Model Interpretation Considering Both Time and Frequency Axes Given Time Series Data
Abstract
1. Introduction
- We propose the novel approach to interpret a deep learning classifier given medical signals such as EEG considering both aspects of time and frequency while physicians consider both of them to diagnose in practice.
- In the experiments, we confirm the suggestion from this work with the real-world dataset, which is EEG signals recorded from patients during polysomnogrphic studies.
- We show that our suggestion captures the probable explanations such as K-complexes and delta waves, which are considered strong evidence of the second sleep stage and the third sleep stage, respectively.
2. Related Work
2.1. Model Interpretation in General Domain
2.2. Model Interpretation in Medical Domain
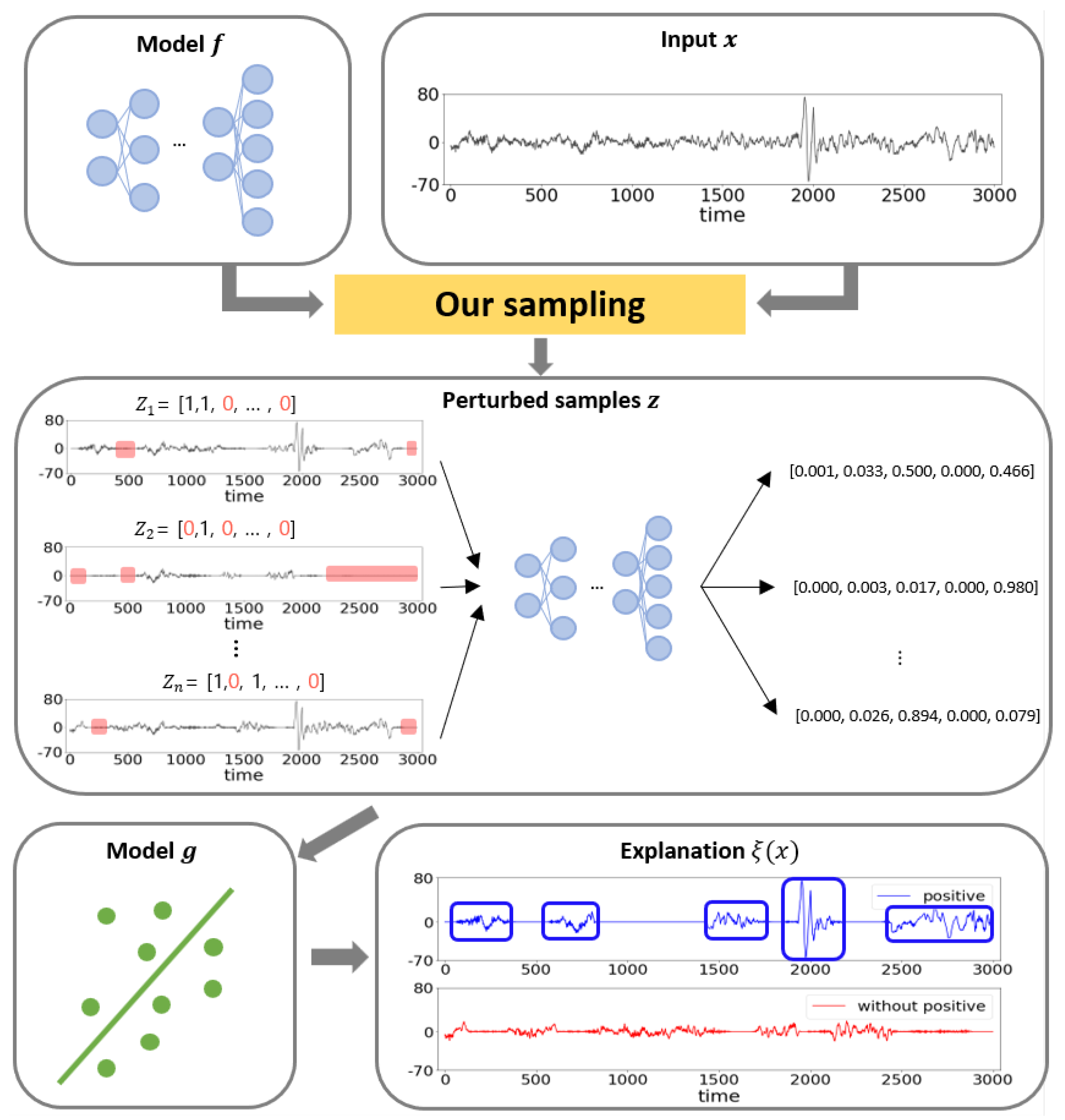
3. Model Interpretation for Signal Classifier
3.1. Prerequisites
3.2. Perturbed Sample Generation for Time Series Data
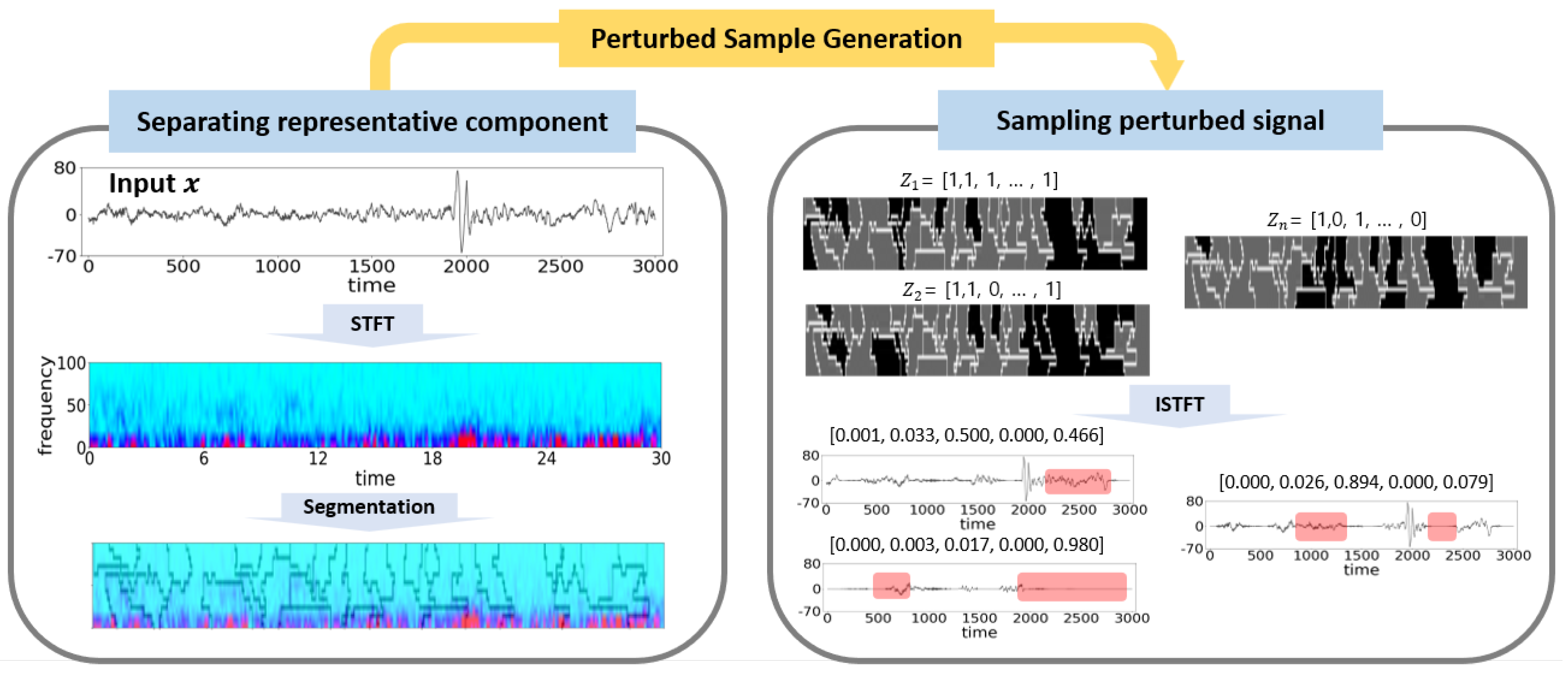
3.2.1. Separating Representative Component of Signal Data Using STFT and Super-Pixel
3.2.2. Restoration the Signal Data from Separated Representations
3.3. Data Description
4. Experiments
4.1. Implementation Methods
- Target model: Since our main goal is to interpret a deep learning classifier, we train a deep learning model, which is to be explained, by the proposed method. The target model is proposed for automatic sleep stage classification [39]. We train the model using the SleepEDF dataset, which is described in Section 3.3. The model consists of convolutional neural networks (CNNs) and long short-term memory networks (LSTMs). CNNs and LSTMs in the model are designed to capture features from a given epoch of an electroencephalography (EEG) signal and from sequential epochs of EEG signals, respectively. Because only CNN layers, which are called FeatureNet, achieve comparative performance with the full model in [39], our target model is FeatureNet.
- LIMESegment [25]: We compare our method with LIMESegment. They also exploit LIME to interpret a classifier for time series. They focus on detecting the change point given the time series data, which are considered the boundary of segments. Once the change points of the input signal are determined comparing the similarity of window slides, it generates perturbed samples. To create the perturbed samples, the intuition is that the segment in the input signal is filtered based on the threshold frequency. The threshold is determined by the highest frequency value over time with minimal variance. After that, anything lower than the threshold is filtered out. We exploit the imeplementation given by the authors from their GitHub repository.
- Ours: We also implement our interpretation method, which is described in Section 3.
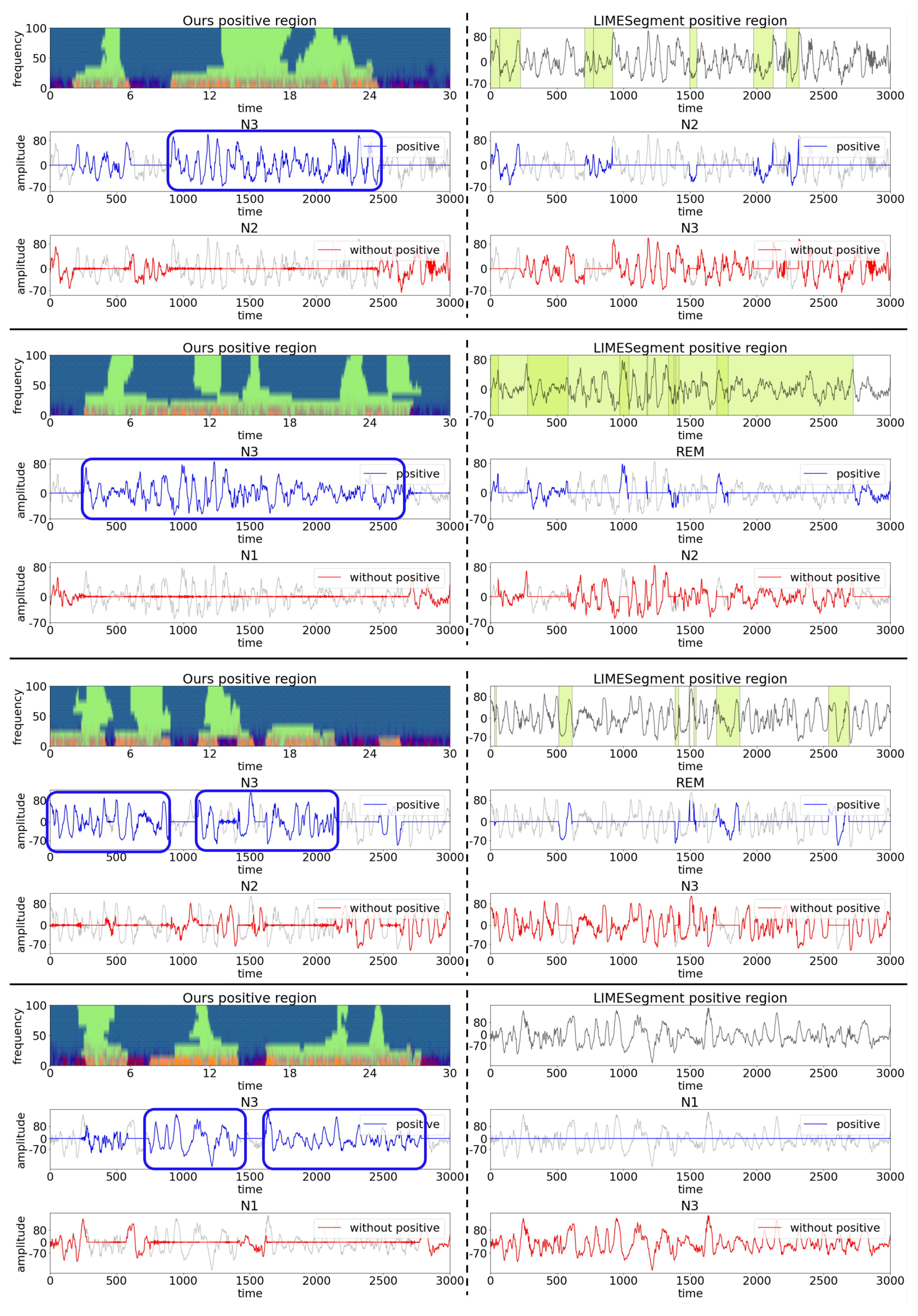
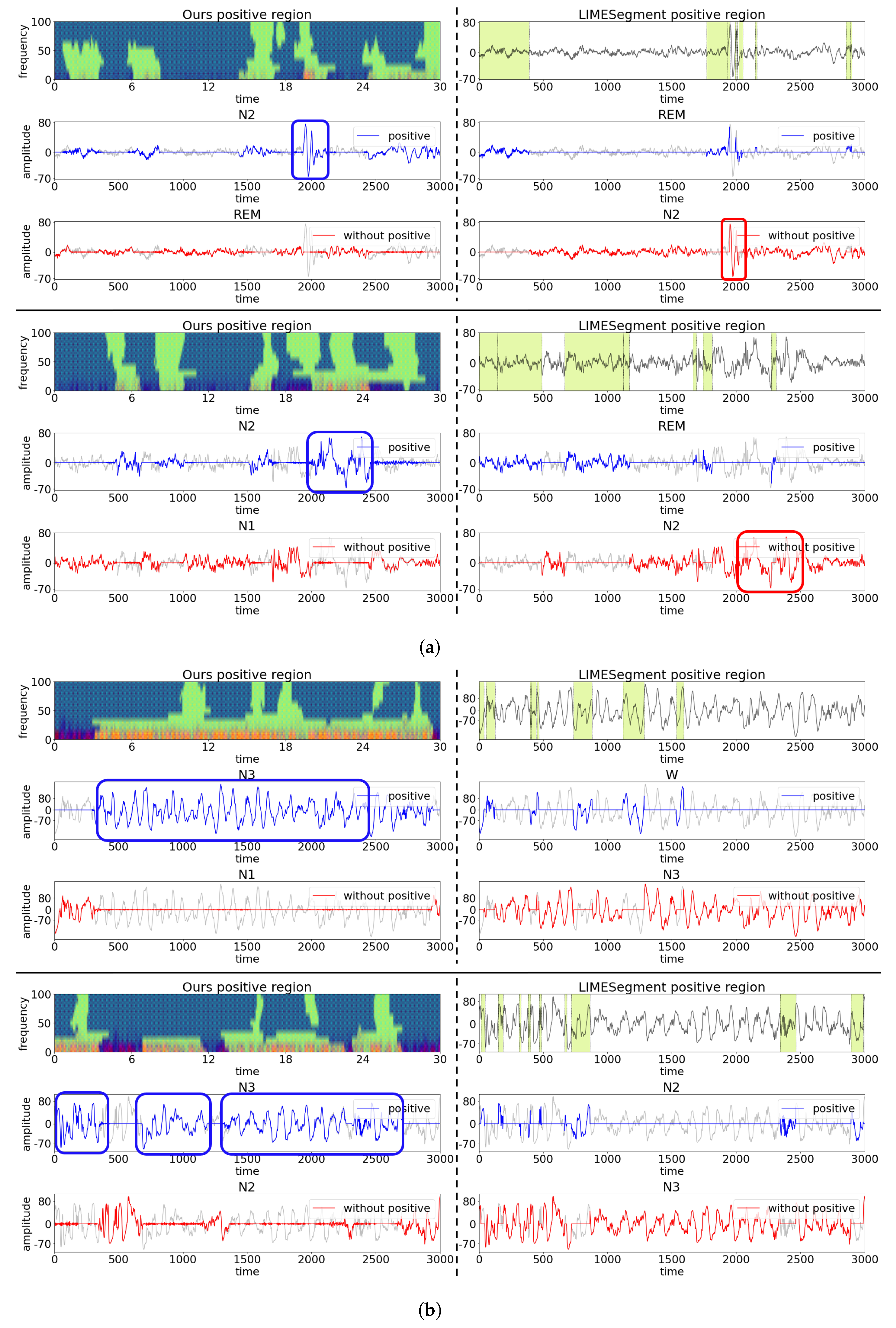
4.2. Qualitative Results
- Null hypothesis: The representations obtained by ours and LIMESegment are same.
- Alternative hypothesis: The representation obtained by ours is more critical for the model decision than the baseline.
4.3. Case Study
5. Discussion
5.1. Analysis of Results
5.2. Possible Application of Proposed Method
5.3. Limitations and Future Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Additional Representations

References
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Granada, Spain, 2018; pp. 3–11. [Google Scholar]
- Zhou, Y.; Huang, W.; Dong, P.; Xia, Y.; Wang, S. D-UNet: A dimension-fusion U shape network for chronic stroke lesion segmentation. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 18, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Kickingereder, P.; Wick, W.; Bendszus, M.; Maier-Hein, K.H. Brain tumor segmentation and radiomics survival prediction: Contribution to the brats 2017 challenge. In Proceedings of the International MICCAI Brainlesion Workshop, Quebec City, QC, Canada, 14 September 2017; pp. 287–297. [Google Scholar]
- Kayalibay, B.; Jensen, G.; van der Smagt, P. CNN-based segmentation of medical imaging data. arXiv 2017, arXiv:1701.03056. [Google Scholar]
- Razavian, N.; Marcus, J.; Sontag, D. Multi-task prediction of disease onsets from longitudinal laboratory tests. In Proceedings of the Machine Learning for Healthcare Conference. PMLR, Los Angeles, CA, USA, 19–20 August 2016; pp. 73–100. [Google Scholar]
- Kachuee, M.; Fazeli, S.; Sarrafzadeh, M. Ecg heartbeat classification: A deep transferable representation. In Proceedings of the 2018 IEEE International Conference on Healthcare Informatics (ICHI), New York, NY, USA, 4–7 June 2018; pp. 443–444. [Google Scholar]
- Yamaç, M.; Duman, M.; Adalıoğlu, İ.; Kiranyaz, S.; Gabbouj, M. A Personalized Zero-Shot ECG Arrhythmia Monitoring System: From Sparse Representation Based Domain Adaption to Energy Efficient Abnormal Beat Detection for Practical ECG Surveillance. arXiv 2022, arXiv:2207.07089. [Google Scholar]
- Li, R.; Zhang, X.; Dai, H.; Zhou, B.; Wang, Z. Interpretability analysis of heartbeat classification based on heartbeat activity’s global sequence features and BiLSTM-attention neural network. IEEE Access 2019, 7, 109870–109883. [Google Scholar] [CrossRef]
- Bin Heyat, M.B.; Akhtar, F.; Khan, A.; Noor, A.; Benjdira, B.; Qamar, Y.; Abbas, S.J.; Lai, D. A novel hybrid machine learning classification for the detection of bruxism patients using physiological signals. Appl. Sci. 2020, 10, 7410. [Google Scholar] [CrossRef]
- Heyat, M.B.B.; Lai, D.; Khan, F.I.; Zhang, Y. Sleep bruxism detection using decision tree method by the combination of C4-P4 and C4-A1 channels of scalp EEG. IEEE Access 2019, 7, 102542–102553. [Google Scholar] [CrossRef]
- Heyat, M.B.; Akhtar, F.; Khan, M.H.; Ullah, N.; Gul, I.; Khan, H.; Lai, D. Detection, treatment planning, and genetic predisposition of bruxism: A systematic mapping process and network visualization technique. CNS Neurol. Disord.-Drug Targets (Former. Curr. Drug Targets-CNS Neurol. Disord.) 2021, 20, 755–775. [Google Scholar]
- Bin Heyat, M.B.; Akhtar, F.; Ansari, M.; Khan, A.; Alkahtani, F.; Khan, H.; Lai, D. Progress in detection of insomnia sleep disorder: A comprehensive review. Curr. Drug Targets 2021, 22, 672–684. [Google Scholar]
- Sun, J.; Darbehani, F.; Zaidi, M.; Wang, B. Saunet: Shape attentive u-net for interpretable medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 797–806. [Google Scholar]
- Ashtari, P.; Sima, D.; De Lathauwer, L.; Sappey-Marinierd, D.; Maes, F.; Van Huffel, S. Factorizer: A Scalable Interpretable Approach to Context Modeling for Medical Image Segmentation. arXiv 2022, arXiv:2202.12295. [Google Scholar] [CrossRef]
- Margeloiu, A.; Simidjievski, N.; Jamnik, M.; Weller, A. Improving interpretability in medical imaging diagnosis using adversarial training. arXiv 2020, arXiv:2012.01166. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Volume 30. [Google Scholar]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you? ” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- He, T.; Guo, J.; Chen, N.; Xu, X.; Wang, Z.; Fu, K.; Liu, L.; Yi, Z. MediMLP: Using Grad-CAM to extract crucial variables for lung cancer postoperative complication prediction. IEEE J. Biomed. Health Inform. 2019, 24, 1762–1771. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Chen, R.; Wei, W.; Belovsky, M.; Lu, X. Understanding Heart Failure Patients EHR Clinical Features via SHAP Interpretation of Tree-Based Machine Learning Model Predictions. In AMIA Annual Symposium Proceedings; American Medical Informatics Association: San Diego, CA, USA, 2021; Volume 2021, p. 813. [Google Scholar]
- Magesh, P.R.; Myloth, R.D.; Tom, R.J. An explainable machine learning model for early detection of Parkinson’s disease using LIME on DaTSCAN imagery. Comput. Biol. Med. 2020, 126, 104041. [Google Scholar] [CrossRef] [PubMed]
- Neves, I.; Folgado, D.; Santos, S.; Barandas, M.; Campagner, A.; Ronzio, L.; Cabitza, F.; Gamboa, H. Interpretable heartbeat classification using local model-agnostic explanations on ECGs. Comput. Biol. Med. 2021, 133, 104393. [Google Scholar] [CrossRef] [PubMed]
- Sivill, T.; Flach, P. LIMESegment: Meaningful, Realistic Time Series Explanations. In Proceedings of the International Conference on Artificial Intelligence and Statistics. PMLR, Virtual, 28–30 March 2022; pp. 3418–3433. [Google Scholar]
- Berry, R.B.; Brooks, R.; Gamaldo, C.E.; Harding, S.M.; Marcus, C.; Vaughn, B.V. The AASM manual for the scoring of sleep and associated events. Rules Terminol. Tech. Specif. Darien Ill. Am. Acad. Sleep Med. 2012, 176, 2012. [Google Scholar]
- Krahn, L.E.; Silber, M.H.; Morgenthaler, T.I. Atlas of Sleep Medicine; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Moser, D.; Anderer, P.; Gruber, G.; Parapatics, S.; Loretz, E.; Boeck, M.; Kloesch, G.; Heller, E.; Schmidt, A.; Danker-Hopfe, H.; et al. Sleep classification according to AASM and Rechtschaffen & Kales: Effects on sleep scoring parameters. Sleep 2009, 32, 139–149. [Google Scholar] [PubMed]
- Fournier, E.; Arzel, M.; Sternberg, D.; Vicart, S.; Laforet, P.; Eymard, B.; Willer, J.C.; Tabti, N.; Fontaine, B. Electromyography guides toward subgroups of mutations in muscle channelopathies. Ann. Neurol. Off. J. Am. Neurol. Assoc. Child Neurol. Soc. 2004, 56, 650–661. [Google Scholar] [CrossRef]
- Li, Q.; Gao, J.; Huang, Q.; Wu, Y.; Xu, B. Distinguishing Epileptiform Discharges From Normal Electroencephalograms Using Scale-Dependent Lyapunov Exponent. Front. Bioeng. Biotechnol. 2020, 8, 1006. [Google Scholar] [CrossRef]
- Li, Q.; Gao, J.; Zhang, Z.; Huang, Q.; Wu, Y.; Xu, B. Distinguishing epileptiform discharges from normal electroencephalograms using adaptive fractal and network analysis: A clinical perspective. Front. Physiol. 2020, 11, 828. [Google Scholar] [CrossRef]
- Gao, J.; Sultan, H.; Hu, J.; Tung, W.W. Denoising nonlinear time series by adaptive filtering and wavelet shrinkage: A comparison. IEEE Signal Process. Lett. 2009, 17, 237–240. [Google Scholar]
- Shrikumar, A.; Greenside, P.; Kundaje, A. Learning important features through propagating activation differences. In Proceedings of the International Conference on Machine Learning. PMLR, Sydney, Australia, 6–11 August 2017; pp. 3145–3153. [Google Scholar]
- Felzenszwalb, P.F.; Huttenlocher, D.P. Efficient graph-based image segmentation. Int. J. Comput. Vis. 2004, 59, 167–181. [Google Scholar] [CrossRef]
- Guillemé, M.; Masson, V.; Rozé, L.; Termier, A. Agnostic local explanation for time series classification. In Proceedings of the 2019 IEEE 31st International Conference on Tools with Artificial Intelligence (ICTAI), Portland, OR, USA, 4–6 November 2019; pp. 432–439. [Google Scholar]
- Oppenheim, A.V. Discrete-Time Signal Processing; Pearson Education India: Noida, India, 1999. [Google Scholar]
- Kemp, B.; Zwinderman, A.H.; Tuk, B.; Kamphuisen, H.A.; Oberye, J.J. Analysis of a sleep-dependent neuronal feedback loop: The slow-wave microcontinuity of the EEG. IEEE Trans. Biomed. Eng. 2000, 47, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Wolpert, E.A. A Manual of Standardized Terminology, Techniques and Scoring System for Sleep Stages of Human Subjects. Arch. Gen. Psychiatry 1969, 20, 246–247. [Google Scholar] [CrossRef]
- Supratak, A.; Dong, H.; Wu, C.; Guo, Y. DeepSleepNet: A model for automatic sleep stage scoring based on raw single-channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1998–2008. [Google Scholar] [CrossRef] [PubMed]
| Methods\Sleep Stages | W | N1 | N2 | N3 | REM |
|---|---|---|---|---|---|
| LIMESegment | 0.07 | 0.23 | 0.04 | 0.24 | 0.05 |
| Ours | 0.46 | 0.76 | 0.51 | 0.99 | 0.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, W.; Kim, G.; Yu, J.; Kim, Y. Model Interpretation Considering Both Time and Frequency Axes Given Time Series Data. Appl. Sci. 2022, 12, 12807. https://doi.org/10.3390/app122412807
Lee W, Kim G, Yu J, Kim Y. Model Interpretation Considering Both Time and Frequency Axes Given Time Series Data. Applied Sciences. 2022; 12(24):12807. https://doi.org/10.3390/app122412807
Chicago/Turabian StyleLee, Woonghee, Gayeon Kim, Jeonghyeon Yu, and Younghoon Kim. 2022. "Model Interpretation Considering Both Time and Frequency Axes Given Time Series Data" Applied Sciences 12, no. 24: 12807. https://doi.org/10.3390/app122412807
APA StyleLee, W., Kim, G., Yu, J., & Kim, Y. (2022). Model Interpretation Considering Both Time and Frequency Axes Given Time Series Data. Applied Sciences, 12(24), 12807. https://doi.org/10.3390/app122412807









