Electrochemical Control of Biofilm Formation and Approaches to Biofilm Removal
Abstract
1. Introduction
2. Biofilms
2.1. Bacteria Forming Biofilms
2.2. Influence of Environment on Biofilm Formation
Effect of Metal Ions on Biofilm Formation
2.3. Biofilm Structure in Biofilm Systems
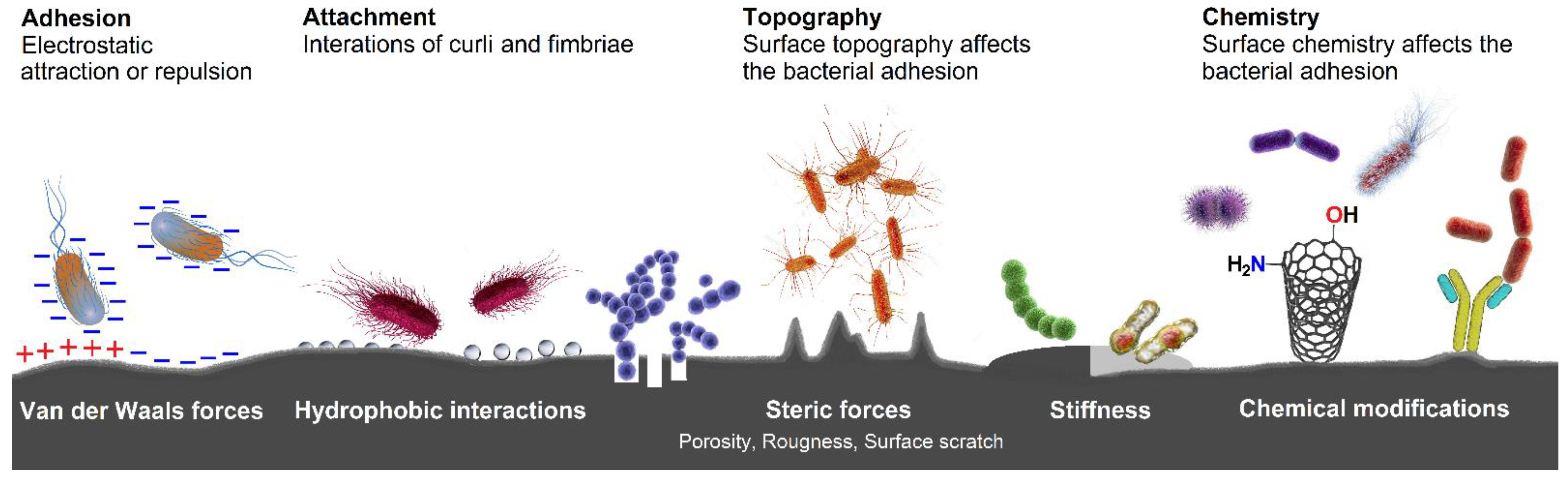
2.4. Bacterial Adhesion and Biofilm Formation
3. Biofilms in Food Industry
3.1. Health Aspects
3.2. Biofilm Control Methods in Food Industry
3.2.1. Surface Modification of Contact Material
3.2.2. Natural Compounds as Biofilm Inhibitors
3.2.3. Microorganisms for Pathogen Biofilm Control
3.3. Detection of Biofilm in Food Processing Plants
3.4. Conventional Approaches to Biofilm Removal
4. Electrochemical Biofilm Control
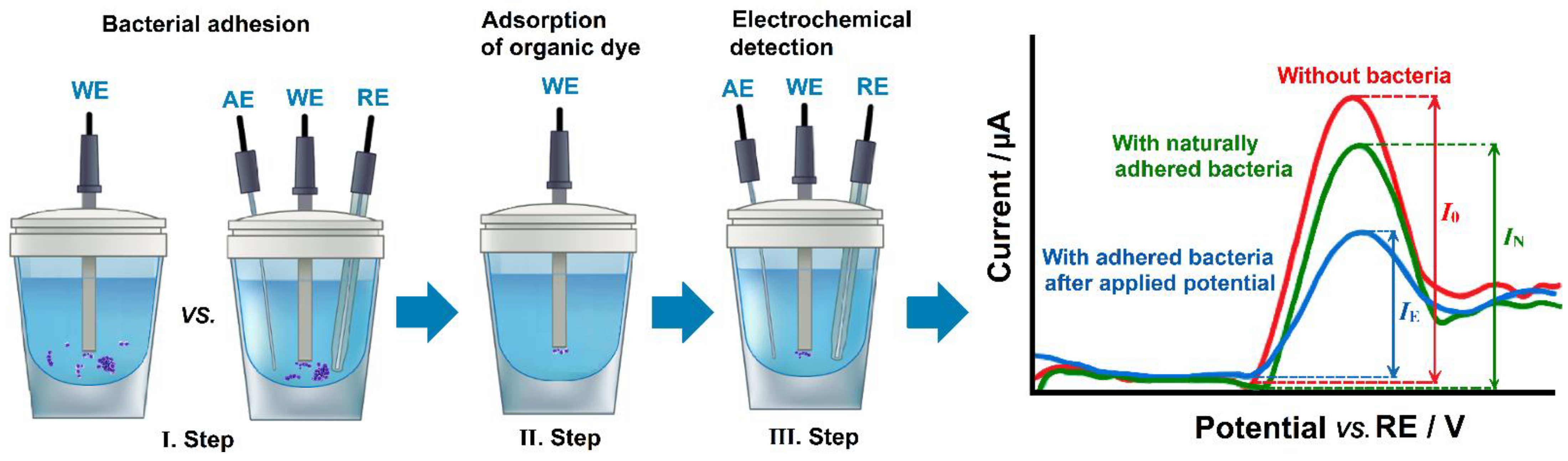
4.1. Electrochemical Control of Bacterial Adhesion
Effect of Divalent Ions on Biofilm Formation
4.2. Electrochemical Communication during Biofilm Formation
4.2.1. Electrochemical Communication between Microbial Cells and Conductive Surfaces
4.2.2. Electrochemical Mapping of Biofilm Location
4.3. Electrochemical Approaches to Biofilm Removal
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Kostakioti, M.; Hadjifrangiskou, M.; Hultgren, S.J. Bacterial biofilms: Development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 2013, 3, a010306. [Google Scholar] [CrossRef]
- Muhammad, M.H.; Idris, L.A.; Fan, X.; Guo, Y.; Yu, Y.; Jin, X.; Qiu, J.; Guan, X.; Huang, T. Beyond risk: Bacterial biofilms and their regulating approaches. Front. Microbiol. 2020, 11, 928. [Google Scholar] [CrossRef]
- Billings, N.; Millan, M.; Caldara, M.; Rusconi, R.; Tarasova, Y.; Stocker, R.; Ribbeck, K. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2013, 9, e1003526. [Google Scholar] [CrossRef]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. J. Pathol. Microbiol. Immunol. 2013, 121, 1–58. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef]
- Stoodley, P.; Hall-Stoodley, L. Evolving concepts in biofilm infections. Cell. Microbiol. 2009, 11, 1034–1043. [Google Scholar] [CrossRef]
- Gebreyohannes, G.; Nyerere, A.; Bii, C.; Sbhatu, D.B. Challenges of intervention, treatment, and antibiotic resistance of biofilm-forming microorganisms. Heliyon 2019, 5, e02192. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef]
- Coughlan, L.M.; Cotter, P.D.; Hill, C.; Alvarez-Ordonez, A. New weapons to fight old enemies: Novel strategies for the (Bio)control of bacterial biofilms in the food industry. Front. Microbiol. 2016, 7, 1641. [Google Scholar] [CrossRef]
- Dobretsov, S.; Dahms, H.U.; Qian, P.Y. Inhibition of biofouling by marine microorganisms and their metabolites. Biofouling 2006, 22, 43–54. [Google Scholar] [CrossRef]
- Bi, Y.; Xia, G.; Shi, C.; Wan, J.; Liu, L.; Chen, Y.; Wu, Y.; Zhang, W.; Zhou, M.; He, H.; et al. Therapeutic strategies against bacterial biofilms. Fundam. Res. 2021, 1, 193–212. [Google Scholar] [CrossRef]
- Galie, S.; Garcia-Gutierrez, C.; Miguelez, E.M.; Villar, C.J.; Lombo, F. Biofilms in the food industry: Health aspects and control methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef]
- Yan, J.; Bassler, B.L. Surviving as a community: Antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef]
- Abee, T.; Kovács, A.T.; Kuipers, O.P.; van der Veen, S. Biofilm formation and dispersal in Gram-positive bacteria. Curr. Opin. Biotechnol. 2011, 22, 172–179. [Google Scholar] [CrossRef]
- Bowler, P.; Murphy, C.; Wolcott, R. Biofilm exacerbates antibiotic resistance: Is this a current oversight in antimicrobial stewardship? Antimicrob. Resist. Infect. Control 2020, 9, 162. [Google Scholar] [CrossRef]
- Han, N.; Mizan, M.F.R.; Jahid, I.K.; Ha, S.D. Biofilm formation by Vibrio parahaemolyticus on food and food contact surfaces increases with rise in temperature. Food Control 2016, 70, 161–166. [Google Scholar] [CrossRef]
- Jahid, I.K.; Mizan, M.F.R.; Ha, A.J.; Ha, S.D. Effect of salinity and incubating time of planktonic cells on biofilm formation, motility, exoprotease production, and quorum sensing of Aeromonas hydrophila. Food Microbiol. 2015, 49, 142–151. [Google Scholar] [CrossRef]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm matrixome: Extracellular components in structured, microbial communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef]
- Mizan, M.F.R.; Jahid, I.K.; Park, S.Y.; Silva, J.L.; Kim, T.J.; Myoung, J.; Ha, S.D. Effects of temperature on biofilm formation and quorum sensing of Aeromonas hydrophila. Ital. J. Food Sci. 2018, 30, 456–466. [Google Scholar]
- Guzmán-Soto, I.; McTiernan, C.; Gonzalez-Gomez, M.; Ross, A.; Gupta, K.; Suuronen, E.J.; Mah, T.F.; Griffith, M.; Alarcon, E.I. Mimicking biofilm formation and development: Recent progress in in vitro and in vivo biofilm models. iScience 2021, 24, 102443. [Google Scholar] [CrossRef]
- Huang, Y.; Flint, S.H.; Palmer, J.S. Bacillus cereus spores and toxins—The potential role of biofilms. Food Microbiol. 2020, 90, 103493. [Google Scholar] [CrossRef]
- Korber, D.R.; Mangalappalli-Illathu, A.K.; Vidoviæ, S. Chapter 6—Biofilm formation by food spoilage microorganisms in food processing environments. In Biofilms in the Food and Beverage Industries; Fratamico, P.M., Annous, B.A., Gunthereds, N.W., Eds.; Woodhead Publishing: Sawston, UK, 2009; pp. 166–199. [Google Scholar]
- Lin, Y.; Briandet, R.; Kovács, Á.T. Bacillus cereus sensu lato biofilm formation and its ecological importance. Biofilm 2022, 4, 100070. [Google Scholar] [CrossRef]
- Park, E.-J.; Hussain, M.S.; Wei, S.; Kwon, M.; Oh, D.-H. Genotypic and phenotypic characteristics of biofilm formation of emetic toxin producing Bacillus cereus strains. Food Control 2019, 96, 527–534. [Google Scholar] [CrossRef]
- Wirtanen, G.; Salo, S. Chapter 5—Biofilm risks. In Handbook of Hygiene Control in the Food Industry, 2nd ed.; Lelieveld, H., Holah, J., Gabriæ, D., Eds.; Woodhead Publishing: San Diego, CA, USA, 2016; pp. 55–79. [Google Scholar]
- Liu, D.; Ge, S.; Wang, Z.; Li, M.; Zhuang, W.; Yang, P.; Chen, Y.; Ying, H. Identification of a sensor histidine kinase (BfcK) controlling biofilm formation in Clostridium acetobutylicum. Chin. J. Chem. Eng. 2021; in press. [Google Scholar] [CrossRef]
- Pantaléon, V.; Bouttier, S.; Soavelomandroso, A.P.; Janoir, C.; Candela, T. Biofilms of Clostridium species. Anaerobe 2014, 30, 193–198. [Google Scholar] [CrossRef]
- Pantaléon, V.; Soavelomandroso, A.P.; Bouttier, S.; Briandet, R.; Roxas, B.; Collignon, M.A.; Janoir, C.; Vedantam, G.; Candela, T. The Clostridium difficile protease Cwp84 modulates both biofilm formation and cell-surface properties. PLoS ONE 2015, 10, e0124971. [Google Scholar] [CrossRef]
- Hu, L. Prevalence of curli genes among Cronobacter species and their roles in biofilm formation and cell-cell aggregation. Int. J. Food Microbiol. 2018, 265, 65–73. [Google Scholar] [CrossRef]
- Ling, N.; Forsythe, S.; Wu, Q.; Ding, Y.; Zhang, Y.; Zeng, H. Insights into Cronobacter sakazakii biofilm formation and control strategies in the food industry. Engineering 2020, 6, 393–405. [Google Scholar] [CrossRef]
- Naziri, Z.; Kilegolan, J.A.; Moezzi, M.S.; Derakhshandeh, A. Biofilm formation by uropathogenic Escherichia coli: A complicating factor for treatment and recurrence of urinary tract infections. J. Hosp. Infect. 2021, 117, 9–16. [Google Scholar] [CrossRef]
- Makled, A.F.; Salem, E.H.; Elbrolosy, A.M. Biofilm formation and antimicrobial resistance pattern of uropathogenic E. coli: Comparison of phenotypic and molecular methods. Egypt. J. Med. Microbiol. 2017, 26, 37–45. [Google Scholar] [CrossRef]
- Sharma, G.; Sharma, S.; Sharma, P.; Chandola, D.; Dang, S.; Gupta, S.; Gabrni, R. Escherichia coli biofilm: Development and therapeutic strategies. J. Appl. Microbiol. 2016, 121, 309–319. [Google Scholar] [CrossRef]
- Tajbakhsh, E.; Ahmadi, P.; Abedpour-Dehkordi, E.; Arbab-Soleimani, N.; Khamesipour, F. Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrob. Resist. Infect. Control 2016, 5, 11. [Google Scholar] [CrossRef]
- Zamani, H.; Salehzadeh, A. Biofilm formation in uropathogenic Escherichia coli: Association with adhesion factor genes. Turk. J. Med. Sci. 2018, 48, 162–167. [Google Scholar] [CrossRef]
- Bonsaglia, E.C.R.; Silva, N.C.C.; Fernades Júnior, A.; Araújo Júnior, J.P.; Tsunemi, M.H.; Rall, V.L.M. Production of biofilm by Listeria monocytogenes in different materials and temperatures. Food Control 2013, 35, 386–391. [Google Scholar] [CrossRef]
- Gandra, T.K.V.; Volcan, D.; Kroning, I.S.; Marini, N.; de Oliveira, A.C.; Bastos, C.P.; da Silva, W.P. Expression levels of the agr locus and prfA gene during biofilm formation by Listeria monocytogenes on stainless steel and polystyrene during 8 to 48 h of incubation 10 to 37 °C. Int. J. Food Microbiol. 2019, 300, 1–7. [Google Scholar] [CrossRef]
- Ramires, T.; Kleinubing, N.R.; Iglesias, M.A.; Vitola, H.R.S.; Núncio, A.S.P.; Kroning, I.S.; Moreira, G.M.S.G.; Fiorentini, Â.M.; da Silva, W.P. Genetic diversity, biofilm and virulence characteristics of Listeria monocytogenes in salmon sushi. Food Res. Int. 2021, 140, 109871. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, L.; Hu, L.; Wang, Z.; Wang, X.; Dong, Q. Adhesion and kinetics of biofilm formation and related gene expression of Listeria monocytogenes in response to nutritional stress. Food Res. Int. 2022, 156, 111143. [Google Scholar] [CrossRef]
- Abd El Galil, K.; Abdelghani, S.M.; Sebak, M.A.; El-Naggar, W. Detection of biofilm genes among clinical isolates of Pseudomonas aeruginosa recovered from some Egyptian hospitals. New Egypt. J. Microbiol. 2013, 36, 86–101. [Google Scholar]
- Abdelraheem, W.M.; Abdelkader, A.E.; Mohamed, E.S.; Mohammed, M.S. Detection of biofilm formation and assessment of biofilm genes expression in different Pseudomonas aeruginosa clinical isolates. Meta Gene 2020, 23, 100646. [Google Scholar] [CrossRef]
- Ambutsi, M.; Okoth, P. Comparative genomic analysis of gene clusters of Pseudomonas aeruginosa that define specific biofilm formation in deciphering target regions for novel treatment options. Sci. Afr. 2021, 13, e00910. [Google Scholar] [CrossRef]
- Ghazalibina, M.; Morshedi, K.; Farahani, R.K.; Babadi, M.; Khaledi, A. Study of virulence genes and related with biofilm formation in Pseudomonas aeruginosa isolated from clinical samples of Iranian patients: A systematic review. Gene Rep. 2019, 17, 100471. [Google Scholar] [CrossRef]
- Kamali, E.; Jamali, A.; Ardebili, A.; Ezadi, F.; Mohebbi, A. Evaluation of antimicrobial resistance, biofilm forming potential, and the presence of biofilm-related genes among clinical isolates of Pseudomonas aeruginosa. BMC Res. Notes 2020, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Laverty, G.; Gorman, S.; Gilmore, B. Biomolecular mechanisms of Pseudomonas aeruginosa and Escherichia coli biofilm formation. Pathogens 2014, 3, 596–632. [Google Scholar] [CrossRef] [PubMed]
- Møretrø, T.; Langsrud, S. Residential bacteria on surfaces in the food industry and their implications for food safety and quality. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1022–1041. [Google Scholar] [CrossRef] [PubMed]
- Giacomodonato, M.N.; Sarnacki, S.H.; Aya Castañeda, M.D.R.; Garófalo, A.N.; Betancourt, D.M.; Cerquetti, M.C.; Llana, M.N. Salmonella enterica se.rovar Enteritidis biofilm lifestyle induces lower pathogenicity and reduces inflammatory response in a murine model compared to planktonic bacteria. Rev. Argent. Microbiol. 2021; in press. [Google Scholar] [CrossRef]
- Obe, T.; Nannapaneni, R.; Schilling, W.; Zhang, L.; Kiess, A. Antimicrobial tolerance, biofilm formation, and molecular characterization of Salmonella isolates from poultry processing equipment. J. Appl. Poult. Res. 2021, 30, 100195. [Google Scholar] [CrossRef]
- Tsai, M.-H.; Liang, Y.-H.; Chen, C.-L.; Chiu, C.-H. Characterization of Salmonella resistance to bile during biofilm formation. J. Microbiol. Immunol. Infect. 2020, 53, 518–524. [Google Scholar] [CrossRef]
- Ghaioumy, R.; Tabatabaeifar, F.; Mozafarinia, K.; Mianroodi, A.A.; Isaei, E.; Monores-Ramírez, J.R.; Afshari, S.A.K.; Kalantar-Neyestanaki, D. Biofilm formation and molecular analysis of intercellular adhesion gene cluster (icaABCD) among Staphylococcus aureus strains isolated from children with adenoiditis. Iran. J. Microbiol. 2021, 13, 458–463. [Google Scholar] [CrossRef]
- Idrees, M.; Sawant, S.; Karodia, N.; Rahman, A. Staphylococcus aureus biofilm: Morphology, genetics, pathogenesis and treatment strategies. Int. J. Environ. Res. Public Health 2021, 18, 7602. [Google Scholar] [CrossRef]
- Khoramrooz, S.S.; Mansouri, F.; Marashifard, M.; Hosseini, S.A.A.M.; Chenarestane-Oli, A.F.; Ganavehei, B.; Charibpour, F.; Shahbazi, A.; Mirzaii, M.; Darban-Sarokhalil, D. Detection of biofilm related genes, classical enterotoxin genes and agr typing among Staphylococcus aureus isolated from bovine with subclinical mastitis in southwest of Iran. Microb. Pathog. 2016, 97, 45–51. [Google Scholar] [CrossRef]
- Nourbakhsh, F.; Namvar, A.E. Detection of genes involved in biofilm formation in Staphylococcus aureus isolates. GMS Hyg. Infect. Control 2016, 11, Doc07. [Google Scholar] [PubMed]
- Shahmoradi, M.; Faridifar, P.; Shapouri, R.; Mousavi, S.F.; Ezzedin, M.; Mirzaei, B. Determining the biofilm forming gene profile of Staphylococcus aureus clinical isolates via multiplex colony PCR method. Rep. Biochem. Mol. Biol. 2019, 7, 181–188. [Google Scholar] [PubMed]
- Bezek, K.; Nipič, D.; Torkar, K.G.; Oder, M.; Dražić, G.; Abram, A.; Žibert, J.; Raspor, P.; Bohinc, K. Biofouling of stainless steel surfaces by four common pathogens: The effects of glucose concentration, temperature and surface roughness. Biofouling 2019, 35, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-B.; Bae, Y.-M.; Lee, S.-Y. Effect of environmental conditions on biofilm formation and related characteristics of Staphylococcus aureus. J. Food Saf. 2016, 36, 412–422. [Google Scholar] [CrossRef]
- Fan, Y.; Qiao, J.; Lu, Z.; Fen, Z.; Tao, Y.; Lv, F.; Zhao, H.; Zhang, C.; Bie, X. Influence of different factors on biofilm formation of Listeria monocytogenes and the regulation of cheY gene. Food Res. Int. 2020, 137, 109405. [Google Scholar] [CrossRef]
- Got, F. Staphylococcus and biofilms. Mol. Microbiol. 2002, 43, 1367–1378. [Google Scholar]
- Moraes, J.O.; Cruz, E.A.; Souza, E.G.; Oliveira, T.C.; Alvarenga, V.O.; Peña, W.E.; Sant’Ana, A.S.; Magnani, M. Predicting adhesion and biofilm formation boundaries on stainless steel surfaces by five Salmonella enterica strains belonging to different serovars as a function of pH, temperature and NaCl concentration. Int. J. Food Microbiol. 2018, 281, 90–100. [Google Scholar] [CrossRef]
- Shi, X.; Zhu, X. Biofilm formation and food safety in food industries. Trends Food Sci. Technol. 2009, 20, 407–413. [Google Scholar] [CrossRef]
- Yin, B.; Zhu, L.; Zhang, Y.; Dong, P.; Mao, Y.; Liang, R.; Niu, L.; Luo, X. The characterization of biofilm formation and detection of biofilm-related genes in Salmonella isolated from beef processing plants. Foodborne Pathog. Dis. 2018, 15, 660–667. [Google Scholar] [CrossRef]
- Lee, K.H.; Lee, J.Y.; Roy, P.K.; Mizan, M.F.R.; Hossain, M.I.; Park, S.H.; Ha, S.D. Viability of Salmonella Typhimurium biofilms on major food-contact surfaces and eggshell treated during 35 days with and without water storage at room temperature. Poult. Sci. 2020, 99, 4558–4565. [Google Scholar] [CrossRef]
- Gomes, I.B.; Lemos, M.; Fernandes, S.; Borges, A.; Simões, L.C.; Simões, M. The effects of chemical and mechanical stresses on Bacillus cereus and Pseudomonas fluorescens single- and dual-species biofilm removal. Microorganisms 2021, 9, 1174. [Google Scholar] [CrossRef] [PubMed]
- Chisti, Y.; Moo-Young, M. Clean-in-place systems for industrial bioreactors: Design, validation and operation. J. Ind. Microbiol. Biotechnol. 1994, 13, 201–207. [Google Scholar] [CrossRef]
- Joseph, B.; Otta, S.K.; Karunasagar, I.; Karunasagar, I. Biofilm formation by Salmonella spp. on food contact surfaces and their sensitivity to sanitizers. Int. J. Food Microbiol. 2001, 64, 367–372. [Google Scholar] [CrossRef]
- Poimenidou, S.V.; Chrysadakou, M.; Tzakoniati, A.; Bikouli, V.C.; Nychas, G.J.; Skandamis, P.N. Variability of Listeria mono-cytogenes strains in biofilm formation on stainless steel and polystyrene materials and resistance to peracetic acid and quater-nary ammonium compounds. Int. J. Food Microbiol. 2016, 237, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; McLandsborough, L.A. Evaluation of the transfer of Listeria monocytogenes from stainless steel and high-density polyethylene to Bologna and American cheese. J. Food Protect. 2007, 70, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Coulier, L.; Orbons, H.G.M.; Rijk, R. Analytical protocol to study the food safety of (multiple) recycled high-density polyeth-ylene (HDPE) and polypropylene (PP) crates: Influence of recycling on the migration and formation of degradation products. Polym. Degrad. Stab. 2007, 92, 2016–2025. [Google Scholar] [CrossRef]
- Moerman, F.; Partington, E. Materials of construction for food processing equipment and services: Requirements, strengths and weaknesses. J. Hyg. Eng. Des. 2014, 6, 10–37. [Google Scholar]
- Srey, S.; Jahid, I.K.; Ha, S.D. Biofilm formation in food industries: A food safety concern. Food Control 2013, 31, 572–585. [Google Scholar] [CrossRef]
- Song, B.; Leff, L.G. Influence of magnesium ions on biofilm formation by Pseudomonas fluorescens. Microbiol. Res. 2006, 161, 355–361. [Google Scholar] [CrossRef]
- Danilova, T.A.; Danilina, G.A.; Adzhieva, A.A.; Vostrova, E.I.; Zhukhovitskii, V.G.; Cheknev, S.B. Inhibitory effect of copper and zinc ions on the growth of Streptococcus pyogenes and Escherichia coli biofilms. Bull. Exp. Biol. Med. 2020, 169, 648–652. [Google Scholar] [CrossRef]
- Simbine, E.O.; Rodrigues, L.C.; Lapa-Guimaraes, J.; Kamimura, E.S.; Corassin, C.H.; Oliveira, C.A.F. Application of silver nanoparticles in food packages: A review. Food Sci. Technol. 2019, 39, 793–802. [Google Scholar] [CrossRef]
- Ashrafi, A.M.; Sýs, M.; Sedláčková, E.; Farag, A.S.; Adam, V.; Přibyl, J.; Richtera, L. Application of the enzymatic electrochemical biosensors for monitoring non-competitive inhibition of enzyme activity by heavy metals. Sensors 2019, 19, 2939. [Google Scholar] [CrossRef]
- Coenye, T.; De Prijck, K.; De Wever, B.; Nelis, H.J. Use of the modified Robbins device to study the in vitro biofilm removal efficacy of NitrAdine™, a novel disinfecting formula for the maintenance of oral medical devices. J. Appl. Microbiol. 2008, 105, 733–740. [Google Scholar] [CrossRef]
- Silvestry-Rodriguez, N.; Bright, K.R.; Slack, D.C.; Uhlmann, D.R.; Gerba, C.P. Silver as a residual disinfectant to prevent biofilm formation in water distribution systems. Appl. Environ. Microbiol. 2008, 74, 1639–1641. [Google Scholar] [CrossRef] [PubMed]
- Petala, M.; Tsiridis, V.; Mintsouli, I.; Pliatsikas, N.; Spanos, T.; Rebeyre, P.; Darakas, E.; Patsalas, P.; Vourlias, G.; Kostoglou, M.; et al. Silver deposition on stainless steel container surfaces in contact with disinfectant silver aqueous solutions. Appl. Surf. Sci. 2017, 396, 1067–1075. [Google Scholar] [CrossRef]
- Hong, S.-H.; Gorce, J.-B.; Punzmann, H.; Francois, N.; Shats, M.; Xia, H. Surface waves control bacterial attachment and formation of biofilms in thin layers. Sci. Adv. 2020, 6, eaaz9386. [Google Scholar] [CrossRef] [PubMed]
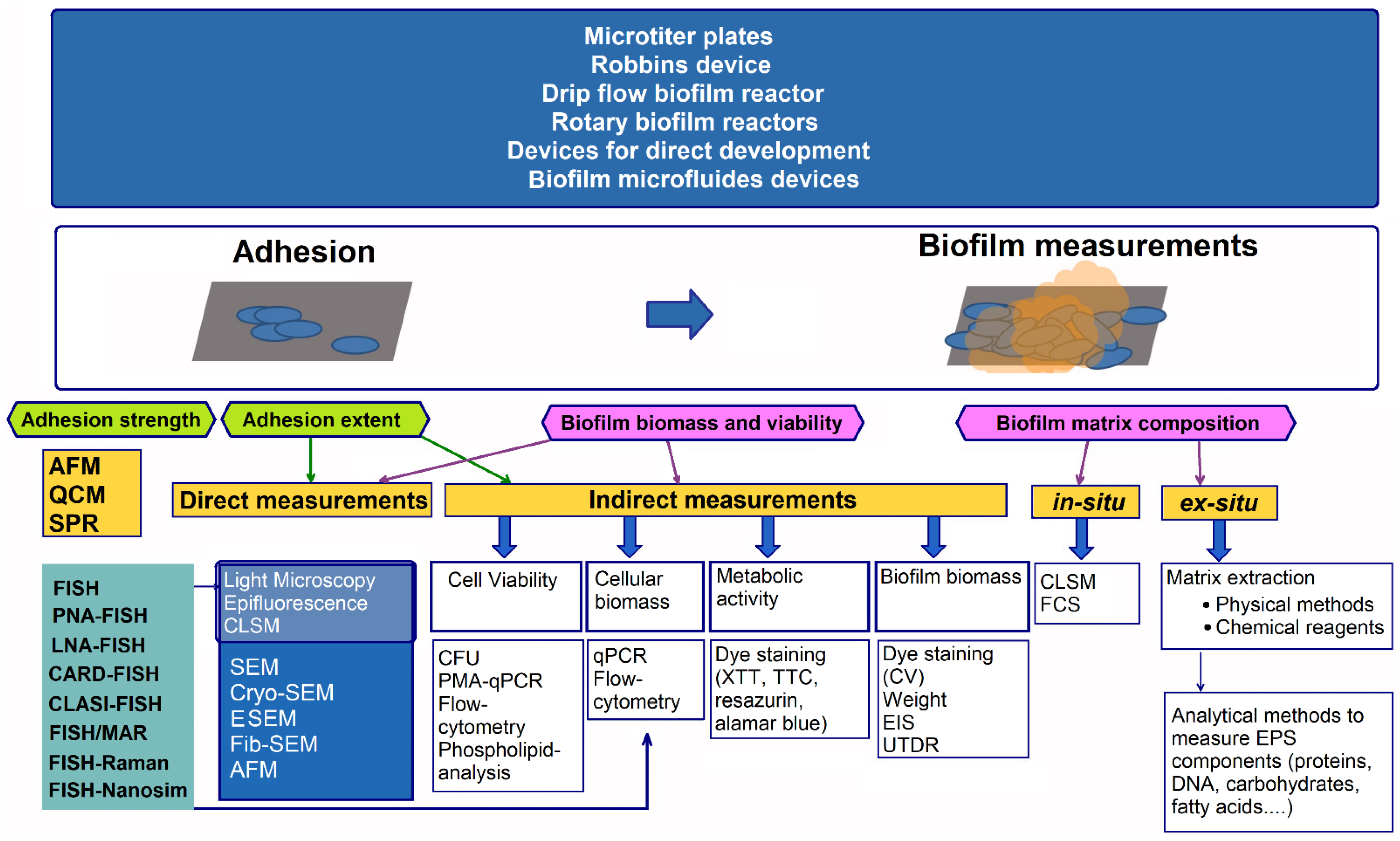
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hébraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef] [PubMed]
- Brindhadevi, K.; LewisOscar, F.; Mylonakis, E.; Shanmugam, S.; Verma, T.N.; Pugazhendhi, A. Biofilm and quorum sensing mediated pathogenicity in Pseudomonas aeruginosa. Process Biochem. 2020, 96, 49–57. [Google Scholar] [CrossRef]
- Brown, S.; Santa Maria, J.P., Jr.; Walker, S. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Vega-Hernández, R.; Ochoa, S.A.; Valle-Rios, R.; Jaimes-Ortega, G.A.; Arellano-Galindo, J.; Aparicio-Ozores, G.; Ibarra, J.A.; Hernández-Castro, R.; Cruz-Córdova, A.; Xicohtencatl-Cortes, J. Flagella, Type I fimbriae and curli of uropathogenic Escherichia coli promote the release of proinflammatory cytokines in a coculture system. Microorganisms 2021, 9, 2233. [Google Scholar] [CrossRef]
- Barnhart, M.M.; Chapman, M.R. Curli biogenesis and function. Annu. Rev. Microbiol. 2006, 60, 131–147. [Google Scholar] [CrossRef]
- Song, F.; Koo, H.; Ren, D. Effects of material properties on bacterial adhesion and biofilm formation. J. Dent. Res. 2015, 94, 1027–1034. [Google Scholar] [CrossRef]
- FAO. Global Food Losses and Food Waste-Extent, Causes and Prevention, Rome. 2011. Available online: www.fao.org/3/i2697e/i2694e.pdf (accessed on 16 May 2022).
- Stenmarck, A.; Jensen, C.; Quested, T.; Moates, G. Estimates of European Food Waste Levels; IVL Swedish Environmental Research Institute: Stockholm, Sweden, 2016; pp. 1–79. [Google Scholar]
- Kwok, T.; Ma, Y.; Chua, S.L. Biofilm dispersal induced by mechanical cutting leads to heightened foodborne pathogen dissemination. Food Microbiol. 2022, 102, 103914. [Google Scholar] [CrossRef]
- Xu, J.G.; Huang, X.N.; Meng, J.; Chen, J.Y.; Han, B.Z. Characterization and comparison of the bacterial community on environmental surfaces through a fresh-cut vegetables processing line in China. Int. Food Res. J. 2022, 155, 111075. [Google Scholar] [CrossRef]
- Wagner, E.M.; Pracser, N.; Thalguter, S.; Fischel, K.; Rammer, N.; Pospíšilová, L.; Alispahic, M.; Wagner, M.; Rychli, K. Identification of biofilm hotspots in a meat processing environment: Detection of spoilage bacteria in multi-species biofilms. Int. J. Food Microbiol. 2020, 328, 108668. [Google Scholar] [CrossRef]
- Rather, M.A.; Gupta, K.; Bardhan, P.; Borah, M.; Sarkar, A.; Eldiehy, K.S.H.; Bhuyan, S.; Mandal, M. Microbial biofilm: A matter of grave concern for human health and food industry. J. Basic Microbiol. 2021, 61, 380–395. [Google Scholar] [CrossRef]
- Lai, H.; Tang, Y.; Ren, F.; Li, Z.; Li, F.; Cu, C.; Jiao, X.; Huang, J. An investigation into the critical factors influencing the spread of Campylobacter during chicken handling in commercial kitchens in China. Microorganisms 2021, 9, 1164. [Google Scholar] [CrossRef]
- Ashrafudoulla, M.; Na, K.W.; Byun, K.-H.; Kim, D.H.; Yoon, J.W.; Mizan, M.F.R.; Kang, I.; Ha, S.-D. Isolation and characterization of Salmonella spp. From food and food conctat surfaces in chicken processing factory. Poult. Sci. 2021, 100, 101234. [Google Scholar] [CrossRef]
- Visvalingam, J.; Ells, T.C.; Yang, X. Impact of persistent and nonpersistent generic Escherichia coli and Salmonella sp. recovered from a beef packing plant on biofilm formation by E. coli O157. J. Appl. Microbiol. 2017, 123, 1512–1521. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union summary report on trends and sources of zoonozes, zoonotc agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, 5500. [Google Scholar]
- Borges, K.A.; Furian, T.Q.; de Souza, S.N.; Menezes, R.; Salle, C.T.P.; de Souza Moraes, H.L.; Tondo, E.C.; do Nascimento, V.P. Phenotypic and molecular characterization of Salmonella Enteritidis SE86 isolated from poultry and salmonellosis outbreaks. Foodborne Pathog. Dis. 2017, 14, 742–754. [Google Scholar] [CrossRef]
- Castro, H.; Jaakkonen, A.; Hakakorpi, A.; Hakkinen, M.; Isidro, J.; Korkeala, H.; Lindström, M.; Hallanvuo, S. Genomic epidemiology and phenotyping reveal on-farm persistent and cold adaptation of raw milk outbreak-associated Yersinia pseudotuberculosis. Front. Microbiol. 2019, 10, 1049. [Google Scholar] [CrossRef]
- Etter, A.J.; West, A.M.; Burnett, J.L.; Wu, S.T.; Veenhuizen, D.R.; Ogas, R.A.; Oliver, H.F. Salmonella enterica subsp. enterica serovar Heidelberg food isolates associated with a samonellosis outbreak have enhanced stress tolarence capabilities. Appl. Environ. Microbiol. 2019, 85, e01065-19. [Google Scholar]
- Jaakkonen, A.; Kivistö, R.; Aarnio, M.; Kalekivi, J.; Hakkinen, M. Persistent contamination of raw milk by Campylobacter jejuni ST-883. PLoS ONE 2020, 15, e0231810. [Google Scholar] [CrossRef]
- Uhlich, G.A.; Sinclair, J.R.; Warren, N.G.; Chmielecki, W.A.; Frantamico, P. Characterization of shiga toxin-producing Escherichia coli isolates associated with two multistate food-borne outbreaks that occurred in 2006. Appl. Environ. Microbiol. 2008, 74, 1268–1272. [Google Scholar] [CrossRef]
- Sinde, E.; Carballo, J. Attachment of Salmonella spp. and Listeria monocytogenes on stainless steel, rubber and polytetrafluorethylene: The influence of free energy and the effect of commercial sanitizer. Food Microbiol. 2000, 17, 439–447. [Google Scholar] [CrossRef]
- Gomes, I.B.; Simões, M.; Simões, L.C. Copper surfaces in biofilm control. Nanomaterials 2020, 10, 2491. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, X.; Yang, C.; Xi, T.; Zhao, J.; Liu, L.; Yang, K. New strategy to delay food spoilace: Application of new food contact material with antibacterial function. J. Mater. Sci. Technol. 2021, 70, 59–66. [Google Scholar] [CrossRef]
- Pontin, K.P.; Borges, K.A.; Furian, T.Q.; Darvalho, D.; Wilsmann, D.E.; Cardoso, H.R.P.; Alves, A.K.; Chitolina, G.Z.; Salle, C.T.P.; de Souza Moraes, H.L.; et al. Antimicrobial activity of copper surfaces against biofilm formation by Salmonella Enteritidis its potential application in the poultry industry. Food Microbiol. 2021, 94, 103654. [Google Scholar] [CrossRef]
- Dula, S.; Ajayeoba, T.A.; Ijabadeniyi, O.A. Bacterial biofilm formation on stainless steel in the food processing environment and its health implications. Folia Microbiol. 2021, 66, 293–302. [Google Scholar] [CrossRef]
- Soule, L.D.; Chomorro, N.P.; Chuong, K.; Mellott, N.; Hammer, N.; Hankenson, K.D.; Chatzistavrou, X. Sol-gel-derived bioactive and antibacterial multi-component thin films by the spin-coating technique. ACS Biomater. Sci. Eng. 2020, 6, 5549–5562. [Google Scholar] [CrossRef]
- Araújo, E.M.; de Andrade, N.J.; da Silva, L.H.M.; Bernardes, P.C.; de Carvalho Teixeira, A.V.N.; Júnior, J.F.Q.F.; de Sá, J.P.N.; Fernandes, P.É. Modification of stainless steel surface hydrophobicity by silver nanoparticles: Strategies to prevent bacterial adhesion in the food processing. J. Adhes. Sci. Technol. 2013, 27, 2686–2695. [Google Scholar] [CrossRef]
- Fang, F.; Kennedy, J.; Dhillon, M.; Flint, S. Antibacterial effect of silver nanofilm modified stainless steel surface. Int. J. Mod. Phys. B 2015, 29, 1540013. [Google Scholar] [CrossRef]
- Shi, L.; Santhanakrishnan, S.; Cheah, Y.S.; Li, M.; Chai, C.L.L.; Neoh, K.G. One-pot UV-triggered o-nitrobenzyl dopamine polymerization and coating for surface antibacterial application. ACS Appl. Mater. Interfaces 2016, 8, 33131–33138. [Google Scholar] [CrossRef]
- Li, N.; Li, X.; Yang, Y.; Liu, Y.; Zhao, L.; Zhou, Z. Adsorption-desorption behaviour of silver ions on stainless steel as a proxy for dissinfection of domestic hot water. Desalin. Water Treat. 2019, 151, 230–241. [Google Scholar] [CrossRef]
- Wong, T.S.; Kang, S.H.; Tang, S.K.; Smythe, E.K.; Hatton, B.D.; Grinthal, A.; Aizenberg, J. Bioinsipired self-repairing slippery surfaces with pressure-stable omniphobicity. Nature 2011, 477, 443–447. [Google Scholar] [CrossRef]
- Awad, T.S.; Asker, D.; Hatton, B.D. Food-safe modification on stainless steel food-processing surfaces to reduce bacterial biofilms. ACS Appl. Mater. Interfaces 2018, 10, 22902–22912. [Google Scholar] [CrossRef]
- Mishra, R.; Panda, A.K.; de Mandal, S.; Shakeel, M.; Bisht, S.S.; Khan, J. Natural anti-biofilm agents: Strategies to control biofilm-forming pathogens. Front. Microbiol. 2020, 11, 566325. [Google Scholar] [CrossRef]
- Zhang, F.; Zhu, J.; Wang, H. Inhibitory activity of tea polyphenols on biofilm development of Shewanella putrefaciens. J. Food Proces. Preserv. 2016, 40, 910–917. [Google Scholar] [CrossRef]
- Walsh, D.J.; Livinghouse, T.; Durling, G.M.; Chase-Bayless, Y.; Arnold, A.D.; Stewart, P.S. Sulfenate esters of simple phenols exhibit enhanced activity against biofilm. ACS Omega 2020, 5, 6010–6020. [Google Scholar] [CrossRef] [PubMed]
- Nazareth, M.S.; Shreelakshmi, S.V.; Shetty, N.P. Identification and characterization of polyphenols from Carissa spinarum fruit and evaluation of their antioxidant and anti-quorum sensing activity. Curr. Microbiol. 2021, 78, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Molina, R.D.I.; Campos-Silva, R.; Díaz, M.A.; Macedo, A.J.; Blázquez, M.A.; Alberto, M.R.; Arena, M.E. Laurel extracts inhibit quorum sensing, virulence factors and biofilm of foodborne pathogens. Lebensm. Wiss. Technol. 2020, 134, 109899. [Google Scholar] [CrossRef]
- Walczak, M.; Michalska-Sionkowska, M.; Olkiewicz, D.; Tarnawska, P.; Warżyńska, O. Potential of carvacrol and thymol in reducing biofilm formation on technical surfaces. Molecules 2021, 26, 2723. [Google Scholar] [CrossRef]
- Minei, C.C.; Gomes, B.C.; Ratti, R.P.; D´Angelis, C.E.M.; de Martinis, E.C.P. Influence of peroxyacetic acid and nisin and coculture with Enterococcus faecium on Listeria monocytogenes biofilm formation. J. Food Prot. 2008, 71, 634–638. [Google Scholar] [CrossRef]
- Seo, H.-J.; Kang, S.-S. Inhibitory effect of bacteriocin produced by Pediococcus acidilactici on the biofilm formation of Salmonella Typhimurium. Food Control 2020, 117, 107361. [Google Scholar] [CrossRef]
- Mulya, E.; Waturangi, D.E. Screening and quantification of anti-quorum sensing and antibiofilm activity of Actinomycetes isolates against food spoilage biofilm-forming bacteria. BMC Microbiol. 2021, 21, 1. [Google Scholar] [CrossRef]
- Santos, R.A.; Oliva-Teles, A.; Pousão-Ferreira, P.; Jerusik, R.; Saavedra, M.J.; Enes, P.; Serra, C.R. Isolation and characterization of fish-gut Bacillus spp. as source of natural antimicrobial compounds to fight aquaculture bacterial diseases. Mar. Biotechnol. 2021, 23, 276–293. [Google Scholar] [CrossRef]
- Alexpandi, R.; Ponraj, J.G.; Swasthikka, R.P.; Abirami, G.; Ragupathi, T.; Jayakumar, R.; Ravi, A.V. Anti-QS mediated anti-infection efficacy of probiotic culture-supernatant against Vibrio campbelli infection and the identification of active compounds through in vitro and in silico analyses. Bioacatal. Agric. Biotechnol. 2021, 35, 102108. [Google Scholar] [CrossRef]
- De Araujo, L.V.; Guimarães, C.R.; da Silva Marquita, R.L.; Santiago, V.M.J.; de Souza, M.P.; Nitschke, M.; Freire, D.M.G. Rhamnolipid and surfactin: Anti-adhesion/antibiofilm and antimicrobial effects. Food Control 2016, 63, 171–178. [Google Scholar] [CrossRef]
- Sterniša, M.; Klančnik, A.; Možina, S.S. Spoilage Pseudomonas biofilm with Escherichia coli protection in fish meat at 5 °C. J. Sci. Food Agric. 2019, 99, 4635–4641. [Google Scholar] [CrossRef] [PubMed]
- Hascoët, A.-S.; Ripolles-Avila, C.; Cervantes-Huamán, B.R.H.; Rodríguez-Jerez, J.J. In vitro performed biofilm of Bacillus safensis inhibit the adhesion and subsequent development of Listeria monocytogenes on stainless-steel surfaces. Biomolecules 2021, 11, 475. [Google Scholar] [CrossRef] [PubMed]
- Achinas, S.; Yska, S.K.; Charalampogiannis, N.; Krooneman, J.; Euverink, G.J.W. A technological understanding of biofilm detection techniques: A review. Materials 2020, 13, 3147. [Google Scholar] [CrossRef] [PubMed]
- Dygico, L.K.; Gaham, C.G.M.; Grogan, H.; Burgess, C.M. Examining efficacy of mushroom industry biocides on Listeria monocytogenes biofilm. J. Appl. Microbiol. 2020, 130, 1106–1116. [Google Scholar] [CrossRef] [PubMed]
- Ríos-Castillo, A.G.; Ripolles-Avila, C.; Rodríguez-Jerez, J.J. Detection of Salmonella Typhimurium and Listeria monocytogenes biofilm cells exposed to different drying and pre-enrichment times using conventional and rapid methods. Int. J. Food Microbiol. 2020, 324, 108611. [Google Scholar] [CrossRef] [PubMed]
- European Chemical Agency, Helsinki, Finland. Information on Biocides. Available online: https://echa.europa.eu/cs/information-on-chemicals/biocidal-active-substances (accessed on 16 May 2022).
- Castro, M.S.R.; da Silva Fernandes, M.; Kabuki, D.Y.; Kuaye, A.Y. Biofilm formation of Enterococcus faecium on stainless steel surfaces: Modelling and control by disinfection agents. J. Food Process. Eng. 2018, 41, e12663. [Google Scholar] [CrossRef]
- Speranza, B.; Monacis, N.; Sinigaglia, M.; Corbo, M.R. Approaches to removal and killing of Salmonella spp. biofilms. J. Food Process. Preserv. 2017, 41, e12758. [Google Scholar] [CrossRef]
- Iñiguez-Moreno, M.; Gutiérrez-Lomelí, M.; Avila-Novoa, M.G. Removal of mixed-species biofilm developed on food contact surfaces with a mixture of enzymes and chemical agents. Antibiotics 2021, 10, 931. [Google Scholar] [CrossRef]
- Vázquez-Sánchez, D.; Galvão, J.A.; Ambrosio, C.M.S.; Gloria, E.M.; Oetterer, M. Sinlge and binary applications of essential oils effectively control Listeria monocytogenes biofilms. Ind. Crops Prod. 2018, 121, 452–460. [Google Scholar] [CrossRef]
- Yu, H.; Liu, Y.; Yang, F.; Xie, Y.; Guo, Y.; Cheng, Y.; Yao, W. Synergistic efficacy of high-intensity ultrasound and chlorine dioxide combination for Staphylococcus aureus biofilm control. Food Control 2021, 122, 107822. [Google Scholar] [CrossRef]
- Hua, Z.; Younce, F.; Tang, J.; Ryu, D.; Rasco, B.; Hanrahan, I.; Zhu, M.-J. Efficacy of saturated steam against Listeria innocua biofilm on common food-contact surfaces. Food Control 2021, 125, 107988. [Google Scholar] [CrossRef]
- Tan, J.; Karwe, M.V. Inactivation and removal of Enterobacter aerogenes biofilm in a model piping system using plasma-activated water (PAW). Innov. Food Sci. Emerg. Technol. 2021, 69, 102664. [Google Scholar] [CrossRef]
- Marino, M.; Maifreni, M.; Baggio, A.; Innocente, N. Inactivation of foodborne bacteria biofilms by aqueous and gaseous ozone. Front. Microbiol. 2018, 9, 2024. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ma, S.; Xie, Y.; Wang, M.; Cai, T.; Li, J.; Guo, D.; Zhao, L.; Xu, Y.; Liang, S.; et al. Inactivation of Pseudomonas aeruginosa biofilms by 405-nanometer-light-emitting diode illumination. Appl. Environ. Microbiol. 2020, 86, e00092-20. [Google Scholar] [CrossRef]
- Bellin, D.L.; Sakhtah, H.; Zhang, Y.; Price-Whelan, A.; Dietrich, L.E.P.; Shepard, K.L. Electrochemical camera chip for simultaneous imaging of multiple metabolites in biofilms. Nat. Commun. 2016, 7, 10535. [Google Scholar] [CrossRef]
- Morisaki, H.; Sugimoto, M.; Shiraishi, H. Attachment of bacterial cells to carbon electrodes. Bioelectrochemistry 2000, 51, 21–25. [Google Scholar] [CrossRef]
- Vu, D.L.; Sýs, M.; Červenka, L. The effect of various potentials on the attachment of Saccharomyces cerevisiae and Staphylococcus epidermidis to carbon paste electrodes. Int. J. Electrochem. Sci. 2011, 6, 5265–5274. [Google Scholar]
- Nostro, A.; Cellini, L.; Di Giulio, M.; D’Arrigo, M.; Marino, A.; Blanco, A.R.; Favaloro, A.; Cutroneo, G.; Bisignano, G. Effect of alkaline pH on staphylococcal biofilm formation. APMIS 2012, 120, 733–742. [Google Scholar] [CrossRef]
- Vu, D.L.; Červenka, L.; Vavřičková, J. The attachment of Staphylococcus epidermidis on the surface of a carbon paste electrode at various positive potentials: The effect of pH, incubation time, and solid-medium type. J. Biomed. Sci. Eng. 2012, 5, 699–704. [Google Scholar] [CrossRef][Green Version]
- Tresse, O.; Lebret, V.; Benezech, T.; and Faille, C. Comparative evaluation of adhesion, surface properties, and surface protein composition of Listeria monocytogenes strains after cultivation at constant pH of 5 and 7. J. Appl. Microbiol. 2006, 101, 53–62. [Google Scholar] [CrossRef]
- Steiger, E.L.; Muelli, J.R.; Braissant, O.; Waltimo, T.; Astasov-Frauenhoffer, M. Effect of divalent ions on cariogenic biofilm formation. BMC Microbiol. 2000, 20, 287. [Google Scholar] [CrossRef] [PubMed]
- Falcón García, C.; Kretschmer, M.; Lozano-Andrade, C.N.; Schönleitner, M.; Dragoŝ, A.; Kovács, Á.T.; Lieleg, O. Metal ions weaken the hydrophobicity and antibiotic resistance of Bacillus subtilis NCIB 3610 biofilms. NPJ Biofilms Microbiomes 2020, 6, 1. [Google Scholar] [CrossRef]
- Guvensen, N.C.; Demir, S.; Ozdemir, G. Effects of magnesium and calcium cations on biofilm formation by Sphingomonas paucimobilis from an industrial environment. Curr. Opin. Biotechnol. 2013, 24, S68. [Google Scholar] [CrossRef]
- Nan, L.; Yang, K.; Ren, G. Anti-biofilm formation of a novel stainless steel against Staphylococcus aureus. Mater. Sci. Eng. C Mater. Biol. Appl. 2015, 51, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, C.; Rajasekharan, S.K.; Reifen, R.; Shemesh, M. Mitigating milk-associated bacteria through inducing zinc ions antibiofilm activity. Foods 2020, 9, 1094. [Google Scholar] [CrossRef]
- Harrison, J.J.; Ceri, H.; Yerly, J.; Rabiei, M.; Hu, Y.; Martinuzzi, R.; Turner, R.J. Metal ions may suppress or enhance cellular differentiation in Candida albicans and Candida tropicalis biofilms. Appl. Environ. Microbiol. 2007, 73, 4940–4949. [Google Scholar] [CrossRef]
- Perrin, C.; Briandet, R.; Jubelin, G.; Lejeune, P.; Mandrand-Berthelot, M.A.; Rodrigue, A.; Dorel, C. Nickel promotes biofilm formation by Escherichia coli K-12 strains that produce curli. Appl. Environ. Microbiol. 2009, 75, 1723–1733. [Google Scholar] [CrossRef]
- Ferrari, A.G.M.; Carrington, P.; Rowley-Neale, S.J.; Banks, C.E. Recent advances in portable heavy metal electrochemical sensing platforms. Environ. Sci. Water Res. Technol. 2020, 6, 2676–2690. [Google Scholar] [CrossRef]
- Milkman, R. An Escherichia coli homologue of eukaryotic potassium channel proteins. Proc. Natl. Acad. Sci. USA 1994, 91, 3510–3514. [Google Scholar] [CrossRef]
- Humphries, J.; Xiong, L.; Liu, J.; Prindle, A.; Yuan, F.; Arjes, H.A.; Tsimring, L.; Süel, G.M. Species-independent attraction to biofilms through electrical signaling. Cell 2017, 168, 200–209. [Google Scholar] [CrossRef]
- Majumdar, S.; Pal, S. Cross-species communication in bacterial world. J. Cell Commun. Signal 2017, 11, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Binepal, G.; Gill, K.; Crowley, P.; Cordova, M.; Brady, L.J.; Senadheera, D.B.; Cvitkovitch, D.G. Trk2 potassium transport system in Streptococcus mutans and its role in potassium homeostasis, biofilm formation, and stress tolerance. J. Bacteriol. 2016, 198, 1087–1100. [Google Scholar] [CrossRef] [PubMed]
- Remis, J.P.; Wei, D.; Gorur, A.; Zemla, M.; Haraga, J.; Allen, S.; Witkowska, H.E.; Costerton, J.W.; Berleman, J.E.; Auer, M. Bacterial social networks: Structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environ. Microbiol. 2014, 16, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Stekolshchikova, A.A.; Radaev, A.V.; Orlova, O.Y.; Nikolaev, K.G.; Skorb, E.V. Thin and flexible ion sensors based on polyelectrolyte multilayers assembled onto the carbon adhesive tape. ACS Omega 2019, 4, 15421–15427. [Google Scholar] [CrossRef] [PubMed]
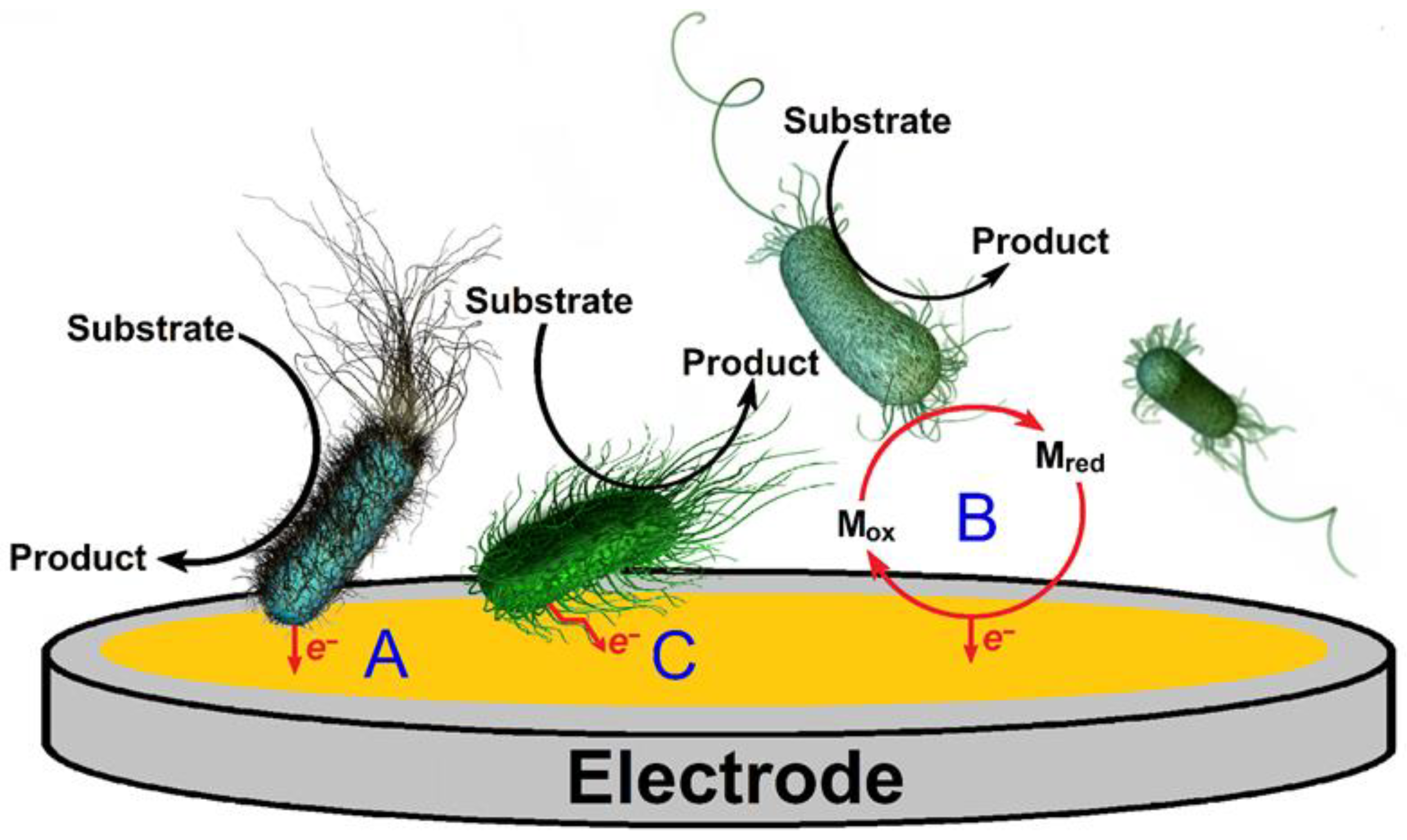
- Steidl, R.; Lampa-Pastirk, S.; Reguera, G. Mechanistic stratification in electroactive biofilms of Geobacter sulfurreducens mediated by pilus nanowires. Nat. Commun. 2016, 7, 12217. [Google Scholar] [CrossRef]
- Kashima, H.; Regan, J.M. Facultative nitrate reduction by electrode-respiring Geobacter metallireducens biofilms as a competitive reaction to electrode reduction in a bioelectrochemical system. Environ. Sci. Technol. 2015, 49, 3195–3202. [Google Scholar] [CrossRef]
- Kitayama, M.; Koga, R.; Kasai, T.; Kouzuma, A.; Watanabe, K. Structures, compositions, and activities of live Shewanella biofilms formed on graphite electrodes in electrochemical flow cells. Appl. Environ. Microbiol. 2017, 83, e00903-17. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, G.; Zhang, H.; Wen, H.; Li, W. Effects of biofilm transfer and electron mediators transfer on Klebsiella quasipneumoniae sp. 203 electricity generation performance in MFCs. Biotechnol. Biofuels 2020, 13, 162. [Google Scholar] [CrossRef]
- Kanga, C.S.; Eaktasang, N.; Kwon, D.Y.; Kim, H.S. Enhanced current production by Desulfovibrio desulfuricans biofilm in a mediator-less microbial fuel cell. Bioresour. Technol. 2014, 165, 27–30. [Google Scholar] [CrossRef]
- Saratale, G.D.; Saratale, R.G.; Shahid, M.K.; Zhen, G.; Kumar, G.; Shin, H.S.; Choi, Y.G.; Kim, S.H. A comprehensive overview on electro-active biofilms, role of exo-electrogens and their microbial niches in microbial fuel cells (MFCs). Chemosphere 2017, 178, 534–547. [Google Scholar] [CrossRef]
- Choi, O.; Sang, B.I. Extracellular electron transfer from cathode to microbes: Application for biofuel production. Biotechnol. Biofuels 2016, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Pankratova, G.; Gorton, L. Electrochemical communication between living cells and conductive surfaces. Curr. Opin. Electrochem. 2017, 5, 193–202. [Google Scholar] [CrossRef]
- Hasan, K.; Patil, S.A.; Leech, D.; Hägerhäll, C.; Gorton, L. Electrochemical communication between microbial cells and electrodes via osmium redox systems. Biochem. Soc. Trans. 2012, 40, 1330–1335. [Google Scholar] [CrossRef]
- Angelaalincy, M.J.; Navanietha, K.R.; Shakambari, G.; Ashokkumar, B.; Kathiresan, S.; Varalakshmi, P. Biofilm engineering approaches for improving the performance of microbial fuel cells and bioelectrochemical systems. Front. Energy Res. 2018, 6, 63. [Google Scholar] [CrossRef]
- Rau, J.; Knackmuss, H.J.; Stolz, A. Effects of different quinoid redox mediators on the anaerobic reduction of azo dyes by bacteria. Environ. Sci. Technol. 2002, 36, 1497–1504. [Google Scholar] [CrossRef] [PubMed]
- Paquete, C.M.; Rosenbaum, M.A.; Bañeras, L.; Rotaru, E.A.; Puig, S. Let’s chat: Communication between electroactive microorganisms. Bioresour. Technol. 2022, 347, 126705. [Google Scholar] [CrossRef]
- Darch, S.E.; Koley, D. Quantifying microbial chatter: Scanning electrochemical microscopy as a tool to study interactions in biofilms. Proc. R. Soc. A 2018, 474, 20180405. [Google Scholar] [CrossRef]
- Darvishi, S.; Pick, H.; Oveisi, E.; Girault, H.H.; Lesch, A. Soft-probe-scanning electrochemical microscopy reveals electrochemical surface reactivity of E. coli biofilms. Sens. Actuators B Chem. 2021, 334, 129669. [Google Scholar] [CrossRef]
- Turick, C.E.; Colon-Mercado, H.; Bagwell, C.E.; Greenway, S.D.; Amoroso, J.W. Non-contact electrochemical evaluation of biofilms. SN Appl. Sci. 2020, 2, 389. [Google Scholar] [CrossRef]
- Pires, L.; Sachsenheimer, K.; Kleintschek, T.; Waldbaur, A.; Schwartz, T.; Rapp, B.E. Online monitoring of biofilm growth and activity using a combined multi-channel impedimetric and amperometric sensor. Biosens. Bioelectron. 2013, 47, 157–163. [Google Scholar] [CrossRef]
- Song, J.; Li, Y.; Ke, D.; Wang, D.; Zhang, X.E. In situ graphene-modified carbon microelectrode array biosensor for biofilm impedance analysis. Electrochim. Acta 2022, 403, 139570. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, X.; Wang, J. Heterogeneous electrochemical characteristics of biofilm/metal interface and local electrochemical techniques used for this purpose. Mater. Corros. 2009, 60, 957–962. [Google Scholar] [CrossRef]
- Lebeaux, D.; Chauhan, A.; Rendueles, O.; Beloin, C. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2013, 2, 288–356. [Google Scholar] [CrossRef]
- Masi, E.; Ciszak, M.; Santopolo, L.; Frascella, A.; Giovannetti, L.; Marchi, E.; Viti, C.; Mancuso, S. Electrical spiking in bacterial biofilms. J. R. Soc. Interface 2015, 12, 20141036. [Google Scholar] [CrossRef]
- Kang, J.; Kim, T.; Tak, Y.; Lee, J.H.; Yoon, J. Cyclic voltammetry for monitoring bacterial attachment and biofilm formation. J. Ind. Eng. Chem. 2012, 18, 800–807. [Google Scholar] [CrossRef]
- Sedki, M.; Hassan, R.Y.A.; Andreescu, S.; El-Sherbiny, I.M. Online-monitoring of biofilm formation using nanostructured electrode surfaces. Mater. Sci. Eng. C 2019, 100, 178–185. [Google Scholar] [CrossRef]
- Tursun, H.; Liu, R.; Li, J.; Abro, R.; Wang, X.; Gao, Y.; Li, Y. Carbon material optimized biocathode for improving microbial fuel cell performance. Front. Microbiol. 2016, 7, 6. [Google Scholar] [CrossRef]
- Sultana, S.T.; Babauta, J.T.; Beyenal, H. Electrochemical biofilm control: A review. Biofouling 2015, 31, 745–758. [Google Scholar] [CrossRef]
- Schneider, S.; Rudolph, M.; Bause, V.; Terfort, A. Electrochemical removal of biofilms from titanium dental implant surfaces. Bioelectrochemistry 2018, 121, 84–94. [Google Scholar] [CrossRef]
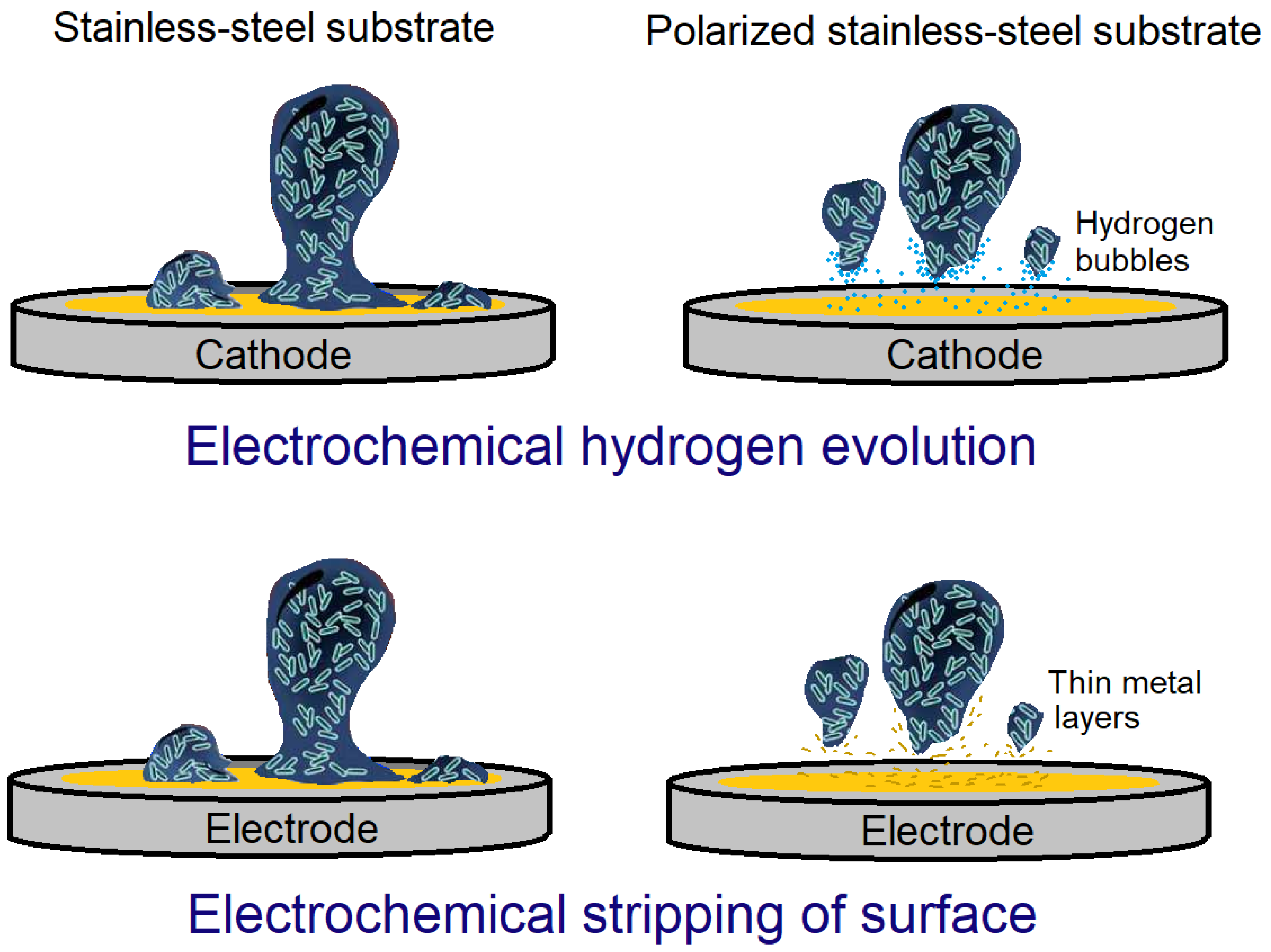
- Dargahi, M.; Hosseinidoust, Z.; Tufenkji, N.; Omanovic, S. Investigating electrochemical removal of bacterial biofilms from stainless steel substrates. Colloids Surf. B 2014, 117, 152–157. [Google Scholar] [CrossRef]
- Wijesinghe, M.S.; Wen, J.; Oh, J.M.; Chow, K.F.; Sun, Y. Demonstration of biofilm removal from type 304 stainless steel using pulsed-waveform electropolishing. Biofouling 2018, 34, 731–739. [Google Scholar] [CrossRef] [PubMed]
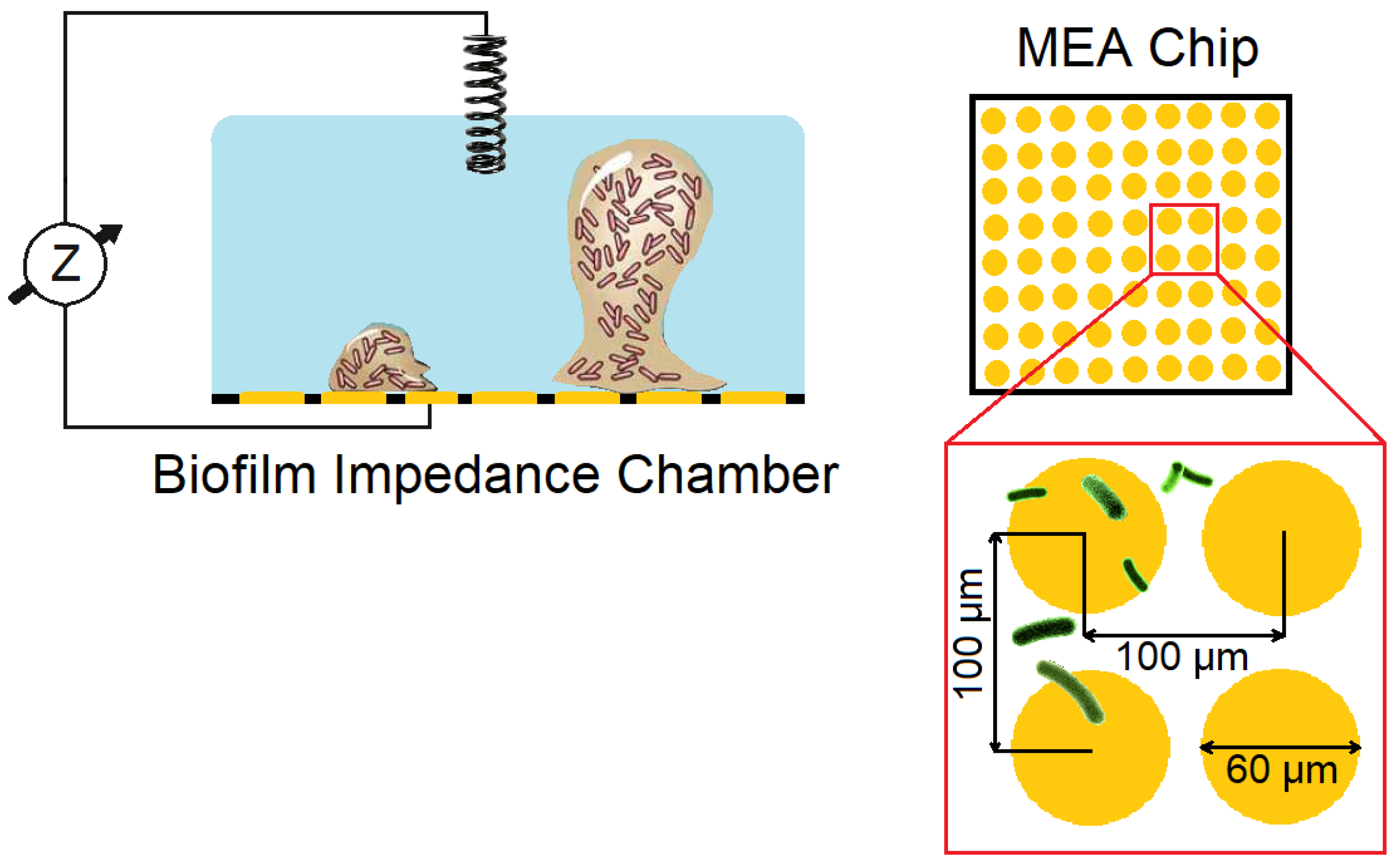
| Foodborne Bacteria | Growing Substrate | Spoiled Food | Genes Related to Biofilm Formation | References |
|---|---|---|---|---|
| Bacillus (Bacillus cereus) | Stainless steel, plastic, soil, and glass wool | Sprouted seeds, fruit juices, fried rice, pasta dishes, meat products, vegetables, and milk products | tasA, galE, eps2, mogR, comER, plcR, rpoN, codY, spo0A, abrB, sinI, sinR and others | [23,24,25,26,27] |
| Clostridium | Multi-species biofilm | Dairy products, fish, cattle meat, poultry, vegetables, honey, and canned food | luxS, spo0A, pilC, pilT, and others | [27,28,29,30] |
| Cronobacter spp. | Powder service and powder packaging rooms, spray-drying areas, and evaporator rooms | Dairy products, vegetables, grains, bread, herbs, sausages, spices, and meat | bcsR, csgA, csgB and others | [27,31,32] |
| Escherichia coli | Stainless steel surfaces, food contact surfaces | Dairy products, fermented meat sausage, meat, poultry, fish products, drinks, and vegetables | fim, pap, bfp, scg, sfa, foc, afa, flu, pgaABCD, bcsABZC, uvrY, csrA and others | [27,33,34,35,36,37] |
| Listeria monocytogenes | Wastewater pipes, floors, conveyor belts, rubber seals, elastomers, and stainless steel | Dairy products, melons, coleslaw, ready-to-eat meat products, and ready-to-eat fish products | luxS, agr (agrABCD), inlA, actA, prfA and others | [27,38,39,40,41] |
| Pseudomonas spp. (Pseudomonas aeruginosa) | Conveyor belts, floors, drains, slicing and milking machines | Dairy products, red meat, and poultry | psl (pslA–pslO), pel (pelA–pelG), algD, algU, algL, ppyR, lasR, lecA, rhlI, pilA, pilT and others | [4,24,42,43,44,45,46,47,48] |
| Salmonella | Stainless steel, elastomers, concrete, glass, and food surfaces (such as lettuce and tomato) | Poultry, pig, cow meats, and dairy products | bapA, csgB, csgD, csgBA, adrA, bcs, fimA, fimH, luxS, flgE and others | [27,49,50,51] |
| Staphylococcus (Staphylococcus aureus) | Stainless steel, plastics (such as polystyrene and polypropylene), and glass | Dairy products, ready-to-eat meat products, ready-to-eat fish and seafood products, and ready-to-eat dairy products | icaA, icaD, icaB, ica, icaR, fib, cna, fnbAB, clfA, clfB, agr (agrA-agrD) and others | [27,52,53,54,55,56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brožková, I.; Červenka, L.; Moťková, P.; Frühbauerová, M.; Metelka, R.; Švancara, I.; Sýs, M. Electrochemical Control of Biofilm Formation and Approaches to Biofilm Removal. Appl. Sci. 2022, 12, 6320. https://doi.org/10.3390/app12136320
Brožková I, Červenka L, Moťková P, Frühbauerová M, Metelka R, Švancara I, Sýs M. Electrochemical Control of Biofilm Formation and Approaches to Biofilm Removal. Applied Sciences. 2022; 12(13):6320. https://doi.org/10.3390/app12136320
Chicago/Turabian StyleBrožková, Iveta, Libor Červenka, Petra Moťková, Michaela Frühbauerová, Radovan Metelka, Ivan Švancara, and Milan Sýs. 2022. "Electrochemical Control of Biofilm Formation and Approaches to Biofilm Removal" Applied Sciences 12, no. 13: 6320. https://doi.org/10.3390/app12136320
APA StyleBrožková, I., Červenka, L., Moťková, P., Frühbauerová, M., Metelka, R., Švancara, I., & Sýs, M. (2022). Electrochemical Control of Biofilm Formation and Approaches to Biofilm Removal. Applied Sciences, 12(13), 6320. https://doi.org/10.3390/app12136320







