1. Introduction
Lactic acid bacteria (LAB) are an economically important group of microorganisms that have utility in the food industry, in clinical settings, and the environment [
1]. The ability to detect and enumerate a range of lactic acid bacteria is relevant to, for example, the identification of beer-spoilage organisms in the brewing industry [
2], in characterizing persistent endodontic infections in dentistry [
3], and in monitoring recreational water for fecal contamination in municipal settings [
4]. Genera belonging to the LAB include
Lactobacillus,
Streptococcus,
Vagococcus, and
Enterococcus. Simple and rapid techniques for enumerating LAB that could be operated by non-specialists would, therefore, find application in numerous settings. Lactic acid bacteria belong to an exclusively Gram-positive phylum, the firmicutes, and are fermentative organisms distinguished by their inability to produce heme [
5]. Even though LAB possess many of the components of respiratory chains, they do not respire on account of their inability to synthesize functional heme-containing cytochromes that typically act as the terminal reductases in oxidative respiratory chains [
6].
Recently, methods for bioelectroanalytical detection of low numbers of organisms in environmental samples have been described [
7]. The technique involves tagging enzyme-specific substrates to electrochemical reporters to achieve specific detection of a target organism by exploiting a phenomenon known as extracellular electron transfer (EET)—the metabolic process that transports electrons from the cytosol to the exterior of a cell [
8,
9]. When the electrochemical reporter, or redox mediator, is released into the medium, it is indicative of the presence of the target organism; the redox mediator is reduced metabolically and subsequently reduces the electrode. EET has been extensively studied, particularly in the model Gram-negative electrogens
Geobacter spp. and
Shewanella spp., and is usually described as a type of anaerobic respiration achieved by heme-containing electron transfer protein complexes [
9,
10]. The environmental significance of EET is in biogeochemical cycling of metal oxides in the subsurface and is achieved biologically, as microbes transport electrons across their membranes and reduce solid terminal electron acceptors, such as Fe (III) or Mn (V), in a process that yields metabolic energy in the form of ATP [
11,
12]. The practical role of respiration in achieving a good detection signal in bioelectroanalytical systems was recently deduced from the protracted detection times for
Escherichia coli (
E. coli) strains with deficient respiratory chains that are only able to grow by fermentation [
13]. Therefore, the utility of bioelectroanalytical systems to
rapidly detect lactic acid bacteria is unknown, as LAB are unable to respire and instead gain their metabolic energy exclusively by fermentation.
EET is well described in Gram-negative organisms but has typically remained more obscure in Gram-positive organisms, except for a few notable exceptions, and despite Firmicutes regularly turning up in the phylogeny of bioelectrochemical systems [
14,
15,
16]. Recent research suggests that EET in Gram-positive bacteria is evolutionarily more ancient than in Gram-negative organisms [
17]. Lately, it has become commonplace to describe EET mechanisms in Gram-positive organisms that include LAB. Light et al. (2019) recently described a flavin-based EET mechanism in
Listeria monocytogenes and electrode-dependent growth that is distinct from more well-described mechanisms of EET [
9]. Similarly, EET has been described in another clinically important LAB,
Enterococcus faecalis (E. faecalis), and although the exact mechanism is yet to be elucidated, its ability to reduce an electrode and Fe (III) has been demonstrated [
8,
18,
19]. In addition, for both
Listeria monocytogenes and
E. faecalis, EET has been implicated in virulence either through increased competitive capabilities, enhanced biofilm potential, or through EET mediated synergistic interactions with commensals. In these systems, however, basic quantitative assessments of the EET process is not always reported.
In light of the recent insights into the EET in LAB, we decided to revisit the idea of their compatibility in bioelectroanalytical detection. The aim of this contribution is to determine whether a clinically relevant member of the LAB,
E. faecalis, is amenable to rapid detection and enumeration in a bioelectroanalytical system (
Figure 1). A secondary aim of this contribution is to comment on the quantitative importance of EET mechanisms in
E. faecalis by looking at EET efficiency. To our knowledge, this is the first report describing the potential for specific and swift bioelectroanalytical LAB detection and to quantitatively assess the efficiency of the EET process and how this relates to EET mechanisms of
E. faecalis in the context of biosensing.
3. Results and Discussion
E. faecalis can easily be detected and enumerated using the proposed detection framework described previously [
7,
13]. Chronoamperometric analysis of
E. facecalis reveals that all test inocula, with the exception of 10
7 CFU mL
−1 where the current onset is almost instant, produce a distinctive curve showing an initial flat period of baseline current representing lag phase growth followed by a sharp increase in current generation resulting from increased metabolic activity (and thus EET) as the culture enters the exponential growth phase (
Figure 2A). A previously reported method defines a detection event when the slope of the chronoamperometric readout exceeds five standard deviations of the baseline current for more than five consecutive time points [
13]. We apply a more conservative definition here and define detection as the time at which the current passes a threshold value (20 µA). This is an objective and robust way to define a detection time and yields a linear calibration curve that inversely correlates with inoculum size. For every log fold increase in inoculum size ranging from 10,000 to 10,000,000 CFU mL
−1, the detection time increased linearly, yielding mean detection times of 235 (±16), 148 (±12), 62 (±12), and 32 (±8) min for 10
4, 10
5, 10
6, and 10
7 CFU mL
−1, respectively, where the values in parentheses represents the standard deviation of three replicates. This observation is easily interpreted from the relationship between inoculum density and the duration of the lag phase in classical growth curve theory. This property can be used to construct a linear standard curve with a regression coefficient (r
2) of 0.96 (
Figure 2B). Additionally, the standard deviation of the mean detection time for three replicates is between 7 and 23%, which is comparable to previous reports and also to commercially available systems for bacterial detection [
4,
7,
20]. In short, the bioelectroanalytical system is an effective means of enumerating
E. faecalis and should be further developed to effect real world detection of lactobacilli in the multitude of settings for which they are relevant. This will require development of selective medium to suppress non-target organisms that are specific to the individual settings.
The detection compounds used here are electroactive glycosides comprising glucose conjugated to resorufin. When the glycosidic bond is cleaved by native glucosidases expressed by
E. faecalis, the electroactive resorufin is liberated from the pyranose ring and subsequently reduced by microbial electron carriers to dihydroresorufin [
21]. Following its reduction by microbes, the mobile dihydroresorufin in turn reduces the electrode generating a current proportional to the metabolic activity of the culture [
22]. Microbial metabolic coupling to an electrode via a mobile redox mediator is well documented and usually described as a component of an energy-yielding type of respiration called mediated EET [
23]. The quantitative importance of respiration compared to fermentation in a similar bioelectroanalytical detector was recently demonstrated by contrasting mediated detection times achieved with wildtype
E. coli vs. a mutant,
E. coli SHSP 18—an
E. coli K12 derivative that is auxotrophic for δ-aminolevulinic acid, a growth factor critical in heme synthesis and therefore respiration [
13,
24]. The wildtype
E. coli detection time, for an inoculum size of 5000 CF, was about 6 h compared with the detection time of around 14 h for the SHSP 18 mutant that was unable to respire. Thus, from this previously reported study, it appears that fermentative metabolism is not ideally compatible with bioelectroanalytical detection even in the presence of mediators.
The detection time for
E. faecalis observed here (
Figure 2A) cannot be directly compared to the previous studies, but it is likely to be similar. For a higher inoculum size of 10,000 CFU, double that reported for the
E. coli SHSP mutant, the detection time is around three hours vs. a six-hour detection time for 5000 CFU of wildtype
E. coli and 14 h for the fermentative strain [
13]. The detection time achieved here for 10,000 CFU
E. faecalis resembles respiratory behavior.
The genome of
E. faecalis is reportedly deficient in heme, but supplementation of this growth factor and or menaquinone induces respiratory behavior in
E. faecalis by activating the redox center of cytrochrome bd and providing a quinone pool to transfer electrons from NADH
+ to terminal reductases. Respiratory behavior is defined in this sense by observations of increased vitality or biomass production accompanied by less acidification of the medium from lactate accumulation [
25].
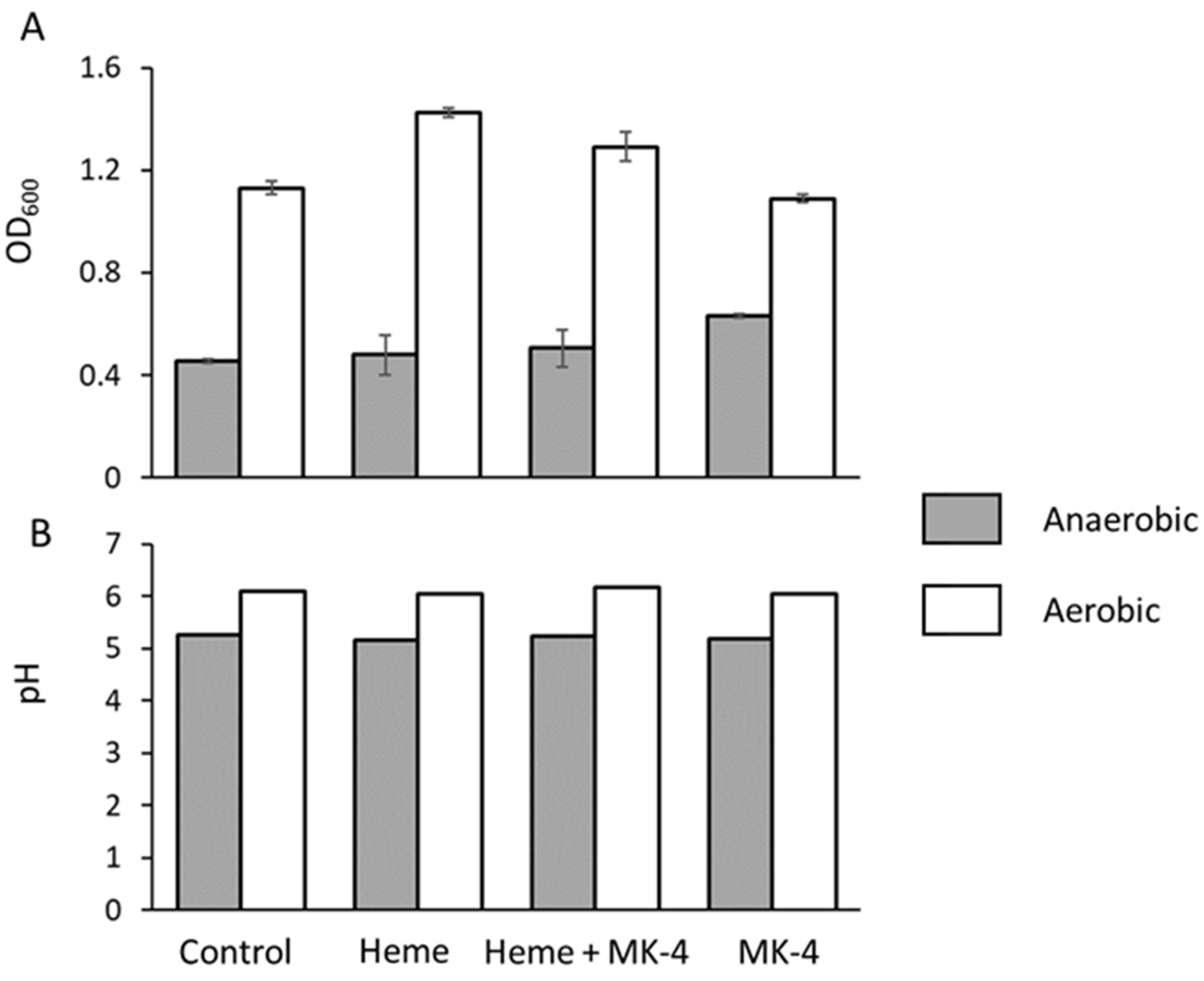
Upon analyzing the growth yields of
E. faecalis in bench top experiments, we observed that the optical density (OD
600) was always higher in aerobic conditions than it was in anaerobic ones (
Figure 3A). This phenomenon was reported previously [
26] and it is suggestive of respiratory behavior and could arise from residual heme or quinoids in the undefined medium that we used as was reported for the Todd Hewitt broth r by Del Papa and Perego (2008). Further analysis shows that the addition of heme, menaquinone, or both does not alter this trend, although menaquinone does give a slight boost in growth, observed by an increase in OD in anaerobic conditions, suggesting a role in managing reactive oxygen species (
Figure 2A). A comparative analysis of Lactobacilli electron transport chain stimulation by heme and menaquinone by Brooijman et al. (2009) [
25] showed similar behavior in
E. faecalis, i.e., that
E. faecalis growth is stimulated by menaquinone and, to greater extent, by the addition of both heme and menaquinone. However, the difference in pH between ‘fermentative’ and ‘respiratory’ growth ranged only between 0.2 pH units (5.62–5.82) [
25], although growth increased by 38%. Under equivalent conditions, we observed a quantitatively lower increase in growth (OD
600) of around 14% and the pH to be similarly stable (
Figure 3B). We observed a pH with a delta (Δ
pH) of only 0.13 pH units but an overall moderately lower pH of 5.14 and 5.27 between the unsupplemented control and supplemented conditions, respectively. The Δ
pH between fermentative LAB and the supplemented treatment thathad been induced to respire was previously reported to be greater what we have observed here, and the observed Δ
pH is usually close to a single pH unit; for example, the Δ
pH between a heme and menaquinone-stimulated
Streptococcus entericus culture compared to an unsupplemented control was 1.07, with the final pH being 4.42 and 5.49 for fermentative and respiratory conditions, respectively [
25]. Taken together, although
E. faecalis exhibits less respiratory stimulation upon the addition of heme in BHI medium relative to other LAB, it is clear that it does derive some growth benefits from medium supplementation with respiratory components. Despite the small difference in pH between respiratory and fermentative growth in
E. faecalis, Broojiman (2009) still concluded that
E. faecalis is both heme and menaquinone stimulated [
25].
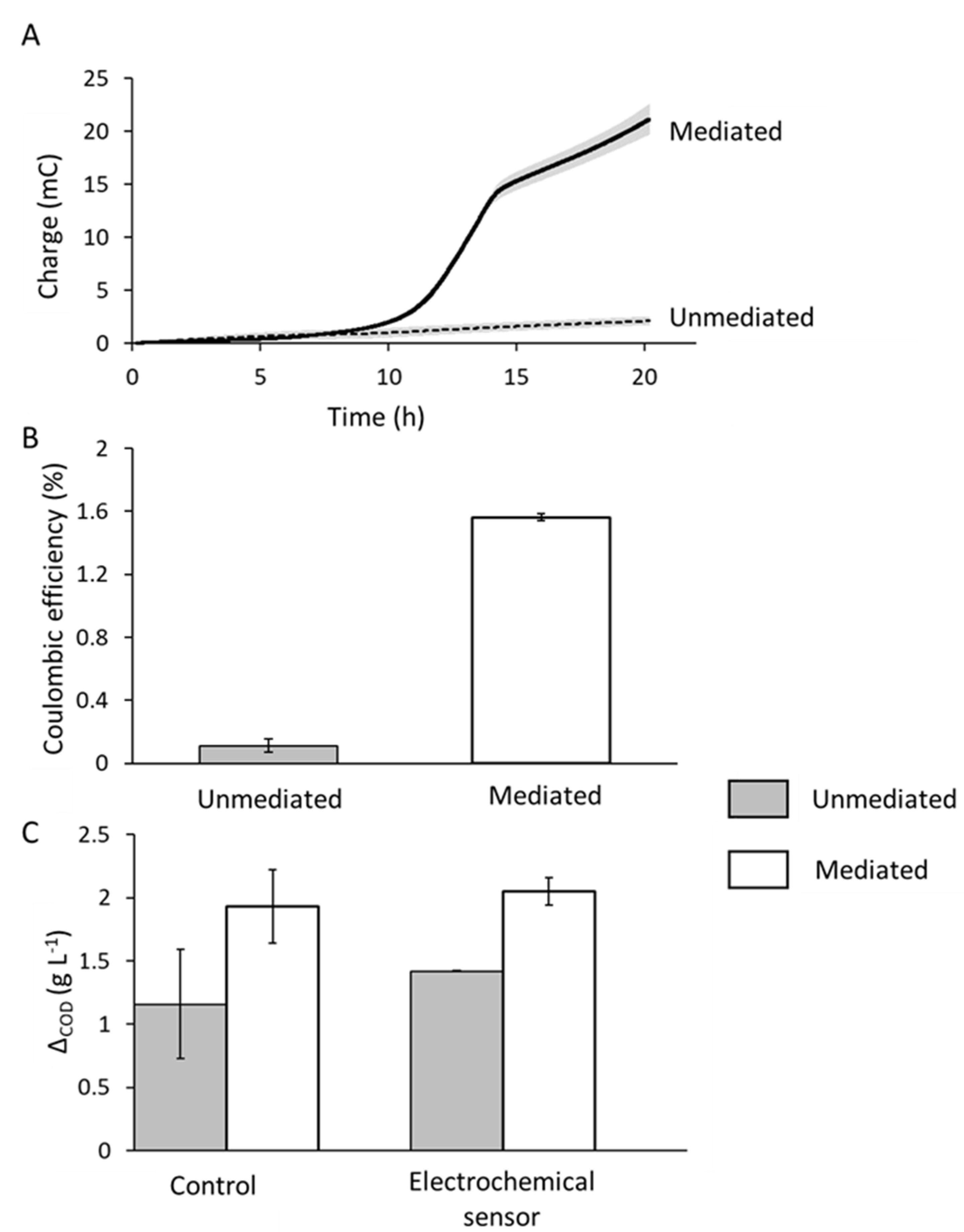
Concluding that
E. faecalis OG1RF will grow vigorously and that its response to additive respiratory components is minimal, we proceeded to examine the quantitative extent of
E. faecalis EET in our microscale detectors using BHI as a medium and the redox active aglycone used in the proposed detection approach, resorufin, to effect electron transport. The charge generated by
E. faecalis was substantially greater (20 mC) when mediated with resorufin than the control current which barely increased above a baseline current of around 2 mC (
Figure 4A). Over a 20 h period, around 600 mg L
−1 COD more was consumed in the mediated system (Δ
COD = 2020 mg L
−1) compared to the unmediated system (Δ
COD = 1420 mg L
−1), which is suggestive of an increase in EET by
E. faecalis upon the addition of the mediator (
Figure 3B). However, the response of
E. faecalis to mediator addition in terms of COD consumption was similar in both benchtop controls and in bioelectrochemical systems. The Δ
COD in equivalent reactors but without the electrode and incubated under identical conditions was 1160 and 1920 mg L
−1 for the unmediated and mediated systems, respectively, equating to 760 mg L
−1 more COD consumed in the benchtop system that was supplemented with resorufin compared with the unsupplemented control. Thus, while the electrochemical behavior of
E. faecalis is stimulated with the addition of redox mediators, so too is a planktonic culture, and this is reflected in the observation in
Figure 4A as well as with the growth studies in
Figure 3A, where the addition of menaquinone only resulted in some growth stimulation under anaerobic conditions. This is also in keeping with observations by other researchers [
25]. Coulombic efficiency (CE) of the electrochemical response of
E. faecalis is low: 1.6% in the mediated system and 0.1% in the unmediated system (
Figure 4B).
EET is increasingly being described in Firmicutes isolated from the gut [
8,
9,
27,
28] and, more recently, in situ directly by inserting electrodes into a mouse gut and comparing the output with germ-free organisms [
29]. These studies, in the main, have lacked a quantitative assessment of the electron flux as a function of the carbon utilization in the system. Where columbic efficiency (CE) is reported, it is low (typically <1%) and of a comparative magnitude to that reported here. For example, similarly diminutive CE observations were made by Naradasu et al. (2019), where a CE of only 0.02% was reported for a gut-isolated
Entrococcacea sp. with a 99% 16 S RNA sequence similarity to
E. avium [
27]. The question therefore remains whether EET in many LAB-based systems, either to an electrode or in the reduction in metal oxides (e.g., the commonly reported Fe (III) reduction), is a respiratory phenomenon or if it is a strategy for Firmicutes to exert redox control over their environment, e.g., to enable nutrient uptake or even mitigate metal toxicity, or even if it is just redox leakage. Since the first option is involved in energy conservation, and the other comes at a metabolic cost, the distinction is an important one. However, the low CE of EET in LAB systems reported to date suggest that such a strategy, if it used to conserve energy, is a minor one.
Mechanisms for EET in Firmicutes have been reported to be flavin dependent [
9,
28], although this does not necessarily mean that the flavins are mobile redox mediators as proposed in
Shewanella spp. Light et al. (2018) and Hederstedt (2020) identified key genes for EET in Firmicutes. In
L. monocytogenes, the important EET gene cluster appears to contain a flavoprotein (PplA) a type-two dehydrogenase (NdH2), and two small proteins, EetA and EetB, as well as genes for quinone synthesis. Orthologous genes in
E. faecalis,
ppl3 and
ndhA, appear to have some role in EET, but alternative dehydrogenases Ndh2 and Ndh3, as well as EetA and EetB, also have a role depending on the type of EET mechanism at play. It appears that Ndh3 and EetA are essential to EET in wildtype
E. faecalis, i.e., where there is no supplementary heme and, hence, they are not respiring (Hederstedt, 2020). Under such conditions, in our systems at least, there appears to be both a growth benefit and an increase in carbon utilization upon introducing a redox mediator. While a useful detection signal (current) from
E. faecalis can only be recorded at an electrode in the presence of a mediator, the growth benefit to
E. faecalis of such an addition appears to be independent of the electrode. Additionally, Hederstedt (2020) and Pankratova (2018) suggest that EET is always promoted by the presence of a mediator and that EET is
curtailed by the activation of cytochromes [
8]. Taken together, the relatively low bioelectricity yield of
E. faecalis, the absence of advanced COD consumption at the electrode, and the fact that assembly of a fully functioning electron transport chain reportedly attenuates EET in
E. faecalis, suggests that the observed current production in our systems is incidental. This may arise from the fact that the quinone pool (which has been shown essential to all EET described in Firmicutes to date) may become charged with electrons and can provide reducing power for other biological processes that are not essential to conserving energy. Alternatively, the current signal in these systems may arise from leakage, a phenomenon well known in mammalian electron transport chains and that produces free radicals [
30]. Nonetheless, the reducing power captured here is sufficient to achieve detection that is likelybe more rapid than traditional techniques.
Accordingly, the question arises of whether it is helpful to describe the bioelectricity production observed here in E. faecalis detection systems as EET. If the phenomenon is not special, i.e., that it applies to most organisms and it is not quantitatively substantial, then the focus should be on the analytical procedure describing EET rather than the biological significance, i.e., the ability to sense the metabolism of a particular organism rather than the ‘special qualities’ of that organism’s metabolism. In this investigation, we observed CE comparable to that attributed to an electrochemically isolated organism in a member of the same genus. Recognizing the need to objectively screen for electrogenicity, Zhou et al. (2015) proposed a rapid colorimetric screening procedure to assign EET capabilities to different microbial genera. They only screened one member of the Firmicutes, and they did not assign to it the ability to carry out EET when it is objectively compared to known EET-capable genera.