How Can a Clinical Data Modelling Tool Be Used to Represent Data Items of Relevance to Paediatric Clinical Trials? Learning from the Conect4children (c4c) Consortium
Abstract
:Featured Application
Abstract
1. Introduction
- (1)
- to enable data from the clinical trial environment to flow seamlessly into point of care health environments;
- (2)
- to validate and improve the content within the data dictionary, to make it semantically and contextually complete.
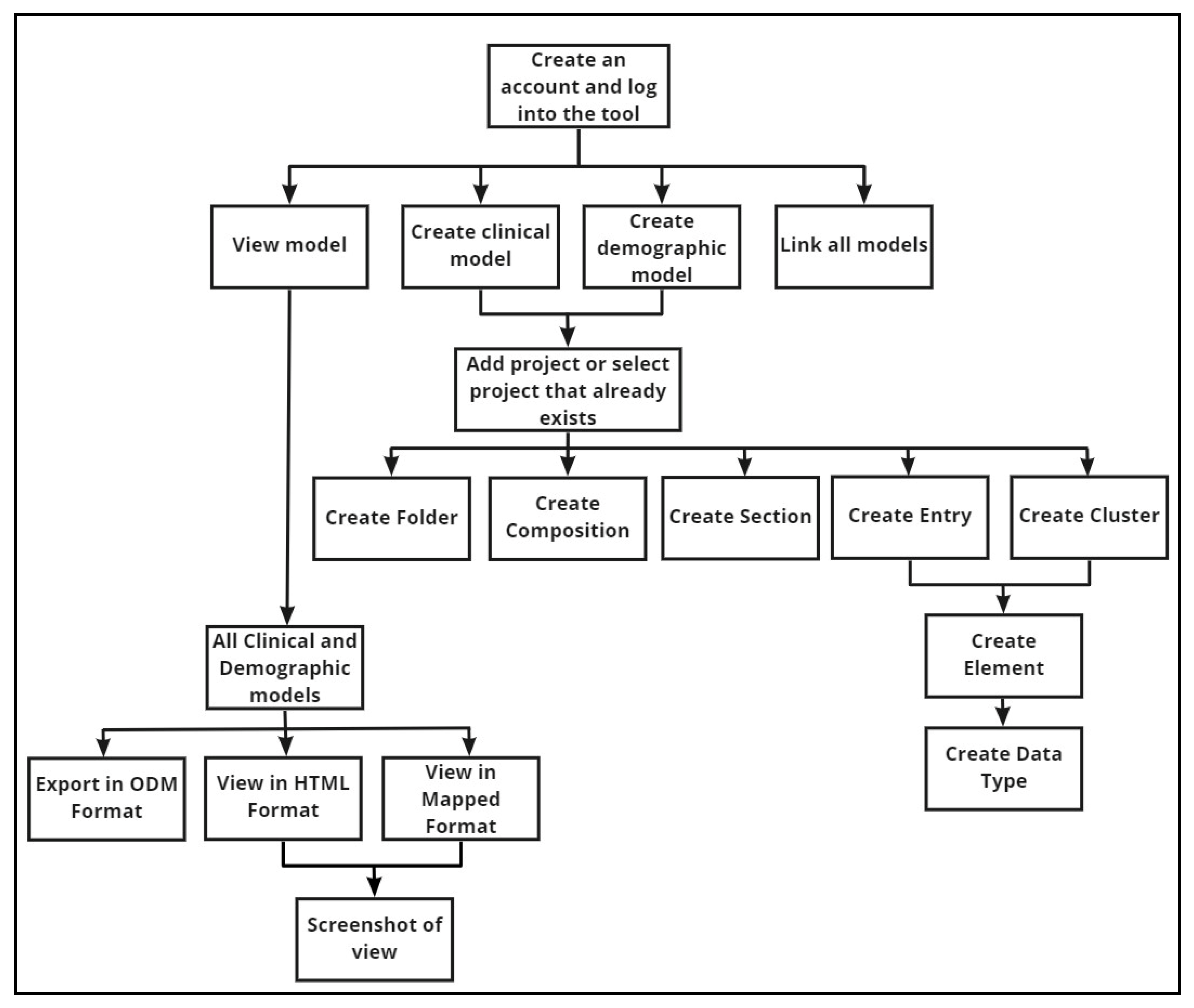
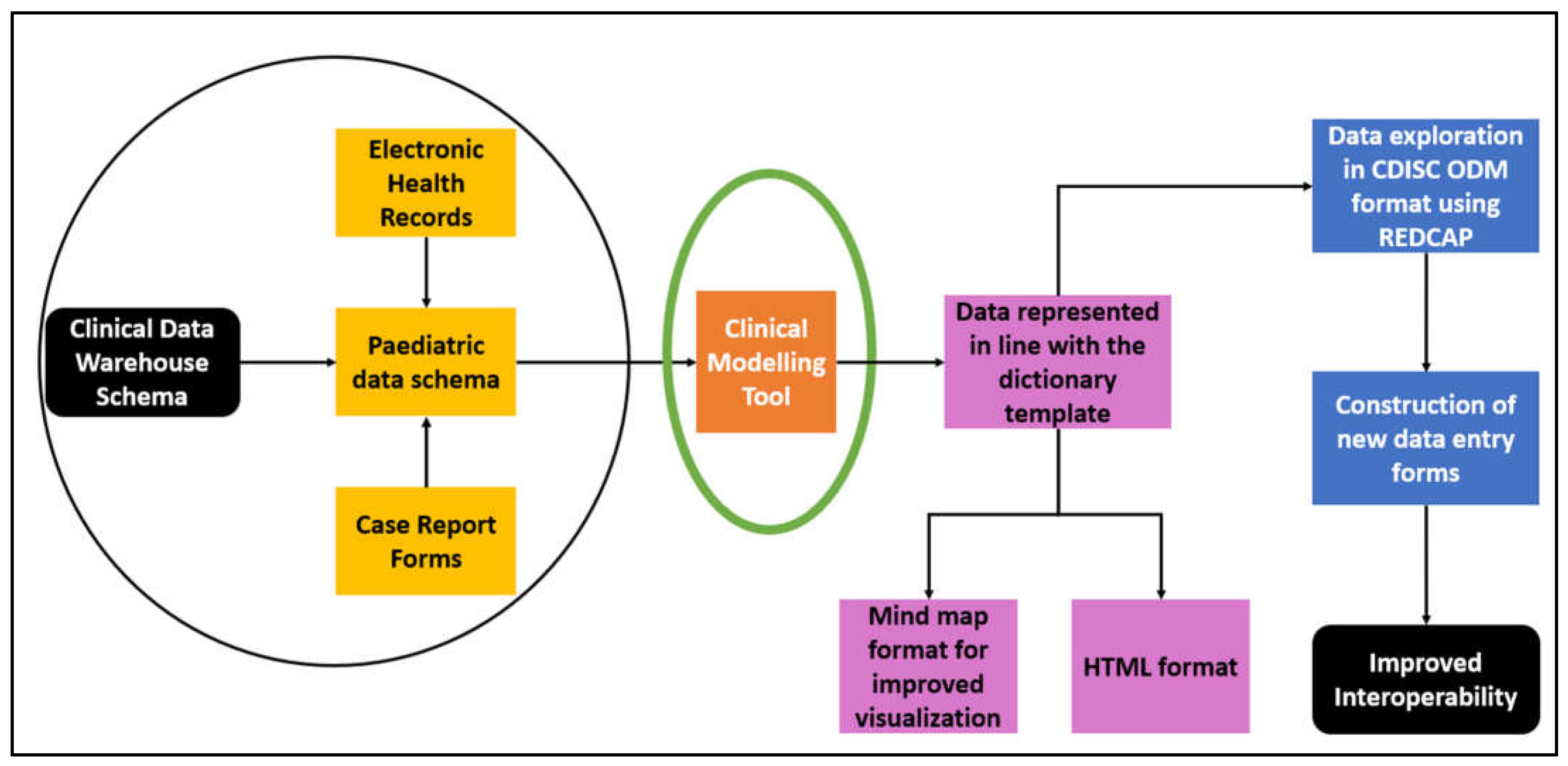
2. Materials and Methods
3. Results
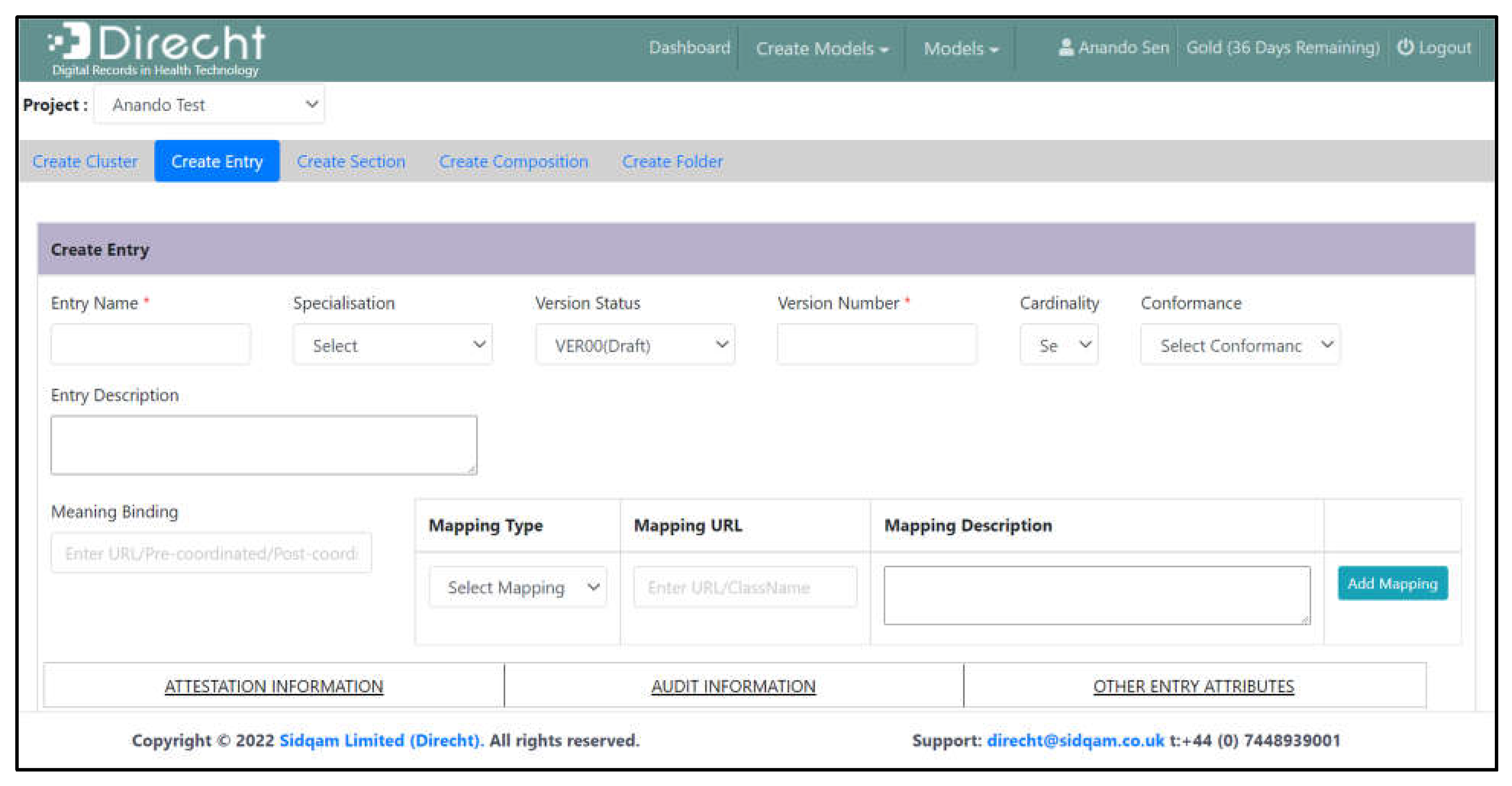
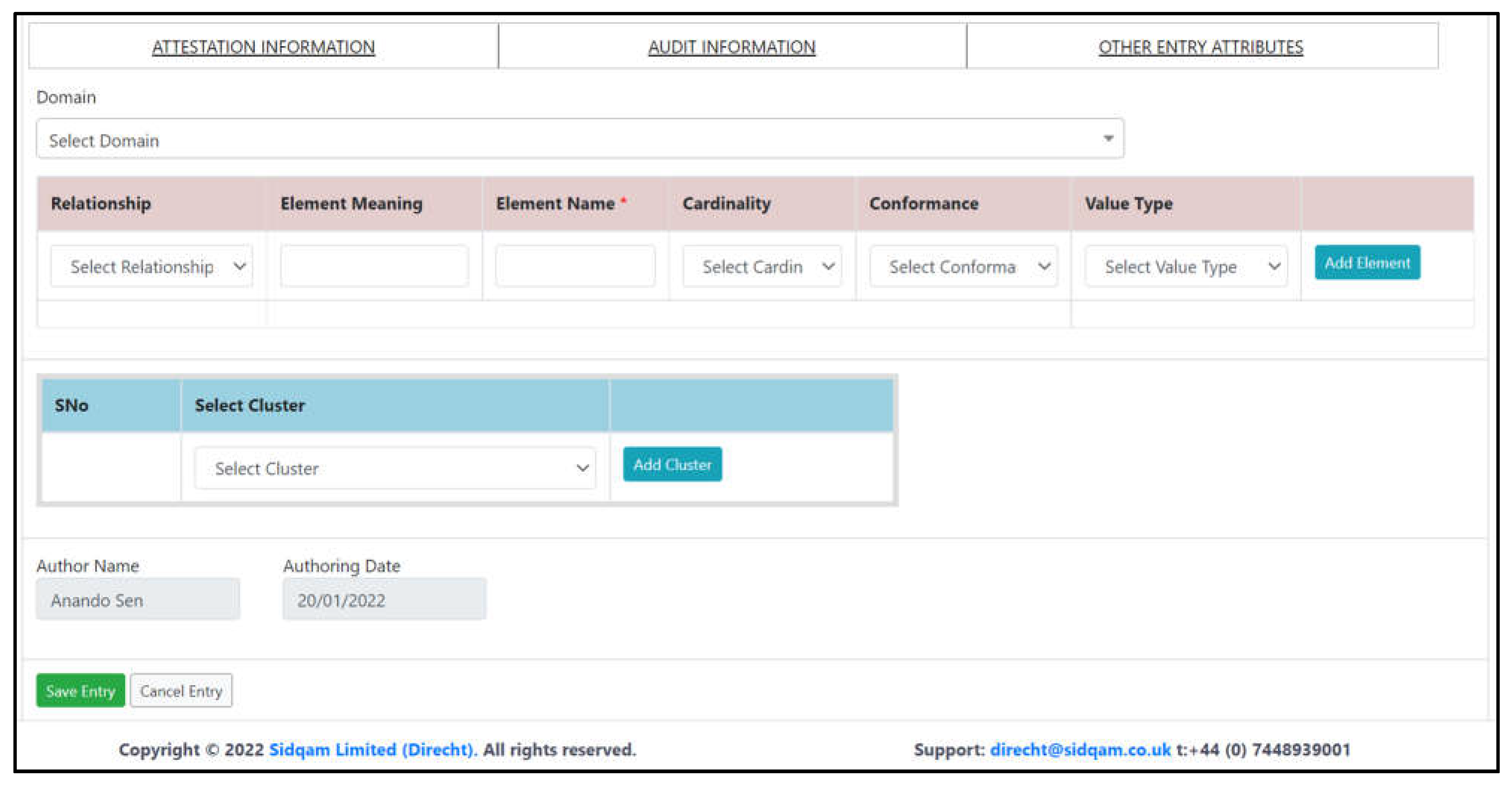
3.1. Facilitation of Good Quality Model Design
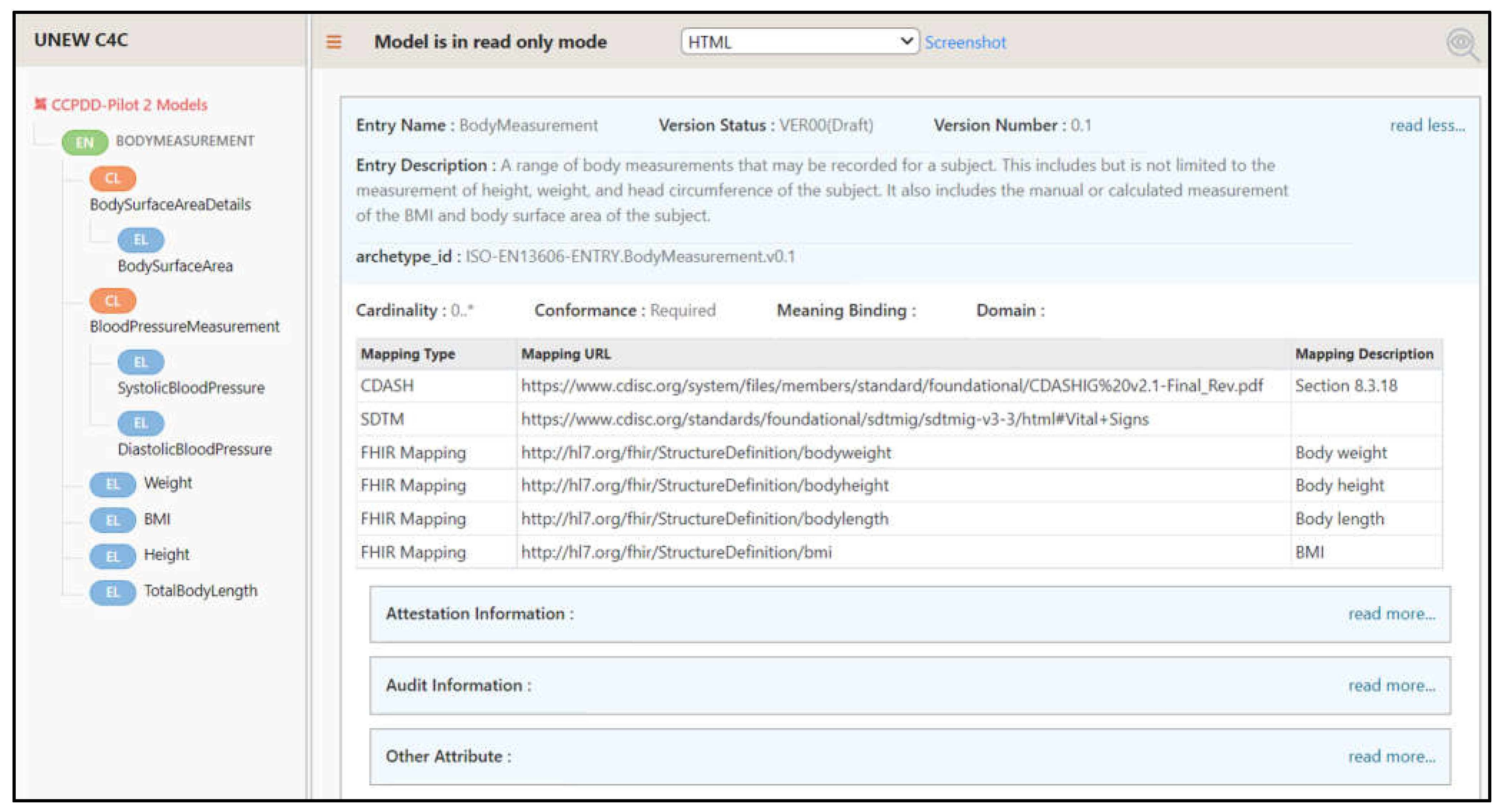
3.2. Data Visualisation
3.3. Export of Data in ODM Format
- The structure of the data model created two CRFs within REDCap, rather than one;
- REDCap took the first variable in the XML file as the Subject ID; however, the model did not have a Subject ID, and as a result, the first field in the data model was not usable in the REDCap project;
- Data were entered into Direcht without spaces, for example, ‘DateOfBirth’. This format was exported into REDCap, affecting the user experience.
3.4. Findings from the First Pilot Study
- A robust, consistent, and systematic approach to modelling items in a non-standardised format can be achieved. This approach also helped in highlighting ambiguity in data definitions, which were otherwise difficult to identify.
- Using modelling tools provided the benefits of following a disciplined manner of structuring data with its attributes, cardinalities, associations, and values. This rigour in data representation benefited downstream tools such as EDC and reporting tools, providing consistent implementations.
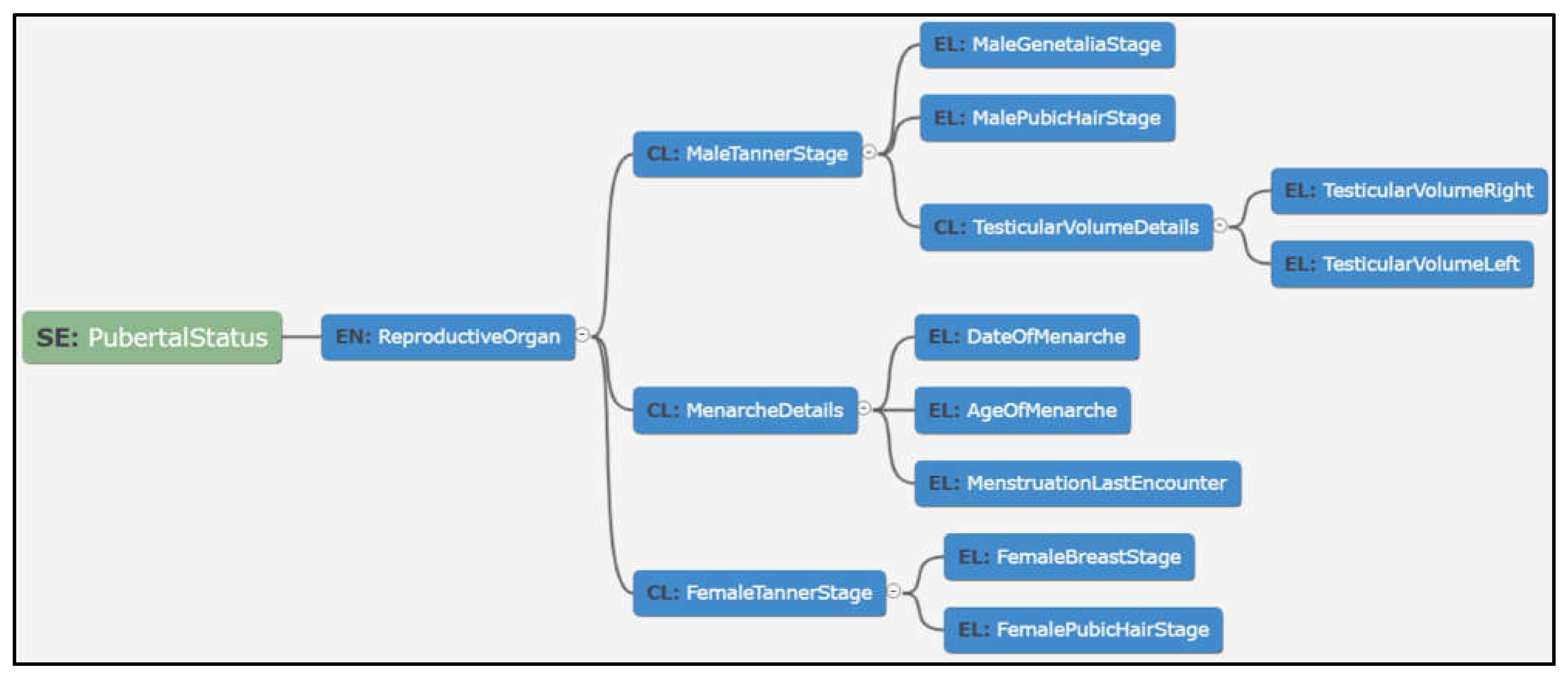
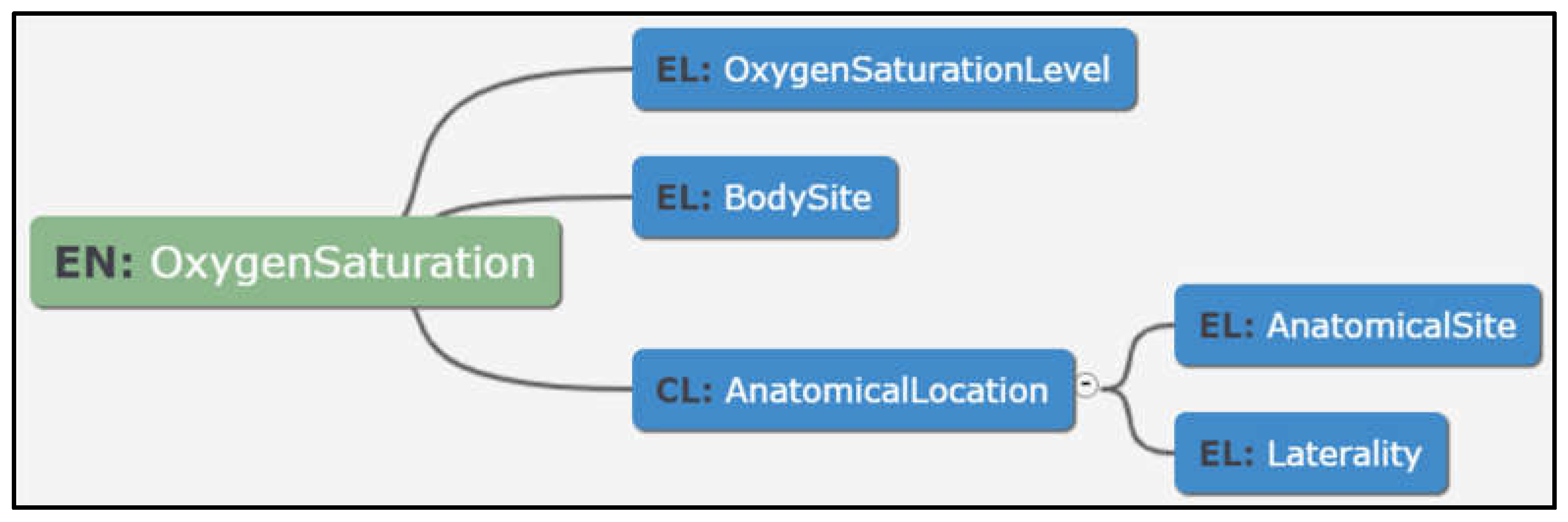
- Modelling in a tool-based environment enabled viewing data in multiple dimensions such as tabular and graphical views to suit the user preference. This resulted in more engagement in reviews and discussions, which ultimately helped in richer and unambiguous data models, as shown in Figure 10 below:
- Transformations to other standard formats including CDISC ODM can be achieved reducing the burden of manual mapping and human errors arising from such a process.
- Representing the data dictionary in the form of models and then exporting them to industry standards such as CDISC ODM enabled various EDC systems, e.g., REDCap, and used them to construct new data entry forms.
3.5. Benefits of Using a Clinical Modelling Approach over Microsoft Excel
- Guided dictionary entry authors as to how to specify new clinical models (data dictionary entries);
- Acted as a collaboration platform for virtual team model creation, capturing design comments and decisions;
- Ensured the quality of each model specification so that all necessary modelling properties were specified and there was internal consistency;
- Enabled seamless linkage to external terminology systems for defining value lists, measurement units, etc.;
- Easily searchable and browsable library of models to avoid the creation of duplicates or overlapping models and to facilitate their use;
- Easy specialisation of models: use one as a base pattern to create a specialised version for a specified purpose, e.g., to adapt an adult model for children or a child model to neonates;
- Models were version managed, distinguishing draft versions and published versions;
- Models can be grouped into sets for convenient reuse and sharing;
- Exportable to CDISC ODM, for import into EDC systems, e.g., REDCap;
- Exportable to formats that can be used within EHR systems and RWD repositories;
- Held in a standards agnostic form, but conforming to ISO modelling standards to avoid vendor lock-in.
4. Discussion
- Enhanced user friendliness and overall usefulness due to visualisation tools. Mind maps enable clinicians and other clinical trial personnel to better understand the step-wise format by which data dictionary items are represented for use in clinical trials [26];
- The HTML summary provides a ‘fact sheet’ of the data dictionary items for enhanced clarity in line with the clinical registry template [27];
- A clearer way of representing a large list of data dictionary items;
- The flexibility to allow generic ‘Clusters’ and ‘Entries’ to be reused for other models.
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Disclaimer
References
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009, 42, 377–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, M.A.; Hildebrand, H.; Fernandes, R.M.; De Wildt, S.N.; Mahler, F.; Hankard, R.; Leary, R.; Bonifazi, F.; Nobels, P.; Cheng, K.; et al. The conect4children (c4c) Consortium: Potential for Improving European Clinical Research into Medicines for Children. Pharmaceut. Med. 2021, 35, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Conect4children (c4c). Available online: https://conect4children.org/ (accessed on 20 January 2022).
- CDISC. Therapeutic Area Data Standards for Type 1 Diabetes—Pediatrics and Device Modules. Available online: https://www.cdisc.org/system/files/members/standard/ta/TAUG- (accessed on 20 January 2022).
- Health Level Seven International. HL7 Standards. Available online: https://www.hl7.org/ (accessed on 20 January 2022).
- Ohmann, C.; Banzi, R.; Canham, S.; Battaglia, S.; Matei, M.; Ariyo, C.; Becnel, L.; Bierer, B.; Bowers, S.; Clivio, L.; et al. Sharing and reuse of individual participant data from clinical trials: Principles and recommendations. BMJ Open 2017, 7, e018647. [Google Scholar] [CrossRef] [PubMed]
- Project, T.Y. Yale University Open Data Access (YODA) Project Procedures to Guide External Investigator Access to Clinical Trial Data. Available online: https://yoda.yale.edu (accessed on 20 January 2022).
- De Moor, G.; Sundgren, M.; Kalra, D.; Schmidt, A.; Dugas, M.; Claerhout, B.; Karakoyun, T.; Ohmann, C.; Lastic, P.Y.; Ammour, N.; et al. Using electronic health records for clinical research: The case of the EHR4CR project. J. Biomed. Inform. 2015, 53, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Claerhout, B.; Kalra, D.; Mueller, C.; Singh, G.; Ammour, N.; Meloni, L.; Blomster, J.; Hopley, M.; Kafatos, G.; Garvey, A.; et al. Federated electronic health records research technology to support clinical trial protocol optimization: Evidence from EHR4CR and the InSite platform. J. Biomed. Inform. 2019, 90, 103090. [Google Scholar] [CrossRef] [PubMed]
- Griffon, N.; Pereira, H.; Djadi-Prat, J.; Garcia, M.T.; Testoni, S.; Cariou, M.; Hilbey, J.; N’Dja, A.; Navarro, G.; Gentili, N.; et al. Performances of a Solution to Semi-Automatically Fill eCRF with Data from the Electronic Health Record: Protocol for a Prospective Individual Participant Data Meta-Analysis. Stud. Health Technol. Inform. 2020, 270, 367–371. [Google Scholar] [CrossRef] [PubMed]
- European Commission, Directorate-General for the Information Society and Media. Semantic Interoperability for Better Health and Safer Healthcare: Deployment and Research Roadmap for Europe; Virtanen, M., Ustun, B., Rodrigues, J., Stroetmann, V., Surjan, G., Rector, A., Stroetmann, K., Lewalle, P., Zanstra, P.E., Kalra, D., Eds.; European Commission: Brussels, Belgium, 2013. [Google Scholar]
- Beale, T. Archetypes: Constraint-based Domain Models for Future-proof Information Systems. 2002. Available online: https://www.researchgate.net/publication/237033734_Archetypes_Constraint-based_Domain_Models_for_Future-proof_Information_Systems (accessed on 20 January 2022).
- Garde, S.; Chen, R.; Leslie, H.; Beale, T.; McNicoll, I.; Heard, S. Archetype-Based Knowledge Management for Semantic Interoperability of Electronic Health Records. Stud. Health Technol. 2009, 150, 1007–1011. [Google Scholar] [CrossRef]
- Moreno-Conde, A.; Moner, D.; Cruz, W.D.; Santos, M.R.; Maldonado, J.A.; Robles, M.; Kalra, D. Clinical information modeling processes for semantic interoperability of electronic health records: Systematic review and inductive analysis. J. Am. Med. Inform. Assoc. 2015, 22, 925–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalra, D.; Tapuria, A.; Austin, T.; De Moor, G. Quality requirements for EHR Archetypes. Qual. Life Through Qual. Inf. 2012, 180, 48–52. [Google Scholar] [CrossRef]
- Ahn, S.; Huff, S.M.; Kim, Y.; Kalra, D. Quality metrics for detailed clinical models. Int. J. Med. Inform. 2013, 82, 408–417. [Google Scholar] [CrossRef] [PubMed]
- ISO. ISO 13606-2:2019. Health Informatics—Electronic Health Record Communication—Part 2: Archetype interchange specification. Available online: https://www.iso.org/standard/62305.html (accessed on 20 January 2022).
- ISO. ISO 13606-1:2019. Health Informatics—Electronic Health Record Communication—Part 1: Reference Model. Available online: https://www.iso.org/standard/67868.html (accessed on 20 January 2022).
- Moreno-Conde, A.; Austin, T.; Moreno-Conde, J.; Parra-Calderon, C.L.; Kalra, D. Evaluation of clinical information modeling tools. J. Am. Med. Inform. Assoc. 2016, 23, 1127–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tapuria, A.; Bruland, P.; Delaney, B.; Kalra, D.; Curcin, V. Comparison and transformation between CDISC ODM and EN13606 EHR standards in connecting EHR data with clinical trial research data. Digit Health 2018, 4, 2055207618777676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Direcht. Available online: https://www.direcht.com/ (accessed on 20 January 2022).
- Martinez-Costa, C.; Menarguez-Tortosa, M.; Fernandez-Breis, J.T. Towards ISO 13606 and openEHR archetype-based semantic interoperability. Stud. Health Technol. Inform. 2009, 150, 260–264. [Google Scholar] [PubMed]
- Austin, T.; Sun, S.; Hassan, T.; Kalra, D. Evaluation of ISO EN 13606 as a result of its implementation in XML. Health Inform. J. 2013, 19, 264–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CDISC. Therapeutic Area User Guides. Available online: https://www.cdisc.org/standards/therapeutic-areas (accessed on 20 January 2022).
- Yamamoto, K.; Ota, K.; Akiya, I.; Shintani, A. A pragmatic method for transforming clinical research data from the research electronic data capture “REDCap” to Clinical Data Interchange Standards Consortium (CDISC) Study Data Tabulation Model (SDTM): Development and evaluation of REDCap2SDTM. J. Biomed. Inform. 2017, 70, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Gregson, C. Clinical trial data visualisation. Trials 2015, 16, P187. [Google Scholar] [CrossRef]
- Hassanzadeh, O.; Lim, L.; Kementsietsidis, A.; Miller, R.J.; Wang, M. LinkedCT: A Linked Data Space for Clinical Trials. arXiv 2009, arXiv:0908.0567. [Google Scholar]
- Meineke, F.A.; Staubert, S.; Lobe, M.; Winter, A. A comprehensive clinical research database based on CDISC ODM and i2b2. Stud. Health Technol. Inform. 2014, 205, 1115–1119. [Google Scholar] [PubMed]



| Issue | Explanation |
|---|---|
| Version control | As the Cross Cutting Paediatric Data Dictionary (CCPDD) grew and new versions were developed, there was a risk that obsolete versions of it were used. The size of the conect4children (c4c) consortium made this more likely. |
| Complexity | As the CCPDD grew, it became more cumbersome to manage. Adding new worksheets to the spreadsheet made the document less user-friendly. |
| Modelling errors | Risk of data dictionary concepts being modelled differently in different domains by different teams. |
| Linkage | Due to the flat structure of Microsoft Excel, it was challenging to show nested structures and interlinking items. |
| Semantic detail | Not all structure and semantic specifications can be captured, and there was no way of prompting authors about missing data properties. |
| Usability | As the CCPDD grew, visualisation became increasingly important to help users to understand what the dictionary contains, and what they were now in the process of authoring. This functionality was not possible in Microsoft Excel. |
| Only one standard used (CDISC) | Predominantly focussed on one standard (Clinical Data Interchange Standards Consortium (CDISC)): this reduced opportunities to link items to data from electronic health records. |
| Integration to other IT tools | The Microsoft Excel version required transcription into electronic case report form (eCRF)-generating tools. |
| Feature | Practical Implication |
|---|---|
| Interoperability between standards | Creates a bridge between the clinical and research worlds. Potential to combine standardised data from different sources. |
| User friendliness | Heightened understanding of the representation of clinical paediatric data items. Personnel less familiar with data standards can use the models, resulting in a wider pool of standardised data. This could be of particular importance to academic studies. |
| Automated eCRF creation potential | Researchers can export models into their EDC systems. This avoids any human error in applying standards and makes it easier for personnel, particularly those new to implementing data standards at the CRF level. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amadi, C.; Leary, R.; Palmeri, A.; Hedley, V.; Sen, A.; Siddiqui, R.Q.; Kalra, D.; Straub, V. How Can a Clinical Data Modelling Tool Be Used to Represent Data Items of Relevance to Paediatric Clinical Trials? Learning from the Conect4children (c4c) Consortium. Appl. Sci. 2022, 12, 1604. https://doi.org/10.3390/app12031604
Amadi C, Leary R, Palmeri A, Hedley V, Sen A, Siddiqui RQ, Kalra D, Straub V. How Can a Clinical Data Modelling Tool Be Used to Represent Data Items of Relevance to Paediatric Clinical Trials? Learning from the Conect4children (c4c) Consortium. Applied Sciences. 2022; 12(3):1604. https://doi.org/10.3390/app12031604
Chicago/Turabian StyleAmadi, Chima, Rebecca Leary, Avril Palmeri, Victoria Hedley, Anando Sen, Rahil Qamar Siddiqui, Dipak Kalra, and Volker Straub. 2022. "How Can a Clinical Data Modelling Tool Be Used to Represent Data Items of Relevance to Paediatric Clinical Trials? Learning from the Conect4children (c4c) Consortium" Applied Sciences 12, no. 3: 1604. https://doi.org/10.3390/app12031604
APA StyleAmadi, C., Leary, R., Palmeri, A., Hedley, V., Sen, A., Siddiqui, R. Q., Kalra, D., & Straub, V. (2022). How Can a Clinical Data Modelling Tool Be Used to Represent Data Items of Relevance to Paediatric Clinical Trials? Learning from the Conect4children (c4c) Consortium. Applied Sciences, 12(3), 1604. https://doi.org/10.3390/app12031604






