Featured Application
The preliminary study showed that paraprobiotics-containing moisturizers could improve skin hydration and positively alter the skin microbiome in healthy adults. The findings might support the development of paraprobiotics-based cosmetics to improve skin health.
Abstract
The skin is a physical barrier to protect the human body and is rich in microbial niches. Skin is damaged due to several factors, including poor nutrition and exposure to harsh environments resulting in dryness, acidic skin, and infections. Studies have shown that probiotics and their derivatives could protect the skin. Skin care products with probiotic components are the latest approach to developing cosmetic products with health benefits. The current study aimed to examine the moisturizing effect of paraprobiotics (moist heat-inactivated Bifidobacterium lactis (B. lactis), Lactobacillus plantarum (L. plantarum))-containing moisturizer (APM) and its influences on the skin microbiome of healthy subjects. Fifty healthy subjects were randomly divided into treatment (n = 25) and control (n = 25) groups. The APM or placebo (without paraprobiotics; PM) was applied on the skin of the right forehand of subjects, and the changes in transepidermal water loss (TEWL) and stratum corneum moisture (SCM) levels every 1 h for 4 h and after 4 weeks of treatment were observed. Skin swab samples were collected before and after the treatments (4 weeks) and subjected to microbiome analysis through next-generation sequencing technology. The results indicated that the APM treatment significantly reduced the TEWL and increased the SCM values compared to the respective baseline values and controls. The sequencing study showed significant changes in Cutibacterium (p = 0.0431), Corynebacterium (p = 0.0431), and Acinetobacter (p = 0.0431) in the treatment group. The changes in phylum were not statistically significant. Still, based on the relative frequency, the abundance of phylum Proteobacteria and Firmicutes and Cyanobacterial was decreased, and the abundance of Planctomycetes, Chloroflexi, Verrucomicrobia, and Gemmatimonadetes was increased after treatment. Additionally, the APM treatment suppressed C. tuberculostearicum in healthy subjects. The results suggested that APM could improve skin hydration and skin-beneficial microbial composition. The study has limitations such as a small sample size and treatment period, so further extensive studies are required to confirm the findings of the current study, which could aid in developing paraprobiotics-based skin care formulations.
1. Introduction
Skin is the first line of defense against external intruders and serves as a haven for various bacteria. Skin bacteria’s degree of richness and diversity is varied and related to distinct skin locations and their physiological properties [1]. Some skin parts have higher microbial diversity than the gut and dental cavities. Within the community investigated, individual variation differed widely, while the temporal stability of the assessed skin microbial populations was reasonably consistent [2,3]. They were considered commensals (non-pathogenic permanent inhabitants) or transients (temporary residents) [4,5]. Most skin pathogens are opportunistic, i.e., living on the skin as commensals; in their favorable conditions, including microbial dysbiosis or microbial imbalance, antibiotic use, and host immune status, they become pathogens [6].
Skin functionality was severely affected by reduced skin moisture, age-related dermal and epidermal junction modification, degeneration of vital skin structural protein, microbial alteration, and fragility resulting in fine lines and wrinkles, uneven skin tone, and dryness [7]. Consequently, maintaining the balance of the skin microbiome with the dermal and epidermal is essential for skin health [8].
In recent years, skin disorders have received much attention. People have been concerned about skin aging, wrinkles, pigmentation, and dryness [9]. Researchers are focusing on developing skin care products such as an anti-aging facial moisturizer with a neutral skin pH (4.5–5.5) and other active ingredients, such as Astragalus membranaceus root extract, ursolic acid, and tetrahexyldecyl ascorbate, rhamnose, silanetriol, and other natural extracts, to prevent the skin from UV-A radiation and improve skin thickness [10,11].
Cosmetics with chemicals and herbal extracts may be toxic or cause side effects and are less effective for skin health [12]. Nowadays, micro-ecological skincare is preferable, and using probiotics to improve skin health is a choice of several studies [9,13]. Parameters such as moisture retention, transepidermal water loss (TEWL), elasticity, and wrinkles are the determinants of the efficacy of orally ingested probiotics in maintaining healthy skin by reducing the roughness and dryness on the skin [14,15,16].
Probiotics are reported for their role in preventing, treating, and managing skin aging and diseases [17,18,19]. Lactobacillus spp. and Bifidobacterium spp. are the most used probiotics to treat skin diseases [17,20]. Furthermore, research on topical probiotics in skincare and dermatological therapy is still in its infancy. Further investigation is warranted to establish their security, effectiveness, and mode of action in various skin conditions [13].
The paraprobiotics (non-viable cells; intact or broken) and postbiotics (non-viable bacterial products or metabolic byproducts of probiotics) have attracted attention from the food and pharmacological industries recently [21]. Several questions, including the bioactive components of postbiotics, the processes through which they confer health benefits [22,23], and the immunological aspects of postbiotics, still need to be answered completely [21]. However, many researchers have accelerated their research, implementing postbiotics to identify its benefits in areas including the food industry, pharmaceutics, and cosmetics.
Recently, Kim et al. reported that the L. plantarum K8 strain isolated from kimchi could improve skin moisture by regulating the expression of hyaluronic acid synthase-2 and aquaporin-3 in HaCaT cells. The results showed that paraprobiotics of L. plantarum K8 might be used in moisturizing cosmetic products [24].
The studies on paraprobiotics are very limited, and the proficiency of paraprobiotics in skin care products has not been elucidated in detail. Studies on human subjects are necessary to confirm the activity of the paraprobiotics-containing moisturizing product. Thus, the study’s objective was to analyze the impact of paraprobiotics (Bifidobacterium lactis (B. lactis) and Lactobacillus plantarum (L. plantarum)-containing moisturizer on the skin hydration and skin microbiome of healthy subjects.
2. Materials and Methods
2.1. Preparation of Moisturizer
L. plantarum and B. lactis were purchased from LactoMason Co., Ltd., Gyeongsangnam-do, Republic of Korea, as a freeze-dried powder. The probiotic powders were autoclaved (121 °C for 15 min) and used to prepare the moisturizer. The formulations comprised autoclaved L. plantarum and B. lactis cells (1 × 1010 CFU each), xanthan gum (1% w/w), and water. First, xanthan gum was dispersed into water and stirred until the dispersion was complete. Then, the autoclaved probiotics were combined into the mixture (APM). The placebo formulation was prepared without autoclaved probiotics (PM).
2.2. Study Design and Analysis
Fifty healthy subjects, either men or women, were included in the study. The inclusion and exclusion criteria of the subjects are as follows. Inclusion criteria: (1) Aged between 20–60 years old; (2) No history of skin diseases, tattoos, or scars; (3) No history of cosmetics allergy; (4) No psychiatric illness; (5) No hospitalization during the past 6 months before the study; (6) No history of immune-related diseases; (7) No antibiotic use during the past 14 days before the study. Exclusion criteria: (1) Pregnancy or breastfeeding mother. The subjects were asked to stop using other moisturizing cosmetics for 3 days before the study. The subjects were randomly allocated into two groups (Placebo (P); n = 25 and treatment (T); n = 25). Each group received either APM or PM.
Skin hydration was measured using MoistureMeterSC (Delfin Technologies Ltd., Kuopio, Finland) and VapoMeter® (Delfin Technologies Ltd., Kuopio, Finland). There were two courses of study, namely short-term and long-term studies. For the short-term study, subjects’ skin moisture was measured and recorded at 0 h (baseline) and then measured again every hour after the moisturizer application for 4 h. For the long-term study, the subjects were asked to apply their respective moisturizers twice a day on their skin for 4 weeks. The changes in their skin were assessed after 4 weeks.
The study protocol was approved by the ethical committee of the Faculty Pharmacy, Chiang Mai University (Approval no. 11/2563 dated 28 February 2020).
2.3. Next-Generation Sequencing
Due to the insufficient quality and quantity of skin metagenomes, we sequenced only 10 samples (Control; n = 5; treatment; n = 5) to determine the microbiome in the present study. The baseline and after four weeks of treatment samples were denoted as pre- and post-samples of control (Cpre and Cpost) and treatments (Tpre and Tpost). The samples codes are denoted as SK11-1-Cpre/post, SK12-1-Cpre/post, SK16-1-Cpre/post, SK19-1-Cpre/post, SK9-1-Cpre/post for the control group, and SK10-1-Tpre/post, SK14-1-Tpre/post, SK2-1-Tpre/post, SK6-1-Tpre/post, SK8-1-Tpre/post for the treatment group.
The sequencing of metagenomics isolated from the skin swab was performed as detailed in our previous study. In short, the 16S rRNA gene was amplified using 16S V3-V4 primers and sparQ HiFi PCR master mix (Quantabio, Beverly, MA, USA). Subsequently, the 16S amplicon was purified using sparQ PureMag Beads (Quantabio, Beverly, MA, USA). The amplicon was indexed using Nextera XT index primer and amplified. Then, the amplicon was cleaned, pooled, and diluted to the final loading concentration (4 pM). Cluster generation and 250-bp paired-end read sequencing were performed on an Illumina MiSeq at Omics Sciences and Bioinformatics Center (Chulalongkorn University, Bangkok, Thailand).
2.4. Microbiome Analyses
The paired-end data for each sample were obtained as forward reads and reverse reads in two separate fastq files, which were paired using QIIME2.0™ using 16S rRNA workflow. Phylogenetic diversity (PD) was annotated from the rarefaction curve. The Shannon diversity index was calculated for the control and treatment samples. A separate comparison between post-samples of control and treatment groups was performed to estimate species diversity.
The Shannon diversity index was used to calculate the significant differences between the groups by implementing the Kruskal–Wallis (pairwise) test. The relation between samples was identified using PCoA and visualized in QIIME 2 View. Herein, the first three main coordinates were used to create PCoA plots and labeled according to their variance. The taxonomy (phylum to species) was assigned to the samples. The intra (Cpro vs. Cpost; Tpro vs. Tpost) and inter-group comparisons (Cpro vs. Tpro; Cpost vs. Tpost) were also performed.
2.5. Statistical Analysis
All the statistical analysis was performed using STATA/IC 14.0 (StataCorp L.L.C., College Station, TX, USA). The demographic parameters such as gender, age, and BMI were assessed by implementing the exact Fisher test, rank-sum, and t-test with a statistical significance of p ≤ 0.05.
The differences in TEWL and SCM were calculated by paired t-test and Wilcoxon’s sign rank test and expressed as the mean differences. Additionally, the linear regression analysis was performed to obtain the coefficients for the TEWL and SCM at the 4th h and 4th weeks (p ≤ 0.05). Significant changes in the microbiome at the phylum, genus, and species in the control and treatment groups were analyzed using Wilcoxon’s sign rank and rank-sum tests (p ≤ 0.05). Power analysis was performed to confirm the validity of the statistical analysis.
3. Results
3.1. Demographic Parameters
The gender, age, and body mass index (BMI) of the study subjects were tabulated (Table 1). Fisher’s exact, Wilcoxon rank-sum, and t-tests were employed to confirm the indifference in the recruited subjects. There were no statistically significant differences (p ≥ 0.05) among the subjects in the treatment and placebo groups (Table 1).

Table 1.
Grouping and demographic data.
3.2. Skin Moisture
The transepidermal water loss (TEWL) was slightly reduced in the placebo from the first hour to the fourth hour and in the fourth week. The stratum corneum moisture (SCM) values were flexible during analysis in the placebo group. The changes in TEWL and SCM values were not statistically significant in the placebo. The treatment group showed a statistically significant reduction in TEWL values and increased SCM values compared to the respective baseline values (Table 2). The TEWL and SCM values between the placebo and treatment groups significantly differed. The TEWL value was significantly reduced, and SCM values were high in the treatment group compared to the placebo (p ≤ 0.05) (Table 3).

Table 2.
The changes in the studied parameters within the group at different times. The values are expressed as the mean ± SE.

Table 3.
The comparison of the changes in the studied parameters between the groups at different times is expressed as the mean difference.
The reduction in TEWL in the treatment group was −2.494 mg(water)/m2/hour after four hours of APM application on the skin (Table 4), and an increase in the SCM was 13.190 Au in the treatment group after four hours of APM application on the skin (Table 5). The changes were significant compared to the placebo group (p < 0.0001) (Table 4 and Table 5). Similarly, the reduction in TEWL in the treatment group was −1.251 mg(water)/m2/hour after four weeks of treatment (p = 0.010) (Table 6), and an increase in the SCM was 4.615 Au in the treatment group after four hours of APM application on the skin (p = 0.041) (Table 7). The changes were significant compared to the placebo group (Table 6 and Table 7).

Table 4.
Linear regression analysis of changes in the transepidermal water loss (TEWL) after 4 h of treatment.

Table 5.
Linear regression analysis of changes in the stratum corneum moisture (SCM) after 4 h of treatment.

Table 6.
Linear regression analysis of changes in the transepidermal water loss (TEWL) after 4 weeks of treatment.

Table 7.
Linear regression analysis of changes in the stratum corneum moisture (SCM) after 4 weeks of treatment.
3.3. Microbiome Analysis
The total sequences read in QIIME2 were 1,109,656, 1,248,103, 1,045,814, and 372,775 for Cpre, Cpost, Tpre, and Tpost, respectively. After filtering, denoising, and merging the non-chimeric sequences, about 96,781, 73,948, 65,699, and 43,193 read were obtained for Cpre, Cpost, Tpre, and Tpost, respectively (Table S1; Supplementary File S1).
3.3.1. Rarefaction Curve Analysis
The skin microbial diversity of the control and treatment groups was assessed. The microbial richness was increased in Cpost and Tpost samples compared to their respective baseline samples (Figure 1A,B).
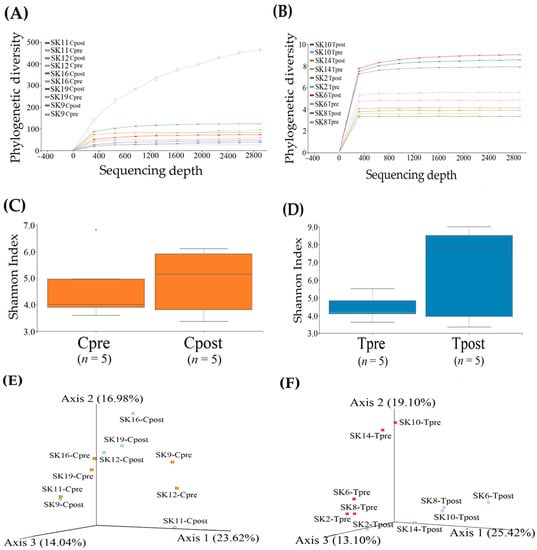
Figure 1.
Refraction analysis for the pre-and post-samples of the control and treatment groups. The rarefaction curves of the phylogenetic diversity were computed and displayed for the control (A) and treatment (B) samples. Shannon H-index alpha diversity of the control (C) and treatment (D) samples. Comparison of similarity and dissimilarity for the control (E) and treatment (F) samples using PCoA represent unweighted unifrac distances.
Microbial Richness
The diversity richness of the skin microbiome was estimated using the Shannon index. The Shannon index revealed that the microbial diversity richness was increased in the Cpost samples with a median value of 5.16 (lower quartile (LQ) = 3.37; upper quartile (UQ) = 6.11) compared to the Cpre samples (LQ = 3.59; Median = 4.00; UQ = 6.82) (Figure 1C). Likewise, the diversity richness was significantly increased in the Tpost sample with a median value of 7.91 (LQ = 3.35; UQ = 9.00) compared to the Tpre samples (LQ = 3.62; Median = 4.17; UQ = 5.51) (Figure 1D). The Kruskal–Wallis (pairwise) test identified the Shannon group significance of control (p = 0.92815) and treatment (p = 0.464702) groups. The results revealed that there could be a significant microbial difference in the treatment group compared to the control group.
Similarity and Variations in the Microbiome
The similarity and variations in the microbiome were performed using principal coordinate analysis (PCoA) with the representation of unweighted UniFrac distances between the groups. The two groups were completely separated from each other in 3D space.
PCoA plot axis 1, axis 2, and axis 3 explained the variances in the microbial abundances in the samples as 23.62%, 16.98%, and 14.04%. The microbial load of SK-11 (Cpre) and SK-9 (Cpost) samples were found in nearly the same place in the PCoA plot, indicating their similarity (Figure 1E). It is likely that the clusters are formed between the pre-and post-samples of SK-19, SK-16, and SK-12, indicating the existence of common species among them.
The PCoA plot for the treatment groups showed significant differences in the microbial abundances and diversity between Tpre and Tpost samples. PCoA plot axis 1, axis 2, and axis 3 explained the variances in microbial abundances in the samples as 25.42%, 19.10%, and 13.10%. SK-2Tpre sample was scattered near the Tpost samples of SK-2 and SK-8, which indicated the subsided microbial similarity between them. Overall, pre- and post-treatment samples were scattered, indicating the significant microbial differences between them (Figure 1F).
3.3.2. Taxonomy Assignment
The taxonomy of the skin microbiome from the control and treated samples are displayed (Figure 2A,B).
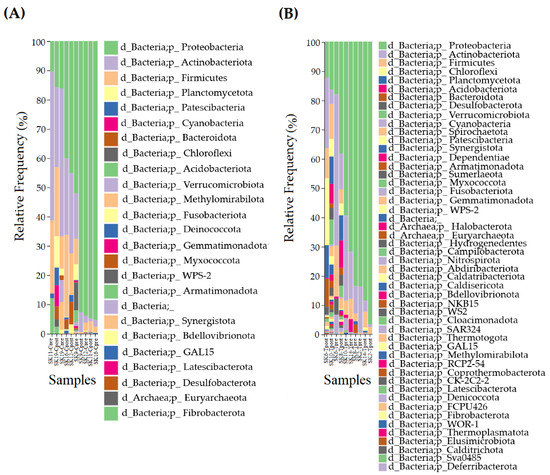
Figure 2.
Taxonomical assignment for the control and treatment samples. The comparison of phylum in the control (A) treatment (B) samples.
Estimation of the Phylum
The relative frequency (RF) of the phylum in control (Cpre and Cpost) (Figure 3A) and treatment (Tpre and Tpost) (Figure 4A) samples were computed and displayed. The phylum such as Proteobacteria (Cpre: RF = 56.40%; Cpost: RF = 58.63%), Firmicutes (Cpre: RF = 11.22%; Cpost: RF = 14.85%), Actinobacteriota (Cpre: RF = 27.59%; Cpost: RF = 18.74%), Cyanobacteria (Cpre: RF = 0.46%; Cpost: RF = 2.00%), Planctomycetota (Cpre: RF = 1.89%; Cpost: RF = 2.50%), Bacteroidota (Cpre: RF = 0.42%; Cpost: RF = 3.27%), and Verrucomicrobiota (Cpre: RF = 2.02%; Cpost: RF = 0.02%) was dominantly detected in the control group (Figure 3A).
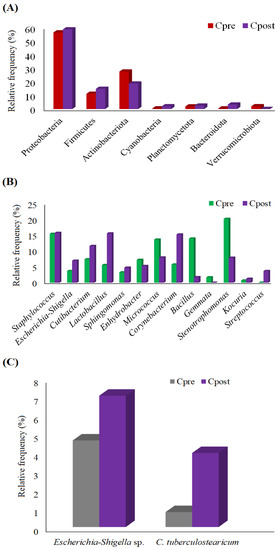
Figure 3.
The graph shows the detected phylum (A), genus (B), and species (C) in the control group.
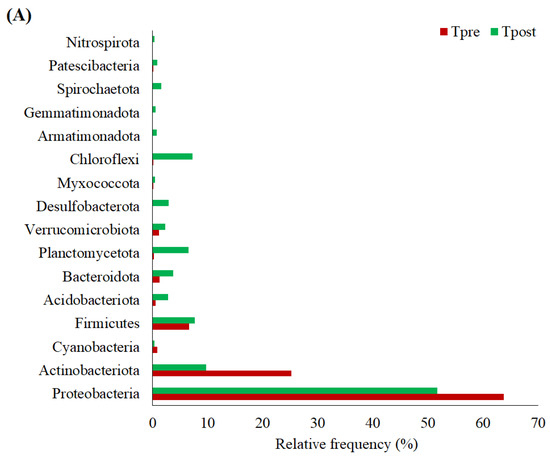

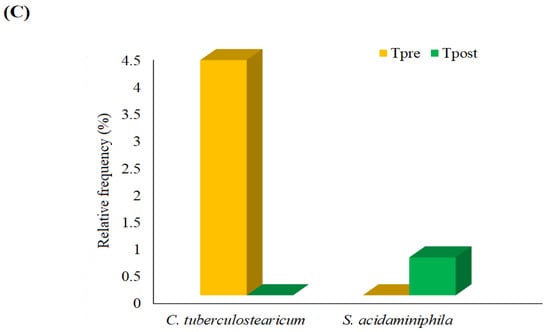
Figure 4.
The graph shows the detected phylum (A), genus (B), and species (C) in the treatment group. * Statistically significant changes in the abundance of the respective genus.
Likely, the phylum, Proteobacteria (Tpre: RF = 63.74%; Tpost: RF = 51.71), Actinobacteriota (Tpre: RF = 25.18%; Tpost: RF = 9.74%), Cyanobacteria (Tpre: RF = 0.92%; Tpost: RF = 0.32%), Firmicutes (Tpre: RF = 6.60%; Tpost: RF = 7.65%), Acidobacteriota (Tpre: RF = 0.61%; Tpost: RF = 2.88%), Bacteroidota (Tpre: RF = 1.25%; Tpost: RF = 3.73%), Planctomycetota (Tpre: RF = 0.21%; Tpost: RF = 6.54%), Verrucomicrobiota (Tpre: RF = 1.17%; Tpost: RF = 2.33%), Desulfobacterota (Tpre: RF = 0%; Tpost: RF = 2.98%), Myxococcota (Tpre: RF = 0.09%; Tpost: RF = 0.48%), Chloroflexi (Tpre: RF = 0.01%; Tpost: RF = 7.25%), Armatimonadota (Tpre: RF = 0%; Tpost: RF = 0.76%), Gemmatimonadota (Tpre: RF = 0%; Tpost: RF = 0.55%), Spirochaetota (Tpre: RF = 0%; Tpost: RF = 1.57%), Patescibacteria (Tpre: RF = 0.03%; Tpost: RF = 0.88%), and Nitrospirota (Tpre: RF = 0%; Tpost: RF = 0.36%) was mainly detected in the treatment samples (Figure 4A).
The significant inter and intra-group differences in phylum were estimated in the control and treatment samples using the non-parametric Wilcoxon matched-paired signed-ranks test. The results indicated no significant differences in phylum within and between the groups (Tables S3 and S4). However, based on the RF, the phylum, such as Proteobacteria, Firmicutes, Cyanobacteria, Planctomycetota, and Bacteroidota, were increased in the Cpost samples. Also, Actinobacteriota and Verrucomicrobiota were decreased in the Cpost samples (Figure 3A and Table S2). Similarly, the phylum, such as Firmicutes, Acidobacteriota, Bacteroidota, Planctomycetota, Verrucomicrobiota, Myxococcota, Chloroflexi, Patescibacteria, was increased in the Tpost samples compared to the Tpre samples. Additionally, the Proteobacteria, Actinobacteriota, and Cyanobacteria were decreased in the Tpost samples compared to the Tpre samples. Likewise, Desulfobacterota, Armatimonadota, Gemmatimonadota, Spirochaetota, and Nitrospirota were detected only in the Tpost samples (Figure 4A and Table S3).
Estimation of the Genus
The RF of the genus was calculated for control and treatment samples and is displayed (Figure 3B and Figure 4B). The genus includes Staphylococcus, Escherichia-Shigella, Cutibacterium, Lactobacillus, Sphingomonas, Enhydrobacter, Micrococcus, Corynebacterium, Bacillus, Gemmata Stenotrophomonas, Kocuria, and Streptococcus were detected in the control group (Figure 3B).
The genus such as Ochrobactrum, Micrococcus, Enhydrobacter, Cutibacterium, Xanthomonas, Corynebacterium, Staphylococcus, Mycobacterium, Sphingomonas, Roseomonas, Paracoccus, Finegoldia, Kocuria, Microbacterium, Chryseobacterium, Haemophilus, Acinetobacter, Allorhizobium, Pla1_lineage, Streptomyces, JG30-KF-CM45, Pseudomonas, Neochlamydia, WD2101, Treponema, Stenotrophomonas, Pseudogracilibacillus, Bacillus, Ruminiclostridium, WPS-2, 67-14, Lysinibacillus, Paenibacillus, Aquisphaera, Bryobacter, IMCC26256, Nitrospira, Mesorhizobium, TK10, Legionella, SJA-28, SC-I-84, and Anaeromyxobacter genus was detected in the treatment group (Figure 4B).
Non-parametric statistical analysis was performed to predict the changes in the detected genus in the control and treatment samples. The Wilcoxon matched-paired signed-ranks test was implemented with the statistical significance of p ≤ 0.05. The statistical outcomes indicated no significant differences between the Cpre and Cpost samples. Cutibacterium (p-value = 0.0431), Corynebacterium (p-value = 0.0431), and Acinetobacter (p-value = 0.0431) showed significant changes in the Tpost samples compared to the Tpre samples (Table S3).
Streptomyces, JG30-KF-CM45, Neochlamydia, WD2101, Treponema, Pseudogracilibacillus, Ruminiclostridium, 67–14, Lysinibacillus, Aquisphaera, Bryobacter, IMCC26256, Nitrospira, Mesorhizobium, TK10, Legionella, SJA-28, SC-I-84, and Anaeromyxobacter were detected only in the Tpost samples (Figure 4B and Table S4).
Estimation of the Species
The relative frequency of species detected in the control and treatment samples was estimated (Figure 3C and Figure 4C). The non-parametric statistical analysis was performed using the Wilcoxon matched-paired signed-ranks test to identify the significant differences among the species (Tables S2 and S3). No significant changes were noted between the control and treatment samples at the species level. However, some species, such as Escherichia-Shigella sp. and Corynebacterium tuberculostearicum (C. tuberculostearicum), were slightly higher in the Cpost samples compared to the Cpre samples (Figure 3C). The predominant changes in RF of C. tuberculostearicum and Stenotrophomonas acidaminiphila were detected in the treatment groups (Figure 4C).
4. Discussion
4.1. Moisturizing Effects of APM
A significant decrease and increase in TEWL and SCM were observed in the treatment group. The mean change in the TEWL and SCM was higher in the treatment group than in the control group, indicating the effect of APM (Table 2 and Table 3).
Iglesia et al. reported that incorporating prebiotics and postbiotics into skin care products might positively impact the facial skin microbiome of healthy subjects [8]. Baldwin et al. reported that selenium-rich postbiotics thermal water and microbial biomass-containing moisturizers could improve barrier function and restore skin homeostasis and microbiome in people with atopic dermatitis [25].
Moreover, the cold creams contained the postbiotics obtained from Bacillus subtilis var. natto and Lactobacillus reuteri accelerated wound healing and helped repair the skin tissues compared to placebo cream in a rat model [26].
The skin care cream containing postbiotics (metabolites, enzymes, organic acids, and peptides) has improved elasticity and moisture in the skin, reduced skin pore size, and wrinkle depth in healthy human subjects significantly, without affecting the sebum production, cleanliness melanin level, and sensitivity [27].
4.2. Changes in Skin Microbiome: Phylum
Grice et al. reported that Proteobacteria, Actinobacteria, Firmicutes, Bacteroidetes, Cyanobacteria, and Acidobacteria are the predominant skin microbiome [28], and the same was observed in the present study. In addition, the phylum Planctomycetota, Bacteroidota, and Verrucomicrobiota was detected in the control samples, and Acidobacteriota, Bacteroidota, Planctomycetota, Verrucomicrobiota, Desulfobacterota, Myxococcota, Chloroflexi, Armatimonadota, Gemmatimonadota, Spirochaetota, Patescibacteria, and Nitrospirota was identified in the treatment samples. The RF values showed the differences in phylum between the control and treatment samples. However, according to the power analysis (two-sample paired means test), statistically, the phylum level changes are not considered significant (Figure 3A and Figure 4A), possibly due to the smaller sample size.
Juge et al. observed that the abundance of Proteobacteria was higher in older women (54 to 69 years old) compared to younger women (21 to 31 years old) on their skin [29]. Compared to healthy skin, firmicutes are predominant in psoriatic lesions [30,31]. The abundance of Proteobacteria was less in Tpost samples and higher in Cpost samples compared to the respective pre-samples. The abundance of Firmicutes was increased in the Cpost samples, and its abundance was controlled in Tpost samples (Figure 3A and Figure 4A).
Burns et al. reported that when the skin is exposed to UV-A and UV-B, the skin’s microbiome might alter, which increases the phylum Cyanobacteria level on the skin [32]. The use of APM reduced the Cyanobacteria compared to the baseline values, indicating that APM might protect the skin against UV (Figure 4A).
Planctomycetes have been reported to produce antibiotics, recognized for their biomedical applications [33]. The abundance of Planctomycetes was increased after APM usage (Figure 4A), indicating that the prepared APM might help develop natural skin antibiotics to prevent other microbial growth.
Chloroflexi was found on the skin of the dominant hand of healthy Chinese people [34]. Chloroflexi load was increased in the Tpost samples (RF = 7.24%), indicating that APM could improve normal skin microbial flora (Figure 4A).
The abundance of Verrucomicrobia was higher in Tpost compared to Cpost samples (Figure 3A and Figure 4A). A lower abundance of Verrucomicrobia was reported in the psoriatic patients [35]. Appropriately, the result of the present study suggested that the APM might prevent the formation of skin lesions.
Somboonna et al. reported that Gemmatimonadetes, Planctomycetes, and Nitrospirae are more prevalent phylum in healthy teenagers [1]. The phylum Gemmatimonadota and Nitrospirota are newly developed in the Tpost samples (Figure 4A and Table S3), which indicates that the APM could improve the healthy skin microbiome.
Some of the predicted phylum Desulfobacterota, Myxococcota, Armatimonadota, Spirochaetota, and Patescibacteria and their role in skin health and diseases are not elucidated.
4.3. Changes in Skin Microbiome: Genus
The microbes belonging to Cutibacterium are the causative agent of “acne vulgaris”, which were uprooted after using APM (Figure 4B and Table S3). Similarly, Acinetobacter, associated with nosocomial infections [36], was uprooted after APM usage.
Corynebacterium had negative correlations with the number of UV spots and positive correlations with TEWL and caused pitted keratolysis, a common plantar infection confined to the thick stratum of corneum [37,38]. The abundance of Corynebacterium was reduced after APM usage (Figure 4B and Table S3). The results indicated that APM could help reduce the stratum corneum’s thickening and UV-induced skin complications.
In thymic epithelial malignancies, Sphingomonas has been found as a dominating genus. These genera comprise the thymoma-specific microbiota [39]. Sphingomonas abundance was reduced in the treatment samples (Figure 4B and Table S3) compared to the control (Figure 3B and Table S2). Haemophilus and Finegoldia are opportunistic pathogens [40,41,42]. Paracoccus was the major genus in leprosy patients [43]. These microbes were not detected after the APM treatment (Figure 4B and Table S3). The results indicated that the APM might reduce skin cancer-associated skin microbiota, inhibit opportunistic pathogens’ growth, and be used as an adjuvant moisturizing agent to manage leprosy.
The bacterial genus, Enhydrobacter, was found in older adults [38] and has been uprooted after APM treatment. The strains belonging to Xanthomonas and Staphylococcus are related to acne [44], slightly painful pimples, and boils on the skin [45] and vanished after APM treatment (Figure 4B and Table S3). The results indicated that APM could reduce the pathogenic skin microbiota associated with skin diseases. Similarly, the abundance of opportunistic pathogens (Roseomonas spp., Haemophilus spp., and Finegoldia spp.) in the skin was reduced or vanished after APM treatment in healthy subjects.
The genus Streptomyces is reported for its antibacterial, antifungal, and antiparasitic activity and has also been used to treat skin infections for millennia [46]. Bacillus, Paenibacillus, and Lysinibacillus have been reported for their antifungal, antibacterial, antitumor, and anti-mycoplastic activities [47,48,49]. These genes were detected in healthy subjects after APM treatment (Figure 4B and Table S3). The results showed that APM treatment could promote the development of healthy skin microbiota in healthy subjects.
Moreover, the abundance of some of the pathogenic and opportunistic pathogenic (Ochrobactrum spp., Mycobacterium spp., Treponema spp., and Stenotrophomonas spp.) [50,51,52,53] bacterial load was increased in Tpost samples (Figure 4B and Table S3). In addition, the genus Chryseobacterium, Allorhizobium, JG30-KF-CM45, Pseudomonas, Neochlamydia, WD2101, Pseudogracilibacillus, 67–14, Aquisphaera, Bryobacter, Mesorhizobium, TK10, Legionella, SJA-28, SC-I-84, and Anaeromyxobacter were detected in the Tpost samples. The significance of these bacterial genera and their association with skin health was unknown.
4.4. Changes in Skin Microbiome: Species
The Wilcoxon matched-paired signed ranks found no significant changes in bacterial species between the control and treatment samples. Thus, the control and treatment groups’ changes were differentiated based on the RF values for the possible discussion.
C. tuberculostearicum induced inflammation in human keratinocytes via TLR2 and activation of IκB kinase and downstream signaling through the canonical NF-κB pathway, which led to chronic skin inflammatory diseases and cutaneous oncology [54]. In the present study, the richness of C. tuberculostearicum was higher in control samples after four weeks compared to baseline (Cpre: RF = 0.80% vs. Cpost: RF = 3.98%), while C. tuberculostearicum was uprooted in the Tpost samples (Figure 3C and Figure 4C and Tables S2 and S3).
Escherichia-Shigella sp. bacteria are notoriously known to exhibit drug resistance and sometimes cause fatal nosocomial infections and were more abundant in the skin of bedridden older age participants [55]. Escherichia-Shigella sp. growth was increased in the Cpost (RF = 7.06%) samples compared to the Cpre (RF = 4.65%) samples (Figure 3C and Figure 4C and Tables S2 and S3). However, Escherichia-Shigella sp. was not detected in the treatment samples (Figure 4C and Table S3). The results showed that the APM treatment could prevent bacterial infections.
4.5. Limitations of the Study
The major limitation of the current study was the smaller sample size and short study period. Additionally, an increase in the abundance of pathogenic strains was detected in treated samples. Therefore, the current study results are preliminary data about how APM affects skin hydration and microbiota in healthy subjects. Further extensive studies are necessary to validate the results.
5. Conclusions
Four weeks of APM treatment significantly improved the TEWL and SCM in healthy subjects compared to the control and respective baseline values. APM treatment increased the abundance of commensal microbes belonging to the phylum Planctomycetes, Chloroflexi, Verrucomicrobia, Gemmatimonadetes, Planctomycetes, and Nitrospirae. The abundance of Proteobacteria and Firmicutes and Cyanobacterial growth was reduced after APM treatment. The beneficial bacterial genera were increased after APM treatment. Meanwhile, some of the pathogenic and opportunistic pathogenic bacteria were detected in the treatment group.
In conclusion, APM treatment improved the skin microbiota associated with skin aging, UV-induced skin damage, acne, skin lesions, and skin diseases. The current study primarily reported the effect of paraprobiotics (B. lactis and L. plantarum)-containing moisturizer on the skin microbiome in healthy Thai subjects. However, as detailed previously, the study has some limitations, so further detailed clinical studies are needed to confirm the beneficial effect of APM on skin hydration and skin microbiome, which could aid in the development of paraprobiotics-based skin care formulations to manage skin diseases.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app122312483/s1. Table S1: The estimated sequences in the control and treatment samples; Table S2: The statistical differences of the phylum, genus, and species level within the Cpre and Cpost samples (Directions based on the RF%); Table S3: The statistical differences of the phylum, genus, and species level within the Tpre and Tpost samples (Directions based on the RF%); Table S4: The statistical differences of the phylum, genus, and species level between control and treatment groups.
Author Contributions
Conceptualization, C.C. and B.S.S.; methodology, C.T., S.P. and C.C.; software, M.B. and C.T.; validation, C.C. and B.S.S.; formal analysis, K.C., M.B. and C.T.; investigation, M.B. and C.C.; resources, C.C.; data curation, M.B.; writing—original draft preparation, M.B., B.S.S. and C.C.; writing—review and editing, M.B., B.S.S. and C.C.; visualization, M.B. and C.T.; supervision, C.C.; project administration, C.C.; funding acquisition, C.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially supported by Chiang Mai University, Chiang Mai, Thailand. The study was supported by the Innovation and Technology Assistance Program (ITAP), Thailand Science Park, Thailand.
Institutional Review Board Statement
The study protocol was approved by the ethical committee of the Faculty Pharmacy, Chiang Mai University (Approval no. 11/2563 dated 28 February 2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available within the article.
Acknowledgments
We thank the Innovation and Technology Assistance Program (ITAP), Thailand Science Park, Thailand, for the support. We thank Chiang Mai University, Chiang Mai, Thailand, for the support. M.B. thankfully acknowledges the CMU Post-doctoral fellowship for its support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Somboonna, N.; Wilantho, A.; Srisuttiyakorn, C.; Assawamakin, A.; Tongsima, S. Bacterial communities on facial skin of teenage and elderly Thai females. Arch. Microbiol. 2017, 199, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.H.; Segre, J.A. Skin microbiome: Looking back to move forward. J. Investig. Dermatol. 2012, 132, 933. [Google Scholar] [CrossRef] [PubMed]
- Suwarsa, O.; Hazari, M.N.; Dharmadji, H.P.; Dwiyana, R.F.; Effendi, R.M.R.A.; Hidayah, R.M.N.; Avriyanti, E.; Gunawan, H.; Sutedja, E. A Pilot study: Composition and diversity of 16S rRNA based skin bacterial microbiome in Indonesian atopic dermatitis population. Clin. Cosmet. Investig. Dermatol. 2021, 14, 1737–1744. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Skowron, K.; Bauza-Kaszewska, J.; Kraszewska, Z.; Wiktorczyk-Kapischke, N.; Grudlewska-Buda, K.; Kwiecińska-Piróg, J.; Wałecka-Zacharska, E.; Radtke, L.; Gospodarek-Komkowska, E. Human skin microbiome: Impact of intrinsic and extrinsic factors on skin microbiota. Microorganisms 2021, 9, 543. [Google Scholar] [CrossRef]
- Findley, K.; Grice, E.A. The skin microbiome: A focus on pathogens and their association with skin disease. PLoS Pathog. 2014, 10, e1004436. [Google Scholar] [CrossRef]
- Roig-Rosello, E.; Rousselle, P. The human epidermal basement membrane: A shaped and cell instructive platform that aging slowly alters. Biomolecules 2020, 10, 1607. [Google Scholar] [CrossRef]
- Iglesia, S.; Kononov, T.; Zahr, A.S. A multi-functional anti-aging moisturizer maintains a diverse and balanced facial skin microbiome. J. Appl. Microbiol. 2022, 133, 1791–1799. [Google Scholar] [CrossRef]
- Yu, J.; Ma, X.; Wang, X.; Cui, X.; Ding, K.; Wang, S.; Han, C. Application and mechanism of probiotics in skin care: A review. J. Cosmet. Dermatol. 2022, 21, 886–894. [Google Scholar] [CrossRef]
- Sevimli-Gür, C.; Onbaşılar, I.; Atilla, P.; Genç, R.; Cakar, N.; Deliloğlu-Gürhan, I.; Bedir, E. In vitro growth stimulatory and in vivo wound healing studies on cycloartane-type saponins of Astragalus genus. J. Ethnopharmacol. 2011, 134, 844–850. [Google Scholar] [CrossRef]
- Pageon, H.; Azouaoui, A.; Zucchi, H.; Ricois, S.; Tran, C.; Asselineau, D. Potentially beneficial effects of rhamnose on skin ageing: An in vitro and in vivo study. Int. J. Cosmet. Sci. 2019, 41, 213–220. [Google Scholar] [CrossRef]
- Dréno, B.; Alexis, A.; Chuberre, B.; Marinovich, M. Safety of titanium dioxide nanoparticles in cosmetics. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 34–46. [Google Scholar] [CrossRef]
- França, K. Topical probiotics in dermatological therapy and skincare: A concise review. Dermatol. Ther. 2021, 11, 71–77. [Google Scholar] [CrossRef]
- Gueniche, A.; Philippe, D.; Bastien, P.; Reuteler, G.; Blum, S.; Castiel-Higounenc, I.; Breton, L.; Benyacoub, J. Randomised double-blind placebo-controlled study of the effect of Lactobacillus paracasei NCC 2461 on skin reactivity. Benef. Microbes 2014, 5, 137–145. [Google Scholar] [CrossRef]
- Lee, D.E.; Huh, C.S.; Ra, J.; Choi, I.D.; Jeong, J.W.; Kim, S.H.; Ryu, J.H.; Seo, Y.K.; Koh, J.S.; Lee, J.H.; et al. Clinical evidence of effects of Lactobacillus plantarum HY7714 on skin aging: A randomized, double blind, placebo-controlled study. J. Microbiol. Biotechnol. 2015, 25, 2160–2168. [Google Scholar] [CrossRef]
- Liu, C.; Tseng, Y.P.; Chan, L.P.; Liang, C.H. The potential of Streptococcus thermophiles (TCI633) in the anti-aging. J. Cosmet. Dermatol. 2022, 21, 2635–2647. [Google Scholar] [CrossRef]
- Sivamaruthi, B.; Kesika, P.; Chaiyasut, C. A review on anti-aging properties of probiotics. Int. J. Appl. Pharm. 2018, 10, 23–27. [Google Scholar] [CrossRef]
- Sharma, D.; Kober, M.M.; Bowe, W.P. Anti-aging effects of probiotics. J. Drugs Dermatol. 2016, 15, 9–12. [Google Scholar]
- Ratanapokasatit, Y.; Laisuan, W.; Rattananukrom, T.; Petchlorlian, A.; Thaipisuttikul, I.; Sompornrattanaphan, M. How microbiomes affect skin aging: The updated evidence and current perspectives. Life 2022, 12, 936. [Google Scholar] [CrossRef]
- Lolou, V.; Panayiotidis, M.I. Functional role of probiotics and prebiotics on skin health and disease. Fermentation 2019, 5, 41. [Google Scholar] [CrossRef]
- Thorakkattu, P.; Khanashyam, A.C.; Shah, K.; Babu, K.S.; Mundanat, A.S.; Deliephan, A.; Deokar, G.S.; Santivarangkna, C.; Nirmal, N.P. Postbiotics: Current trends in food and pharmaceutical industry. Foods 2022, 11, 3094. [Google Scholar] [CrossRef] [PubMed]
- Teame, T.; Wang, A.; Xie, M.; Zhang, Z.; Yang, Y.; Ding, Q.; Gao, C.; Olsen, R.E.; Ran, C.; Zhou, Z. Paraprobiotics and Postbiotics of probiotic lactobacilli, their positive effects on the host and action mechanisms: A review. Front. Nutr. 2020, 7, 570344. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Gonzalez, P.F.; Liceaga, A.M.; Aguilar-Toala, J.E. Postbiotics and paraprobiotics: From concepts to applications. Food Res. Int. 2020, 136, 109502. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jeon, B.; Kim, W.J.; Chung, D. Effect of paraprobiotic prepared from Kimchi-derived Lactobacillus plantarum K8 on skin moisturizing activity in human keratinocyte. J. Funct. Foods 2020, 75, 104244. [Google Scholar] [CrossRef]
- Baldwin, H.; Aguh, C.; Andriessen, A.; Benjamin, L.; Ferberg, A.S.; Hooper, D.; Jarizzo, J.L.; Lio, P.A.; Tlougan, B.; Woolery-Lloyd, H.C.; et al. Atopic dermatitis and the role of the skin microbiome in choosing prevention, treatment, and maintenance options. J. Drugs Dermatol. 2020, 19, 935–940. [Google Scholar] [CrossRef]
- Golkar, N.; Ashoori, Y.; Heidari, R.; Omidifar, N.; Abootalebi, S.N.; Mohkam, M.; Gholami, A. A novel effective formulation of bioactive compounds for wound healing: Preparation, in vivo characterization, and comparison of various postbiotics cold creams in a rat model. Evid.-Based Complement. Altern. Med. 2021, 2021, 8577116. [Google Scholar] [CrossRef]
- Catic, T.; Pehlivanovic, B.; Pljakic, N.; Balicevac, A. The moisturizing efficacy of a proprietary dermo-cosmetic product (cls02021) versus placebo in a 4-week application period. Med. Arch. 2022, 76, 108–114. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C. A diversity profile of the human skin microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef]
- Jugé, R.; Rouaud-Tinguely, P.; Breugnot, J.; Servaes, K.; Grimaldi, C.; Roth, M.P.; Coppin, H.; Closs, B. Shift in skin microbiota of western European women across aging. J. Appl. Microbiol. 2018, 125, 907–916. [Google Scholar] [CrossRef]
- Gao, Z.; Tseng, C.H.; Strober, B.E.; Pei, Z.; Blaser, M.J. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS ONE 2008, 3, e2719. [Google Scholar] [CrossRef]
- Benhadou, F.; Mintoff, D.; Schnebert, B.; Thio, H.B. Psoriasis and microbiota: A systematic review. Diseases 2018, 6, 47. [Google Scholar] [CrossRef]
- Burns, E.M.; Ahmed, H.; Isedeh, P.N.; Kohli, I.; Van Der Pol, W.; Shaheen, A.; Muzaffar, A.F.; Al-Sadek, C.; Foy, T.M.; Abdelgawwad, M.S.; et al. Ultraviolet radiation, both UVA and UVB, influences the composition of the skin microbiome. Exp. Dermatol. 2019, 28, 136–141. [Google Scholar] [CrossRef]
- Kallscheuer, N.; Jogler, C. The bacterial phylum Planctomycetes as novel source for bioactive small molecules. Biotechnol. Adv. 2021, 53, 107818. [Google Scholar] [CrossRef]
- Ling, Z.; Liu, X.; Luo, Y.; Yuan, L.; Nelson, K.E.; Wang, Y.; Xiang, C.; Li, L. Pyrosequencing analysis of the human microbiota of healthy Chinese undergraduates. BMC Genom. 2013, 14, 390. [Google Scholar] [CrossRef]
- Hsu, D.K.; Fung, M.A.; Chen, H. Role of skin and gut microbiota in the pathogenesis of psoriasis, an inflammatory skin disease. Med. Microecol. 2020, 4, 100016. [Google Scholar] [CrossRef]
- Towner, K. The Genus Acinetobacter. In The Prokaryotes, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 746–758. [Google Scholar]
- Blaise, G.; Nikkels, A.F.; Hermanns-Lê, T.; Nikkels-Tassoudji, N.; Piérard, G.E. Corynebacterium-associated skin infections. Int. J. Dermatol. 2008, 47, 884–890. [Google Scholar] [CrossRef]
- Kim, J.H.; Son, S.M.; Park, H.; Kim, B.K.; Choi, I.S.; Kim, H.; Huh, C.S. Taxonomic profiling of skin microbiome and correlation with clinical skin parameters in healthy Koreans. Sci. Rep. 2021, 11, 16269. [Google Scholar] [CrossRef]
- Higuchi, R.; Goto, T.; Hirotsu, Y.; Otake, S.; Oyama, T.; Amemiya, K.; Ohyama, H.; Mochizuki, H.; Omata, M. Sphingomonas and Phenylobacterium as major microbiota in thymic epithelial tumors. J. Pers. Med. 2021, 11, 1092. [Google Scholar] [CrossRef]
- Krzyściak, W.; Jurczak, A.; Kościelniak, D.; Bystrowska, B.; Skalniak, A. The virulence of Streptococcus mutans and the ability to form biofilms. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 499–515. [Google Scholar] [CrossRef]
- Jalalvand, F.; Su, Y.C.; Manat, G.; Chernobrovkin, A.; Kadari, M.; Jonsson, S.; Janousková, M.; Rutishauser, D.; Semsey, S.; Løbner-Olesen, A.; et al. Protein domain-dependent vesiculation of Lipoprotein A, a protein that is important in cell wall synthesis and fitness of the human respiratory pathogen Haemophilus influenzae. Front. Cell. Infect. Microbiol. 2022, 12, 984955. [Google Scholar] [CrossRef]
- Meunier, M.; Scandolera, A.; Chapuis, E.; Lambert, C.; Jarrin, C.; Robe, P.; Chajra, H.; Auriol, D.; Reynaud, R. From stem cells protection to skin microbiota balance: Orobanche rapum extract, a new natural strategy. J. Cosmet. Dermatol. 2019, 18, 1140–1154. [Google Scholar] [CrossRef] [PubMed]
- Bayal, N.; Nagpal, S.; Haque, M.M.; Patole, M.S.; Shouche, Y.; Mande, S.C.; Mande, S.S. Structural aspects of lesional and non-lesional skin microbiota reveal key community changes in leprosy patients from India. Sci. Rep. 2021, 11, 3294. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Kim, H.S.; Lee, S.H.; Kim, S. Characterization and analysis of the skin microbiota in acne: Impact of systemic antibiotics. J. Clin. Med. 2020, 9, 168. [Google Scholar] [CrossRef] [PubMed]
- Karst, M.J. Staphylococcus aureus. In Encyclopedia of Toxicology, 2nd ed.; Wexler, P., Ed.; Academic Press: Cambridge, MA, USA, 2005; pp. 86–87. [Google Scholar]
- Falkinham, J.O.; Wall, T.E.; Tanner, J.R.; Tawaha, K.; Alali, F.Q.; Li, C.; Oberlies, N.H. Proliferation of antibiotic-producing bacteria and concomitant antibiotic production as the basis for the antibiotic activity of Jordan’s red soils. Appl. Environ. Microbiol. 2009, 75, 2735–2741. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Yokota, A.; Yamazoe, A.; Fujiwara, T. Proposal of Lysinibacillus boronitolerans gen. nov. sp. nov., and transfer of Bacillus fusiformis to Lysinibacillus fusiformis comb. nov. and Bacillus sphaericus to Lysinibacillus sphaericus comb. nov. Int. J. Syst. Evol. Microbiol. 2007, 57, 1117–1125. [Google Scholar] [CrossRef]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Fact. 2016, 15, 203. [Google Scholar] [CrossRef]
- Liu, X.; Ma, Z.; Zhang, J.; Yang, L. Antifungal compounds against candida infections from traditional chinese medicine. BioMed Res. Int. 2017, 2017, 4614183. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T. The Genus Ochrobactrum as major opportunistic pathogens. Microorganisms 2020, 8, 1797. [Google Scholar] [CrossRef]
- Gardini, G.; Gregori, N.; Matteelli, A.; Castelli, F. Mycobacterial skin infection. Curr. Opin. Infect. Dis. 2022, 35, 79–87. [Google Scholar] [CrossRef]
- Norris, S.J.; Paster, B.J.; Moter, A.; Göbel, U.B. The Genus Treponema. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; Volume 7, pp. 211–234. [Google Scholar]
- Oya, A.L. Stenotrophomonas, Burkholderia and other related microorganisms. In Encyclopedia of Infection and Immunity, 1st ed.; Rezaei, N., Ed.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 656–661. [Google Scholar]
- Altonsy, M.O.; Kurwa, H.A.; Lauzon, G.J.; Amrein, M.; Gerber, A.N.; Almishri, W.; Mydlarski, P.R. Corynebacterium tuberculostearicum, a human skin colonizer, induces the canonical nuclear factor-κB inflammatory signaling pathway in human skin cells. Immun. Inflamm. Dis. 2020, 8, 62–79. [Google Scholar] [CrossRef]
- Nagase, S.; Ogai, K.; Urai, T.; Shibata, K.; Matsubara, E.; Mukai, K.; Matsue, M.; Mori, Y.; Aoki, M.; Arisandi, D.; et al. Distinct skin microbiome and skin physiological functions between bedridden older patients and healthy people: A Single-center study in Japan. Front. Med. 2020, 7, 101. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).