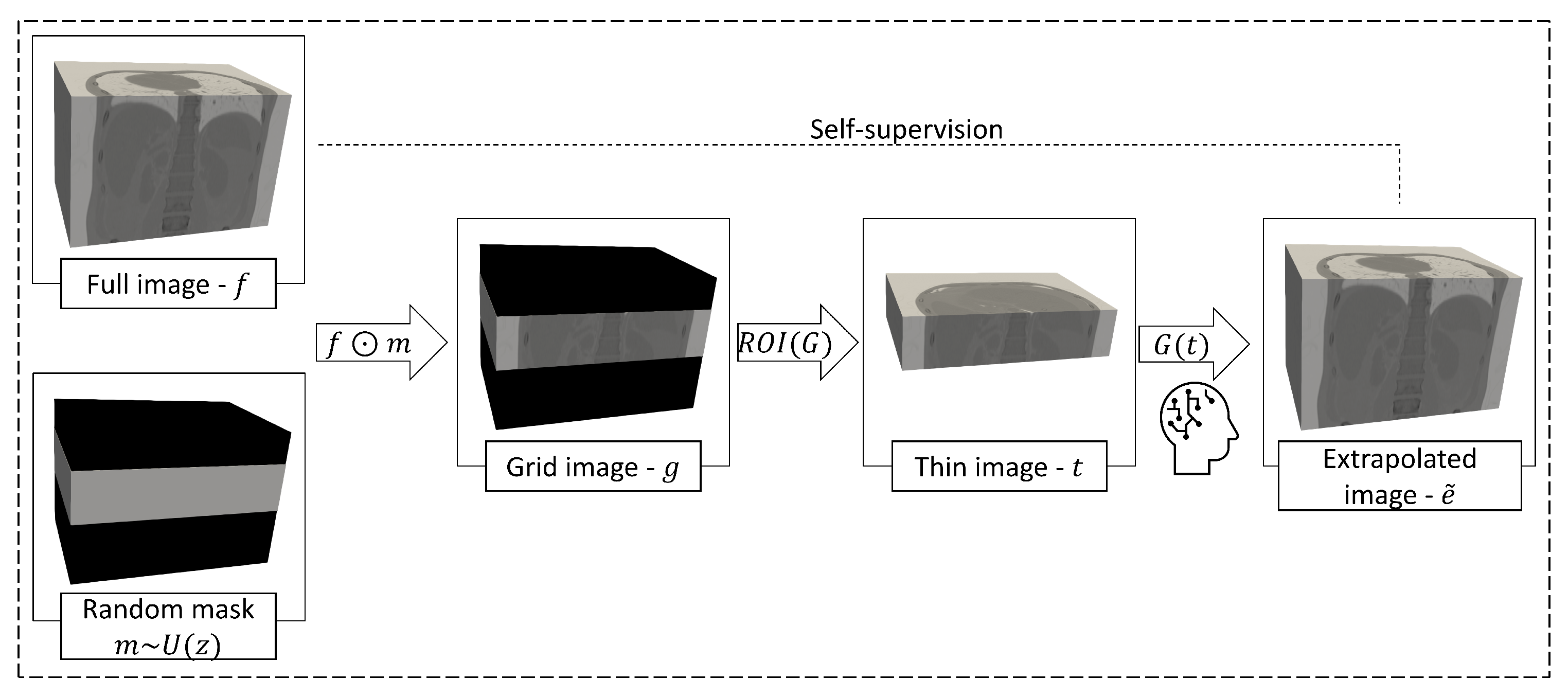
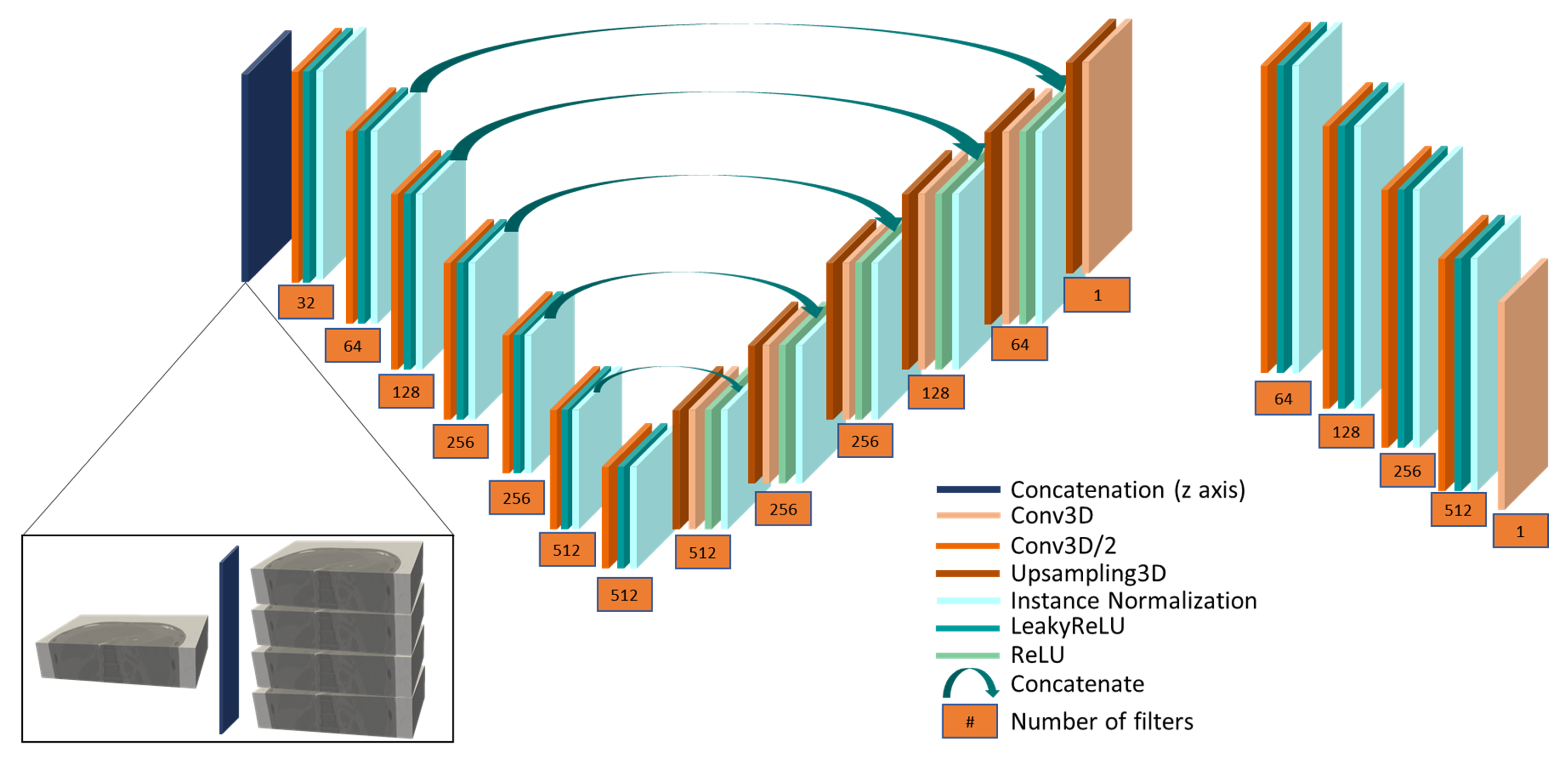
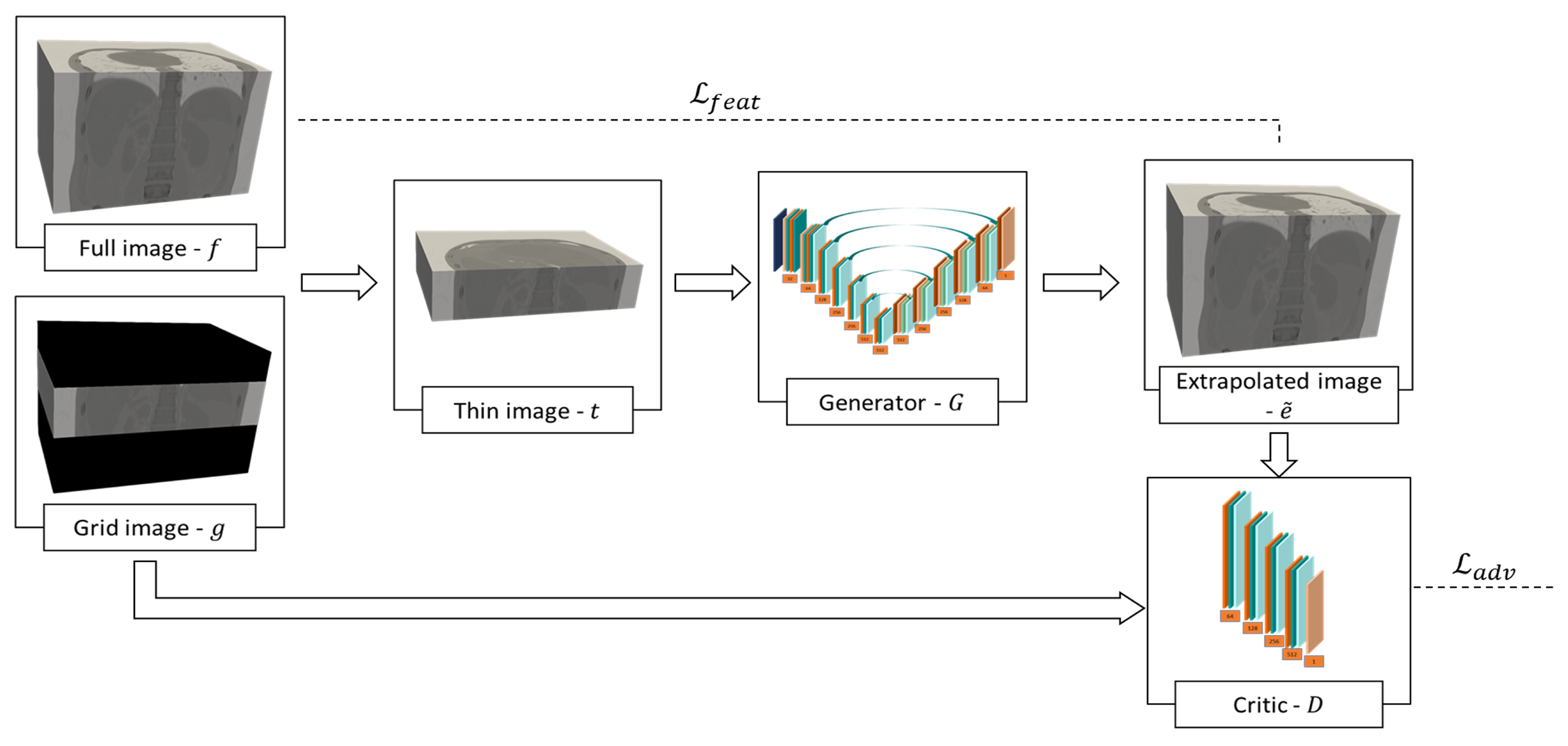
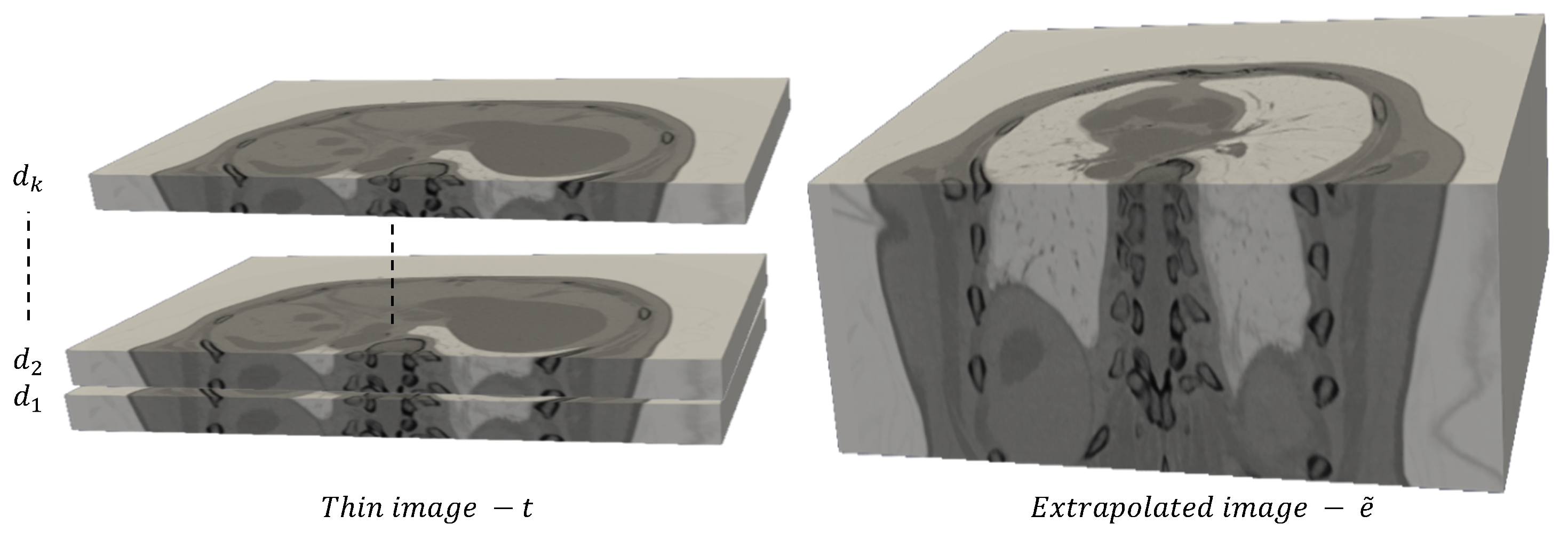
2.2. Proposed Method
2.2.1. Optimization Strategy
2.2.2. Image Metadata Retrieval
2.3. Performance Quantification
3. Results
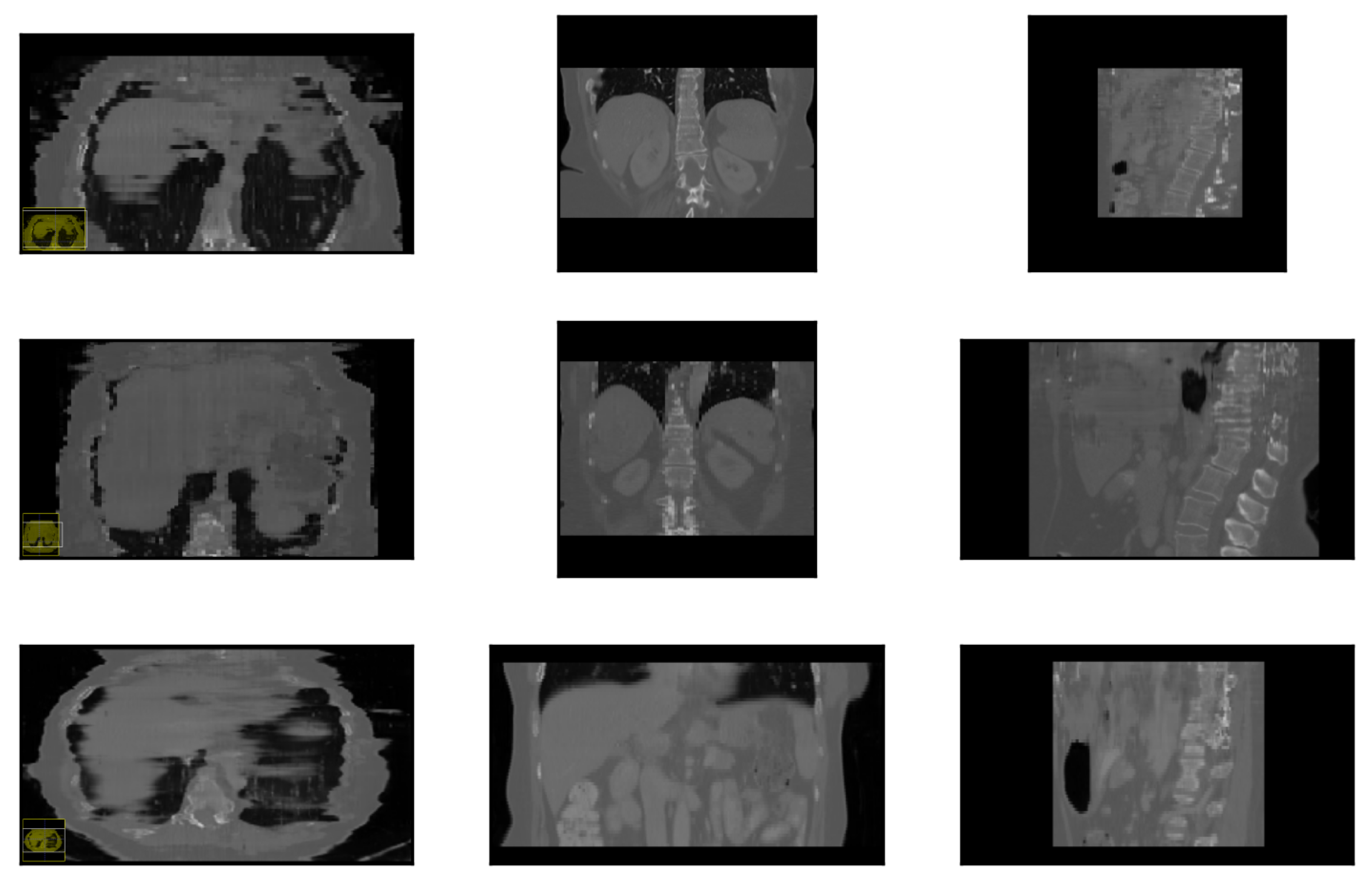
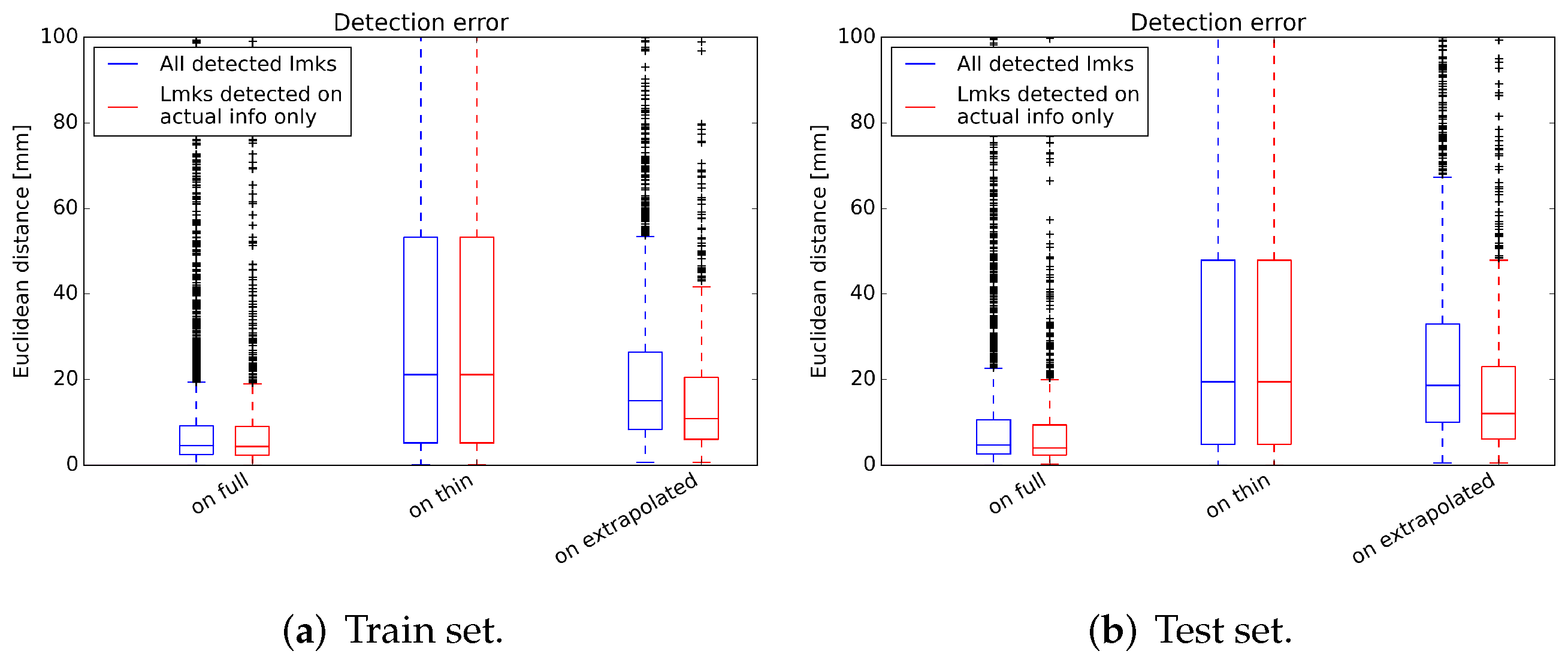
3.1. Landmark Detection Test
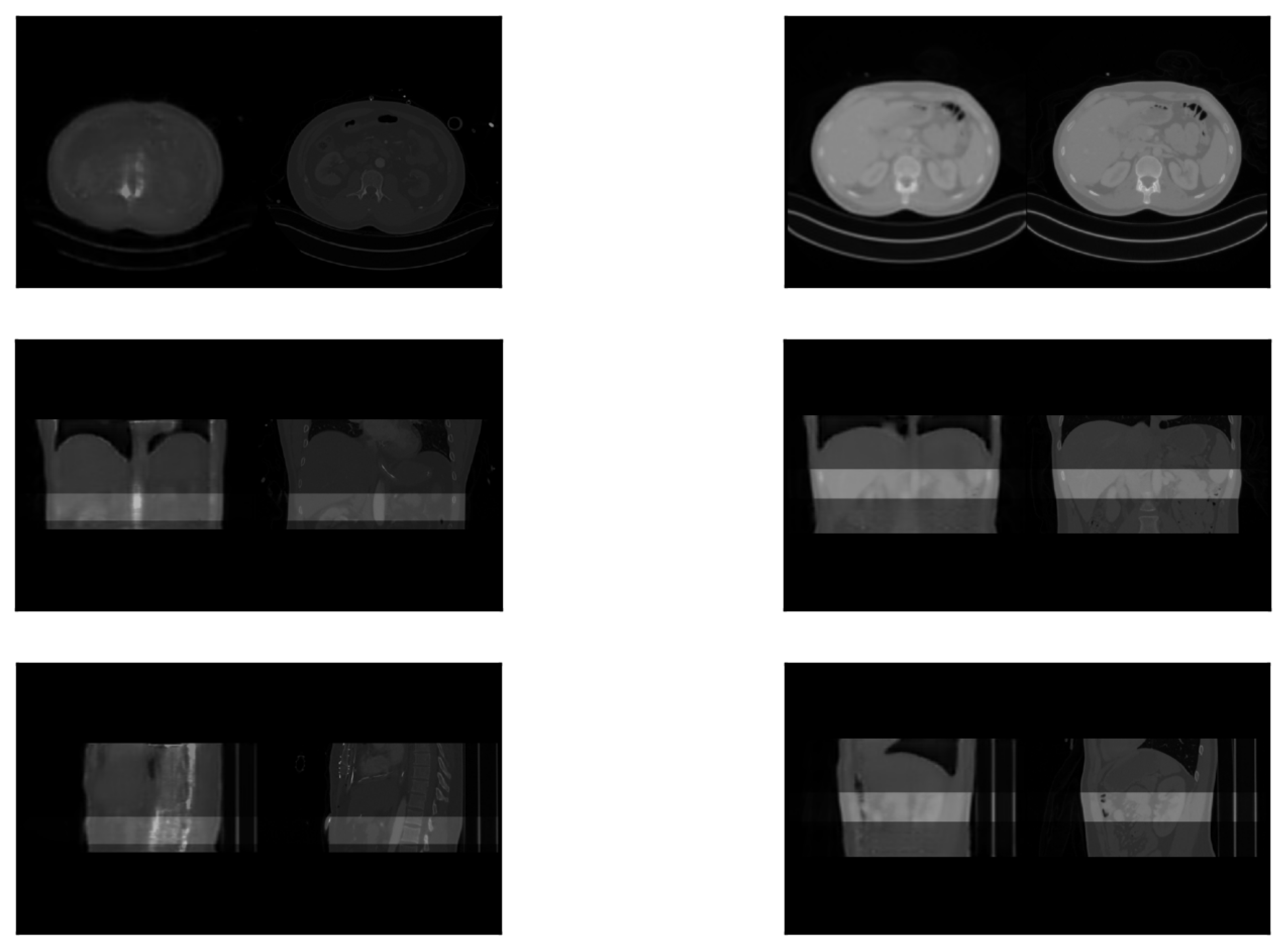
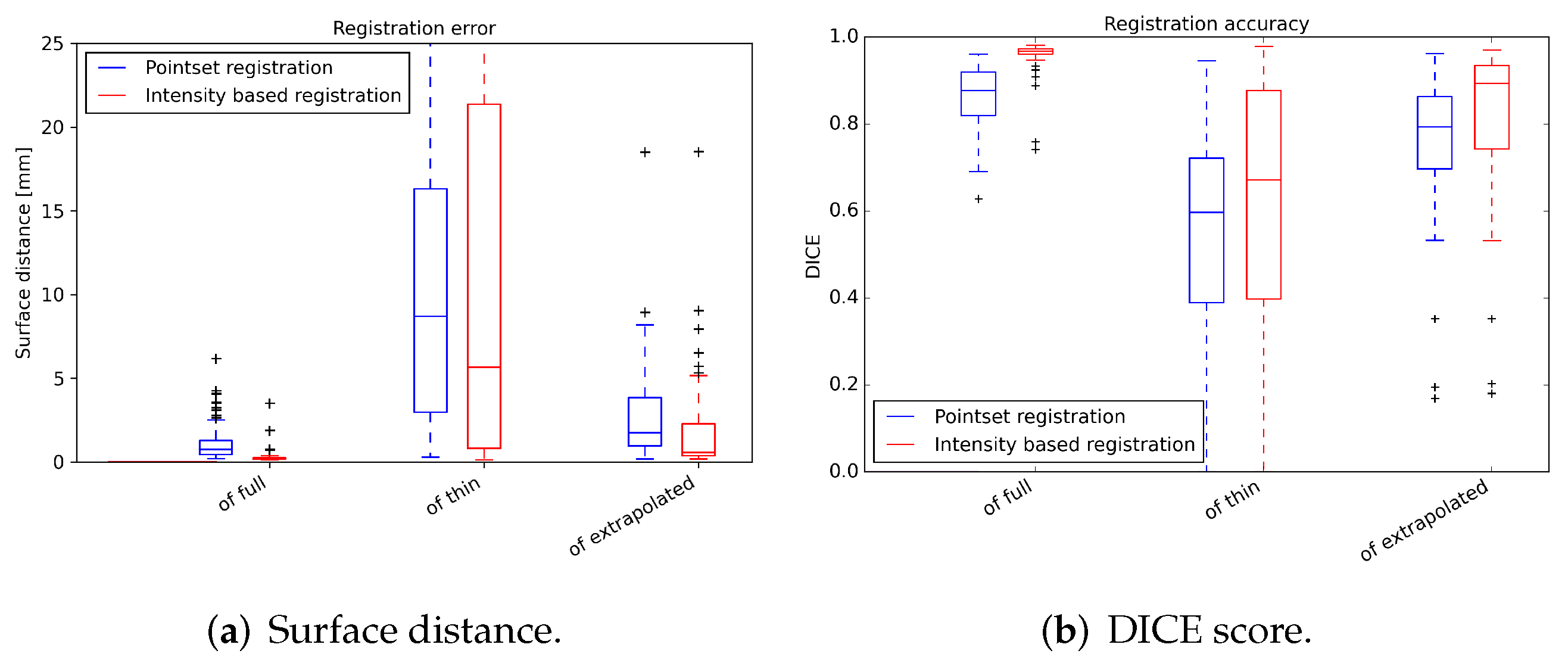
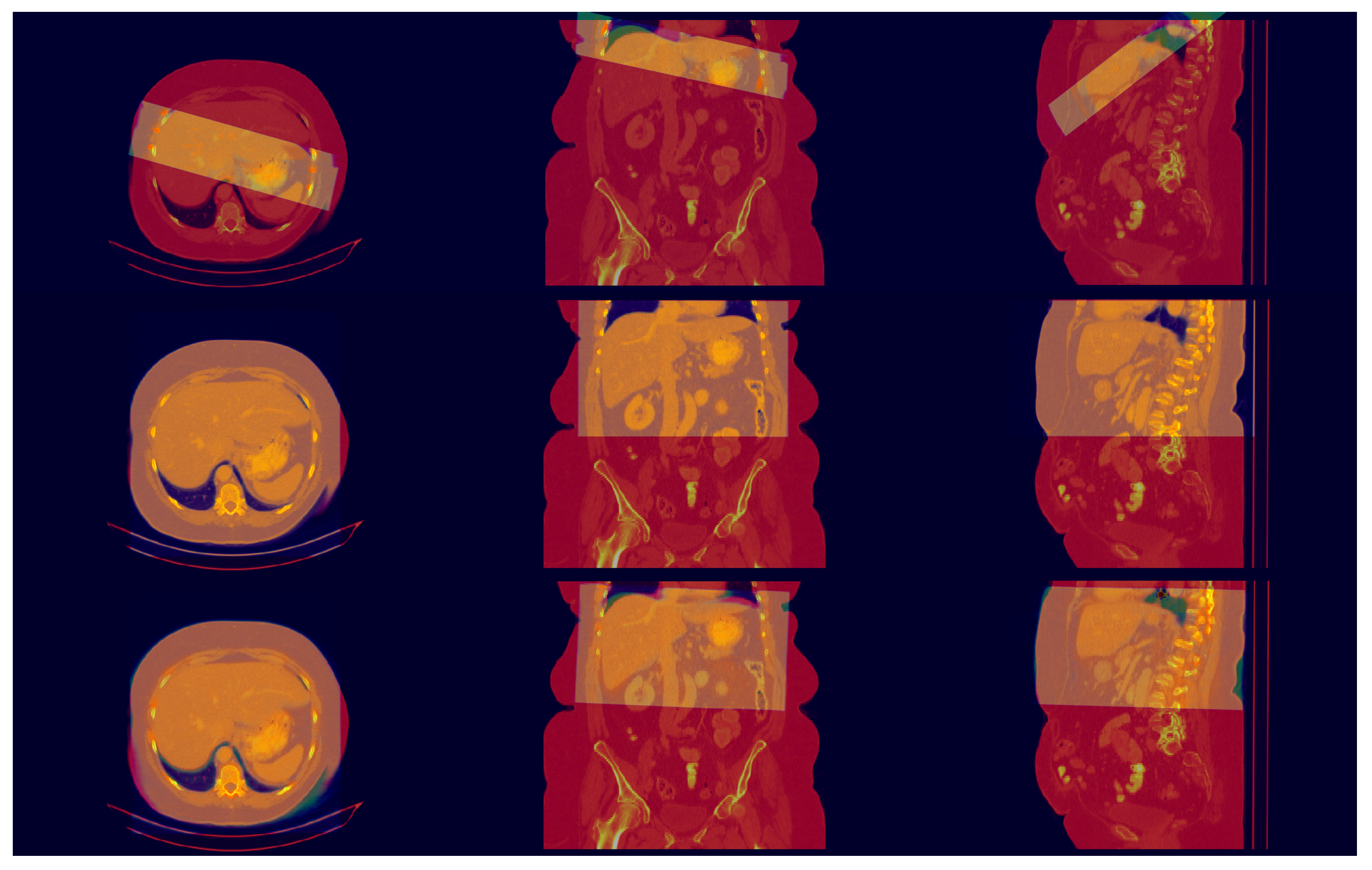
3.2. Registration Test
4. Discussion
4.1. Algorithm Selection
4.2. Overall Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RFA | Radiofrequency ablation |
| CT | Computer Tomograph |
| FOV | Field of view |
| GAN | Generative adversarial network |
| ROI | Region of interest |
| CNN | Convolutional neural network |
| IQR | Interquartile range |
| SSIM | Structural similarity index |
| PSNR | Peak signal to noise ratio |
Appendix A
References
- Mauro, M.; Murphy, K.; Thomson, K.; Venbrux, A.; Morgan, R. Image-Guided Interventions, 3rd ed.; Elsevier, Inc.: Philadelphia, PA, USA, 2020. [Google Scholar]
- Cleary, K.; Peters, T.M. Image-Guided Interventions: Technology Review and Clinical Applications. Annu. Rev. Biomed. Eng. 2010, 12, 119–142. [Google Scholar] [CrossRef] [PubMed]
- Modersitzki, J. Numerical Methods for Image Registration; Oxford University Press Inc.: New York, NY, USA, 2004; pp. 27–44. [Google Scholar]
- Liao, R.; Zhang, L.; Sun, Y.; Miao, S.; Chefd’Hotel, C. A Review of Recent Advances in Registration Techniques Applied to Minimally Invasive Therapy. IEEE Trans. Multimed. 2013, 15, 983–1000. [Google Scholar] [CrossRef]
- Zitová, B.; Flusser, J. Image Registration Methods: A Survey. Image Vis. Comput. 2003, 21, 977–1000. [Google Scholar] [CrossRef] [Green Version]
- Pluim, J.P.W.; Maintz, J.B.A.; Viergever, M.A. Mutual-information-based registration of medical images: A survey. IEEE Trans. Med. Imaging 2003, 22, 986–1004. [Google Scholar] [CrossRef] [PubMed]
- Mansi, T.; Passerini, T.; Comaniciu, D. (Eds.) Artificial Intelligence for Computational Modeling of the Heart, 1st ed.; Academic Press in an Imprint of Elsevier: San Diego, CA, USA, 2019. [Google Scholar]
- Fischer, A.; Klein, P.; Radulescu, P.; Gulsun, M.; Mohamed Ali, A.; Schoebinger, M.; Sahbaee, P.; Sharma, P.; Schoepf, U. Deep Learning Based Automated Coronary Labeling for Structured Reporting of Coronary CT Angiography in Accordance with SCCT Guidelines. J. Cardiovasc. Comput. Tomogr. 2020, 14, S21–S22. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Karras, T.; Aila, T.; Laine, S.; Lehtinen, J. Progressive Growing of GANs for Improved Quality, Stability, and Variation. International Conference on Learning Representations. 2018. Available online: https://openreview.net/forum?id=Hk99zCeAb (accessed on 9 March 2022).
- Karras, T.; Laine, S.; Aila, T. A Style-Based Generator Architecture for Generative Adversarial Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 4217–4228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, T.; Liu, M.Y.; Wang, T.C.; Zhu, J.Y. Semantic Image Synthesis with Spatially-Adaptive Normalization. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 16–20 June 2019; pp. 2332–2341. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Miao, S.; Mansi, T.; Liao, R. Task Driven Generative Modeling for Unsupervised Domain Adaptation: Application to X-ray Image Segmentation. arXiv 2018, arXiv:1806.07201. [Google Scholar]
- You, C.; Li, G.; Zhang, Y.; Zhang, X.; Shan, H.; Li, M.; Ju, S.; Zhao, Z.; Zhang, Z.; Cong, W.; et al. CT Super-resolution GAN Constrained by the Identical, Residual, and Cycle Learning Ensemble (GAN-CIRCLE). IEEE Trans. Med. Imaging 2019, 39, 188–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Yan, P.; Zhang, Y.; Yu, H.; Shi, Y.; Mou, X.; Kalra, M.K.; Zhang, Y.; Sun, L.; Wang, G. Low-Dose CT Image Denoising Using a Generative Adversarial Network with Wasserstein Distance and Perceptual Loss. IEEE Trans. Med. Imaging 2018, 37, 1348–1357. [Google Scholar] [CrossRef] [PubMed]
- Vizitiu, A.; Puiu, A.; Reaungamornrat, S.; Itu, L.M. Data-Driven Adversarial Learning for Sinogram-Based Iterative Low-Dose CT Image Reconstruction. In Proceedings of the 2019 23rd International Conference on System Theory, Control and Computing (ICSTCC), Sinaia, Romania, 9–11 October 2019; pp. 668–674. [Google Scholar] [CrossRef]
- Zhu, J.-Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2242–2251. [CrossRef] [Green Version]
- Karim, A.; Chenming, J.; Marc, F.; Thomas, K.; Tobias, H.; Konstantin, N.; Sergios, G.; Bin, Y. MedGAN: Medical Image Translation using GANs. Comput. Med. Imaging Graph. 2019, 79, 101684. [Google Scholar] [CrossRef]
- Iizuka, S.; Simo-Serra, E.; Ishikawa, H. Globally and Locally Consistent Image Completion. ACM Trans. Graph. 2017, 36, 107. [Google Scholar] [CrossRef]
- Liu, P.; Qi, X.; He, P.; Li, Y.; Lyu, M.R.; King, I. Semantically Consistent Image Completion with Fine-grained Details. arXiv 2017, arXiv:1711.09345. [Google Scholar]
- Yang, C.; Lu, X.; Lin, Z.; Shechtman, E.; Wang, O.; Li, H. High-Resolution Image Inpainting using Multi-Scale Neural Patch Synthesis. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 4076–4084. [Google Scholar] [CrossRef] [Green Version]
- Sabini, M.; Rusak, G. Painting Outside the Box: Image Outpainting with GANs. arXiv 2018, arXiv:1808.08483. [Google Scholar]
- Wang, Y.; Tao, X.; Shen, X.; Jia, J. Wide-Context Semantic Image Extrapolation. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 1399–1408. [Google Scholar] [CrossRef]
- Sumantri, J.S.; Park, I.K. 360 Panorama Synthesis from a Sparse Set of Images with Unknown Field of View. arXiv 2019, arXiv:1904.03326. [Google Scholar]
- Knuth, D.E. The Art of Computer Programming, 3rd ed.; Addison-Wesley: Reading, MA, USA, 1997; p. 232. [Google Scholar]
- Ulyanov, D.; Vedaldi, A.; Lempitsky, V. Instance Normalization: The Missing Ingredient for Fast Stylization. arXiv 2016, arXiv:1607.08022. [Google Scholar]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-Image Translation with Conditional Adversarial Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5967–5976. [Google Scholar] [CrossRef] [Green Version]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein GAN. arXiv 2017, arXiv:1701.07875. [Google Scholar]
- Gulrajani, I.; Ahmed, F.; Arjovsky, M.; Dumoulin, V.; Courville, A.C. Improved Training of Wasserstein GANs. arXiv 2017, arXiv:1704.00028. [Google Scholar]
- Johnson, J.; Alahi, A.; Fei-Fei, L. Perceptual Losses for Real-Time Style Transfer and Super-Resolution. In European Conference on Computer Vision; ECCV 2016: Computer Vision—ECCV 2016, Lecture Notes in Computer Science; Leibe, B., Matas, J., Sebe, N., Welling, M., Eds.; Springer: Cham, Switzerland, 2016; Volume 9906. [Google Scholar] [CrossRef] [Green Version]
- Isensee, F.; Kickingereder, P.; Wick, W.; Bendszus, M.; Maier-Hein, K. Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge. arXiv 2017, arXiv:1802.10508. [Google Scholar]
- Ghesu, F.C.; Georgescu, B.; Mansi, T.; Neumann, D.; Hornegger, J.; Comaniciu, D. An Artificial Agent for Anatomical Landmark Detection in Medical Images. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016; Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 229–237. [Google Scholar]
- Ghesu, F.C.; Georgescu, B.; Zheng, Y.; Grbic, S.; Maier, A.; Hornegger, J.; Comaniciu, D. Multi-Scale Deep Reinforcement Learning for Real-Time 3D-Landmark Detection in CT Scans. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 41, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Chenoune, Y.; Constantinides, C.; Berbari, R.; Roullot, E.; Frouin, F.; Herment, A.; Mousseaux, E. Rigid registration of Delayed-Enhancement and Cine Cardiac MR images using 3D Normalized Mutual Information. In Proceedings of the 2010 Computing in Cardiology, Belfast, UK, 26–29 September 2010; Volume 37, pp. 161–164. [Google Scholar]
- Mao, X.; Li, Q.; Xie, H.; Lau, R.Y.K.; Wang, Z.; Smolley, S.P. Least Squares Generative Adversarial Networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2813–2821. [Google Scholar] [CrossRef] [Green Version]
| Mean (±std) [mm] | ||
|---|---|---|
| Metric | Train Set | Test Set |
| SSIM | ||
| PSNR | ||
| Mean (±std) [mm] | ||
|---|---|---|
| Image | All Detected Landmarks | Landmarks Detected on Actual Info |
| Full volume | ||
| Thin volume | ||
| Extrapolated volume | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puiu, A.; Reaungamornrat, S.; Pheiffer, T.; Itu, L.M.; Suciu, C.; Ghesu, F.C.; Mansi, T. Generative Adversarial CT Volume Extrapolation for Robust Small-to-Large Field of View Registration. Appl. Sci. 2022, 12, 2944. https://doi.org/10.3390/app12062944
Puiu A, Reaungamornrat S, Pheiffer T, Itu LM, Suciu C, Ghesu FC, Mansi T. Generative Adversarial CT Volume Extrapolation for Robust Small-to-Large Field of View Registration. Applied Sciences. 2022; 12(6):2944. https://doi.org/10.3390/app12062944
Chicago/Turabian StylePuiu, Andrei, Sureerat Reaungamornrat, Thomas Pheiffer, Lucian Mihai Itu, Constantin Suciu, Florin Cristian Ghesu, and Tommaso Mansi. 2022. "Generative Adversarial CT Volume Extrapolation for Robust Small-to-Large Field of View Registration" Applied Sciences 12, no. 6: 2944. https://doi.org/10.3390/app12062944
APA StylePuiu, A., Reaungamornrat, S., Pheiffer, T., Itu, L. M., Suciu, C., Ghesu, F. C., & Mansi, T. (2022). Generative Adversarial CT Volume Extrapolation for Robust Small-to-Large Field of View Registration. Applied Sciences, 12(6), 2944. https://doi.org/10.3390/app12062944






