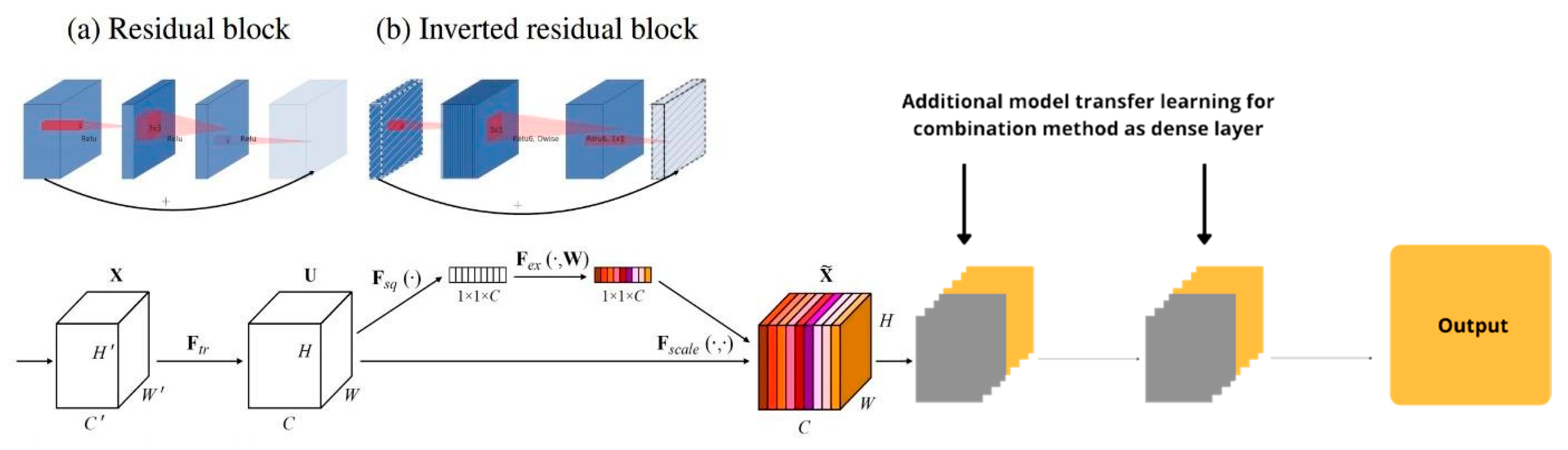
3.1. Independent Model Experiment Results
We used several treatments for each model in a transfer learning procedure. When presented with the same treatment, the differing model designs caused problems in the learning process. The study began with a comparison of two identical models in the same architecture, with the historical correctness of the models being assessed (
Table 2)
VGG19. VGG19 was tested by training a 3 × 3 convolutional layer using an ImageNet classification model using average pooling. The output was assessed by permitting numerous fine-tuning approaches, such as the activation of the rectified linear unit (ReLU) dense layer to achieve a low learning rate, the activation of the soft-max layer and saving checkpoints on the best model [
24]. The period began with an evaluation of the time spent during training, the number of total parameters processed and the storage of a learning process evaluation. With a total parameter of 20,090,435, VGG19 successfully completed the training with an average model accuracy of 0.9361, validated with a high value of 0.9390 and the maximum accuracy value of 0.9981. It is possible to infer that the two VGG models can depict models that are good enough to be classed as medical pictures, based on the findings of the two models. The accuracy of the case suggested by the VGG model was in the range of 0.80 to 0.90 and above. In terms of the value scale, the VGG19 value is an appropriate category to use as an image classification model, as shown in
Table 3.
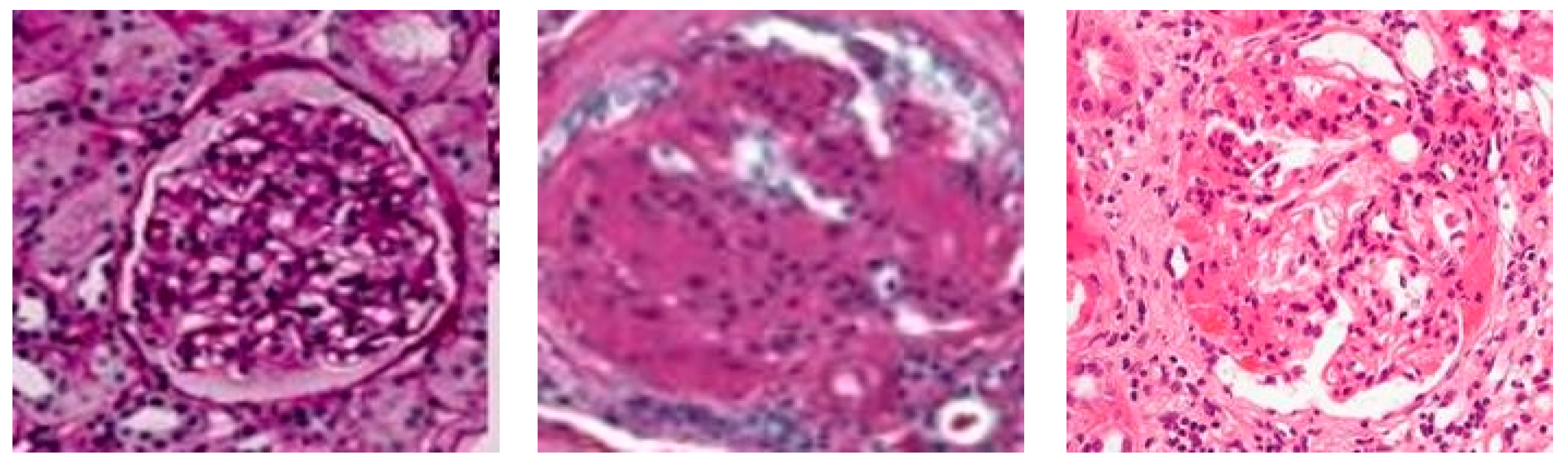
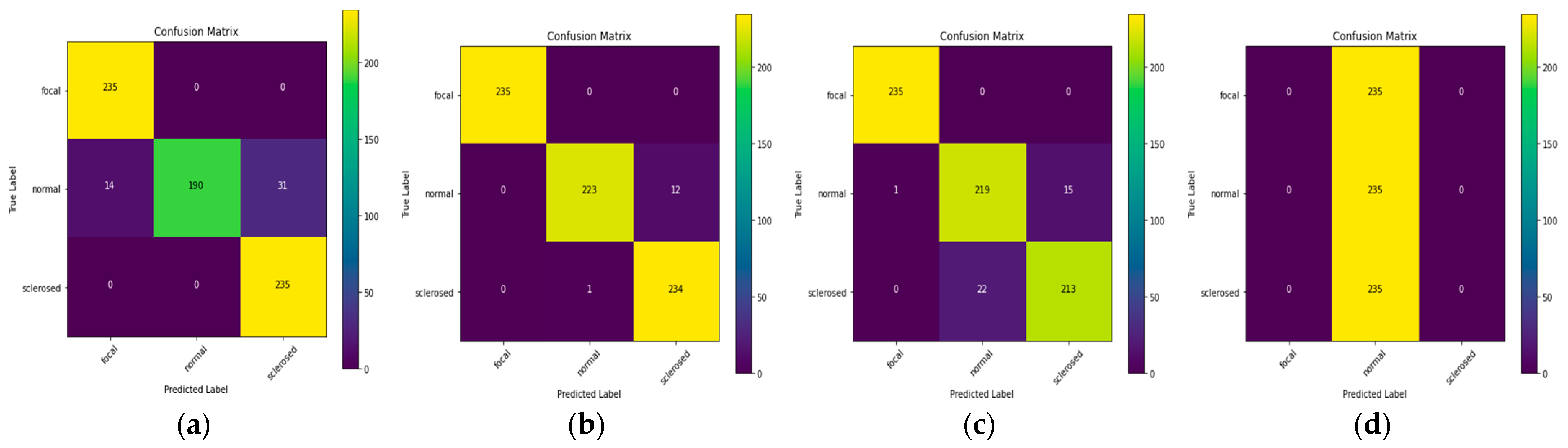
Figure 6a demonstrates that the original value’s correlation with the predicted value shows a high color prediction for the focal and sclerosed classes, while the projected normal class has a sufficient color in the classification.
ResNet101V2. The architectural resemblance between these two models may also result in the same model performance configuration. On the convolutional network, both model sets are constructed with a resolution of 150 × 150 using ImageNet weight. With a logistic regression (LR) value of 0.01 and a momentum value of 0.7, both of these models employed stochastic gradient descent (SGD) optimization for fine-tuning [
25]. This setting is an iterative approach for fine-tuning the objective function. Each high-accuracy validation point was saved in the model checkpoint function and utilized as a weight model in a pre-trained model.
ResNet101V2 runs 55,637,123 parameters in total. After determining these parameters, ResNet101V2 completed the training with an average accuracy of 0.9815, which was confirmed by the maximum value of 0.9801, for which the greatest accuracy score was 1000. The training results, as shown in
Table 3, demonstrate that each model employed in the experiment had the best accuracy.
EfficientNetB7 and EfficientNetL2. The EfficientNet model employed in this study differs significantly from the previous one. EfficientNet-B7 can operate using ImageNet weight, but EfficientNet-L2 requires noisy student weight as a pre-trained model, according to prior research. This occurred because EfficientNet-L2 was incompatible with the ImageNet weights, when the number of layer weight initiations of the model differed. The unevenness of the pre-trained models employed was caused by weight fluctuations, although this is an exception to correctly performing the training process. Even when running on different models, these two models employed the same fine-tuning. The average-pool layer was added to the end of the flattened layer. In addition, the use of the ReLU feature and a dropout layer with a value of 0.2 created a dense layer. A thick layer was seen in the last portion. Furthermore, this approach employed an optimizer in the form of Adam [
26], throughout the compilation process.
With a training parameter of 2,625,539, EfficientNet-B7 achieved an average accuracy of 0.9461. The highest value, 0.9291, was used to confirm this accuracy, and the maximum accuracy was 0.9662. Meanwhile, EfficientNet-L2 utilized a training parameter of 5,640,195, with an average accuracy of 0.3333 as a consequence. The greatest accuracy as 0.3424, and this accuracy was confirmed using the highest value of 0.3333.
The EfficientNet-B7 and EfficientNet-L2 designs had significant variations in their training outcomes. EfficientNet-B7 does a far better job at presenting results than EfficientNet-L2.
Figure 6c,d indicate that the EfficientNet-L2 predictions show that everything is in the usual class, but the EfficientNet-B7 predictions are evenly distributed in each class. These findings demonstrate that the allied model does not generate accurate transfer learning predictions.
Based on the training data, we may infer that the EfficientNet model does not have a high level of accuracy. The inequality found in the EfficientNet-L2 model indicates that the accuracy findings in EfficientNetB7 are inconsistent. The model’s incompatibility with the dataset utilized, on the other hand, prevents it from producing the optimal results.
3.2. Combining Model Experiment Results
After reviewing the results of the experiment, we tried again, this time using many models that were judged unfit for classification. As shown in
Table 3, the findings of Effi-cientNet-L2 do not exhibit a high degree of accuracy, especially when compared to the criteria set by the trials. In a model with low accuracy, a high yield model has a substantial impact on improving accuracy. The experiments on a cognate or unrelated model confirmed this idea.
Combination Allied Model. The accuracy value obtained from the combination of the associated models is dependent on the base model utilized. The value obtained in the combination model was proportional to the model’s independent accuracy value. This experiment was carried out on three family models that is VGG, ResNet, and EfficientNet, all of which have different layer designs despite being related. With VGG16 as the basic model, the VGG model accuracy is 93.62 percent, as shown in
Table 4. According to
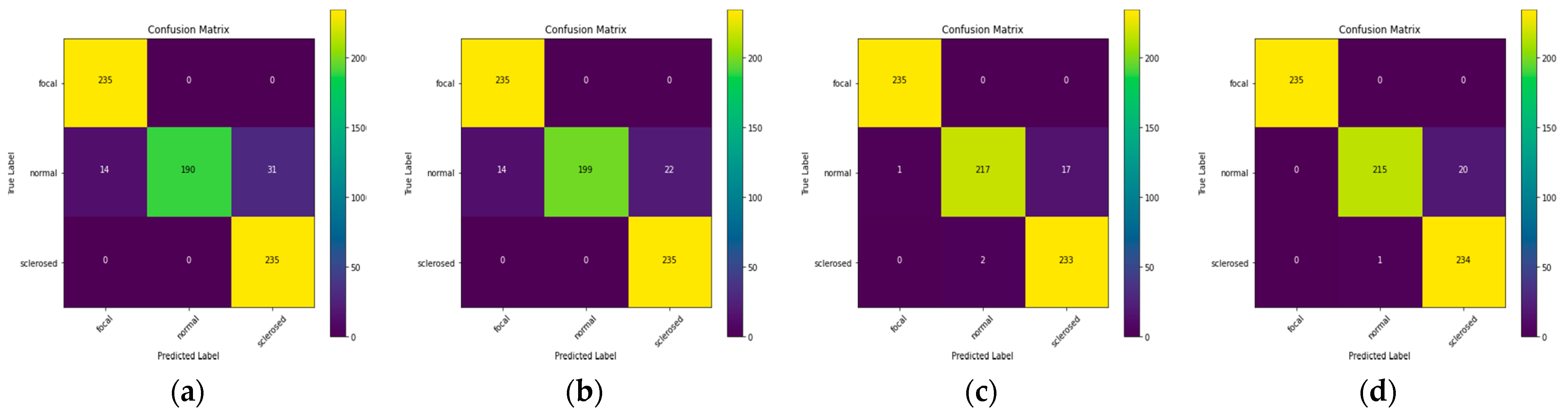
Figure 7, the normal class distribution offers less predictive data than the focused and sclerosed classes.
Combination Cross-Model. Cross-model combination refers to the model that is prioritized in order to enhance its accuracy value, limiting the models that may be combined to those that meet these requirements. EfficientNet-L2 and ResNet 101, for example, confirmed the need to improve. As a result, it is necessary to properly test each of these two models.
According to the base model employed in these tests, there are some varied categorization findings.
Table 4 indicates that the classification results of Res-Net101V2 may enhance the accuracy results of EfficientNet-L2 by approaching the ResNet101V2 value independently, by achieving 97.16 percent. The projected value in
Figure 7c is affected by this finding, with the distributions of each class matching the original value with a few erroneous values.
Combination Multivariate Model. EfficientNet-L2 has risen by up to two times while using ResNet101 as the basic model, compared to the prior trial when the model was conducting independent training. In this situation, ResNet101 has a low accuracy value, but it is still better than EfficientNet-L2; thus, the gain is not substantial. As a result of attempting to add another model with greater accuracy to this combination, two VGG models are used in the classification process as a multi-model combination experiment. The accuracy value from the combination model produces similar results, which are better than the EfficientNet-B7 model independently.
VGG, ResNet and EfficientNet were all used to perform model tests. One model from each model family was selected as the basic model, since it generated high accuracy results. Each model’s accuracy has greatly improved as a result of this setup. This model’s combination experiment takes two to three times as long as the models that conduct experiments individually, depending on the number of models that can be combined. As shown in
Table 4, the time necessary to combine three particular models to obtain the best results, is equal to the training time for each model. This impact does not apply to the combinations inside the same model, since the predicted outcomes do not need numerous iterations, allowing the results to be taken from the same model without having to retrain the model. However, by combining the two models in the same model, it does not significantly enhance the model classification accuracy. As a result, in order to obtain more optimum findings, the time efficiency of this model combination experiment must be addressed.
3.3. Evaluation
We compared and contrasted two categorization studies, each of which had its own set of benefits and drawbacks. This study discovered mixed findings from all of the models evaluated in the different model trials. These outcomes were influenced by the model architecture used, as well as the fine-tuning the model. The projected time for each model was also dependent on the parameters that were utilized during training; the more parameters that are used, the longer the training will take. Furthermore, the training per epoch method produced diverse graphs. The graphs observed did not develope steadily and there was significant irregularity in the graphs acquired. As a result, it is not recommended to use a model with such dramatic findings for classifying medical pictures. The F1-score evaluation parameter was utilized to examine the value of each class and the mean of each model. The accuracy and recall levels in the arithmetic theorem provided this F1-score.
Table 5 indicates that the normal glomeruli class dominates the EfficientNet-L2 model’s prediction results, which is consistent with
Figure 6d, which displays predictions in the normal class. In
Table 6, the class predictions for ResNet101V2 are dispersed in each class with a modest error rate, as shown in
Figure 6b, in which the original and predicted values overlap fairly well.
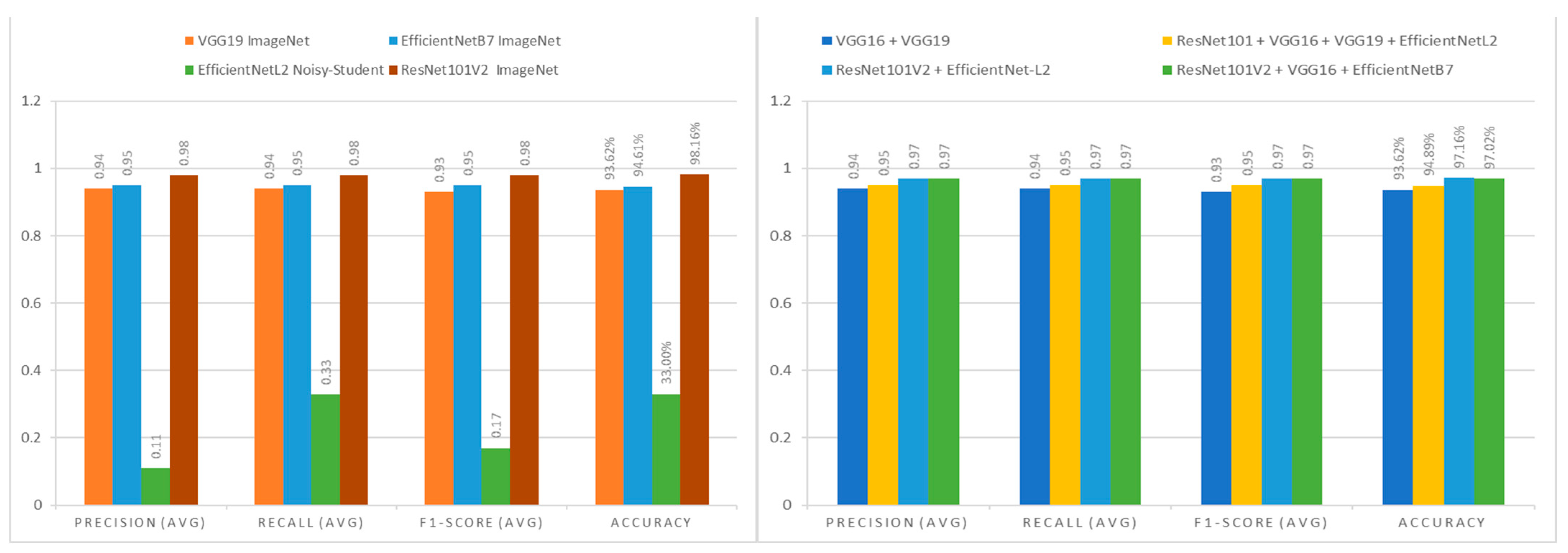
When comparing the independent assessment models displayed in
Figure 8 (left), it is clear that EfficientNet-L2 is not acceptable as a reference model for medical image classification since it has the lowest evaluation value, with just a 0.17 F1-score. With an F1-score of 0.98, ResNet101V2 is the best reference model for medical picture categorization. The findings of the combination technique experiment were identical for each model combination.
Table 4 demonstrates that the model combination experiment yields findings that are more than 90% accurate over time.
Figure 8 shows this outcome, with a predictive value in each class’s distribution that matches the original value and many error values in the incorrect predictions.
Based on these findings, this study assesses each model’s classification prediction class to demonstrate the impact of accuracy on model performance. The three model combination trials revealed a metric assessment value that was relatively steady with a low error rate (
Table 7,
Table 8 and
Table 9). The prediction value of the class distribution, which is near to 1.00, indicates that the forecast value closely follows the actual value.
Figure 8 (right) indicates that all the combination technique trials achieved an assessment value of more than 0.90, with an average F1-score of 0.955. However, the predicted training time for this model combination experiment was very high, with an average of 28 min required, making it less time efficient.