Pseudomonas sp., Strain L5B5: A Genomic and Transcriptomic Insight into an Airborne Mine Bacterium
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, Isolation, and Identification of the Bacterium
2.2. Bacterial Inhibition Assays
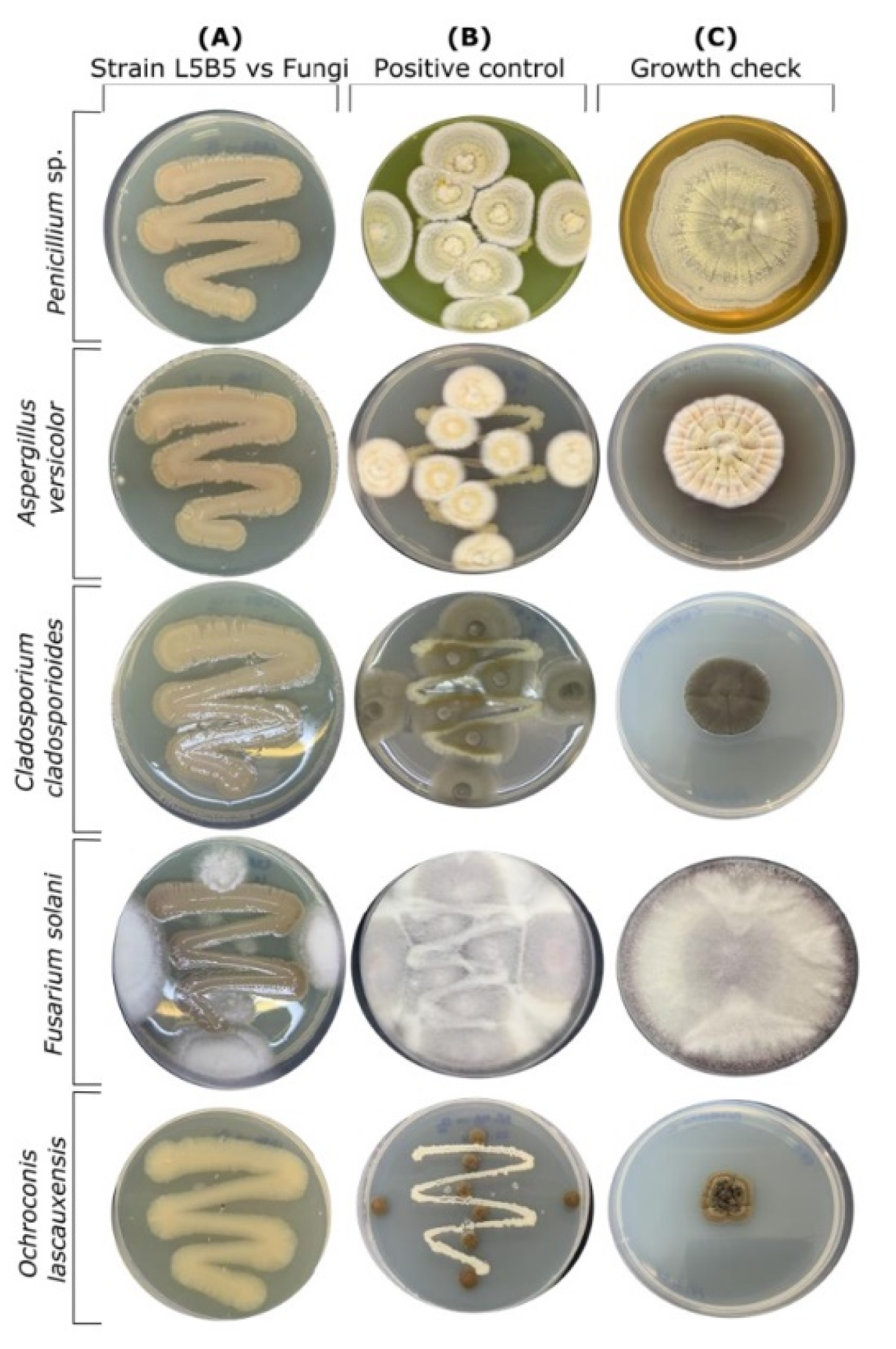
2.3. Fungal Inhibition Assays
2.4. DNA Extraction
2.5. RNA Extraction
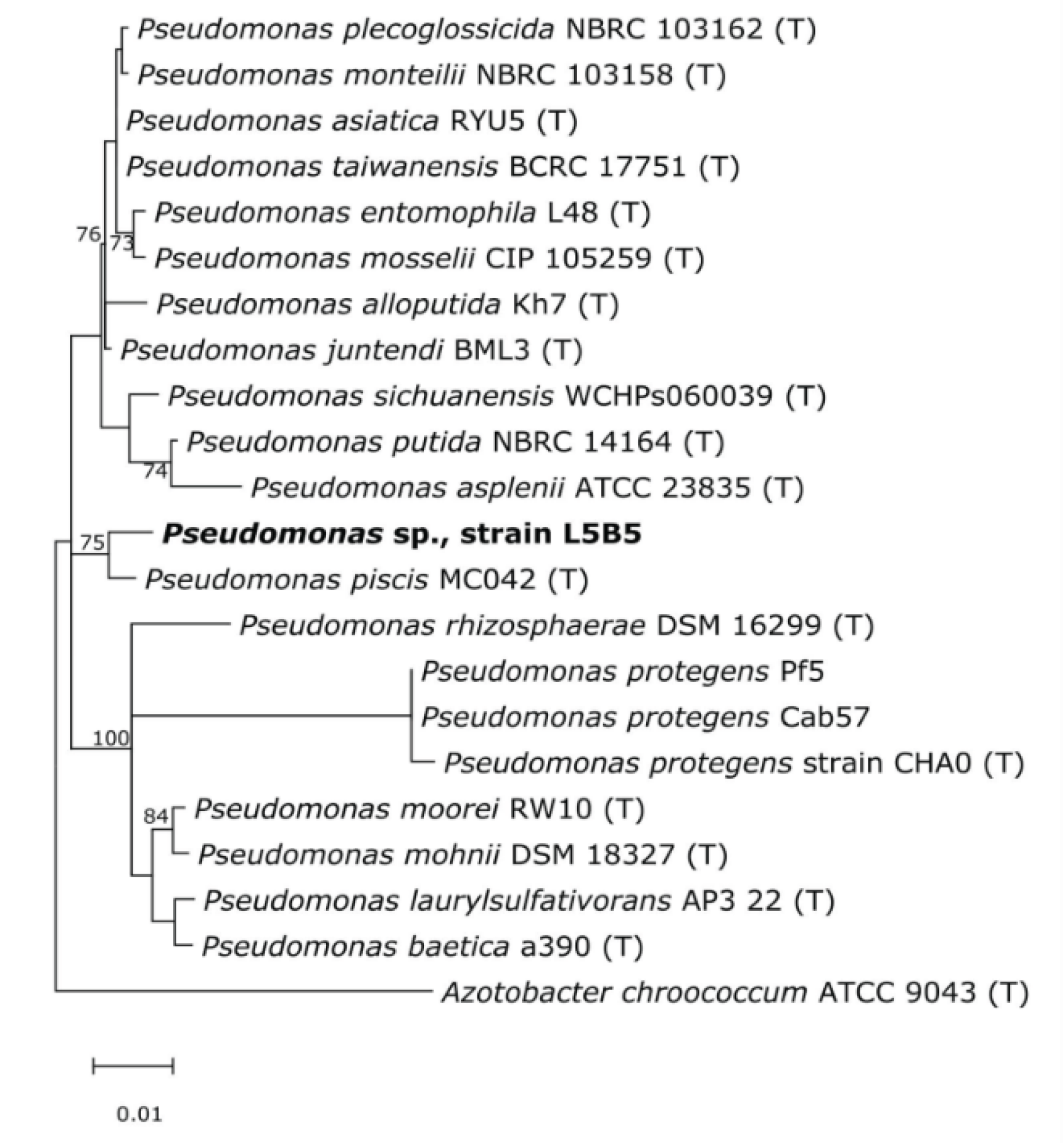
2.6. rRNA 16S Analysis
2.7. Genomic Analysis
2.8. Transcriptomic Analysis
3. Results
3.1. Inhibition Assays
3.2. 16S rRNA Analysis
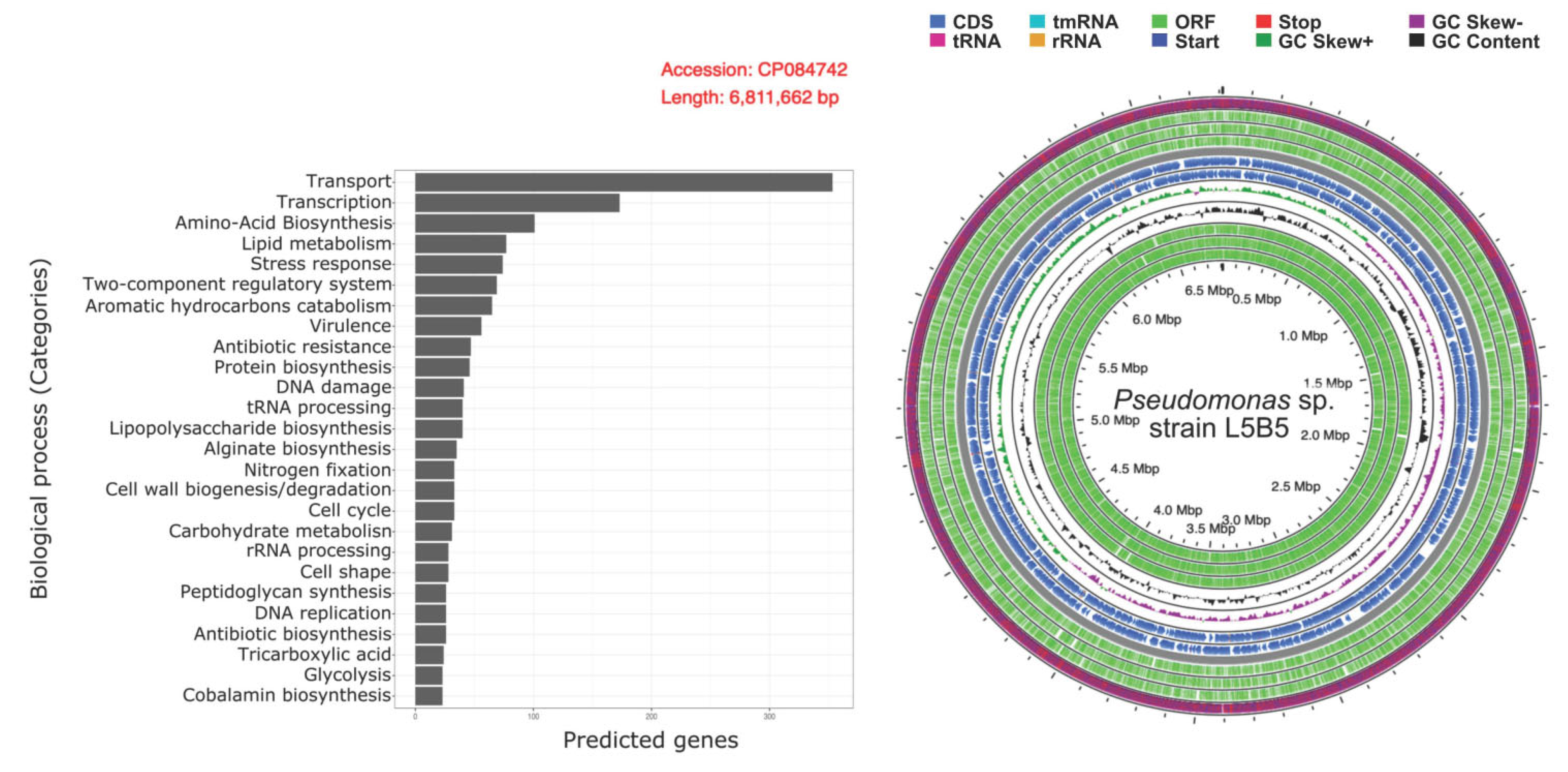
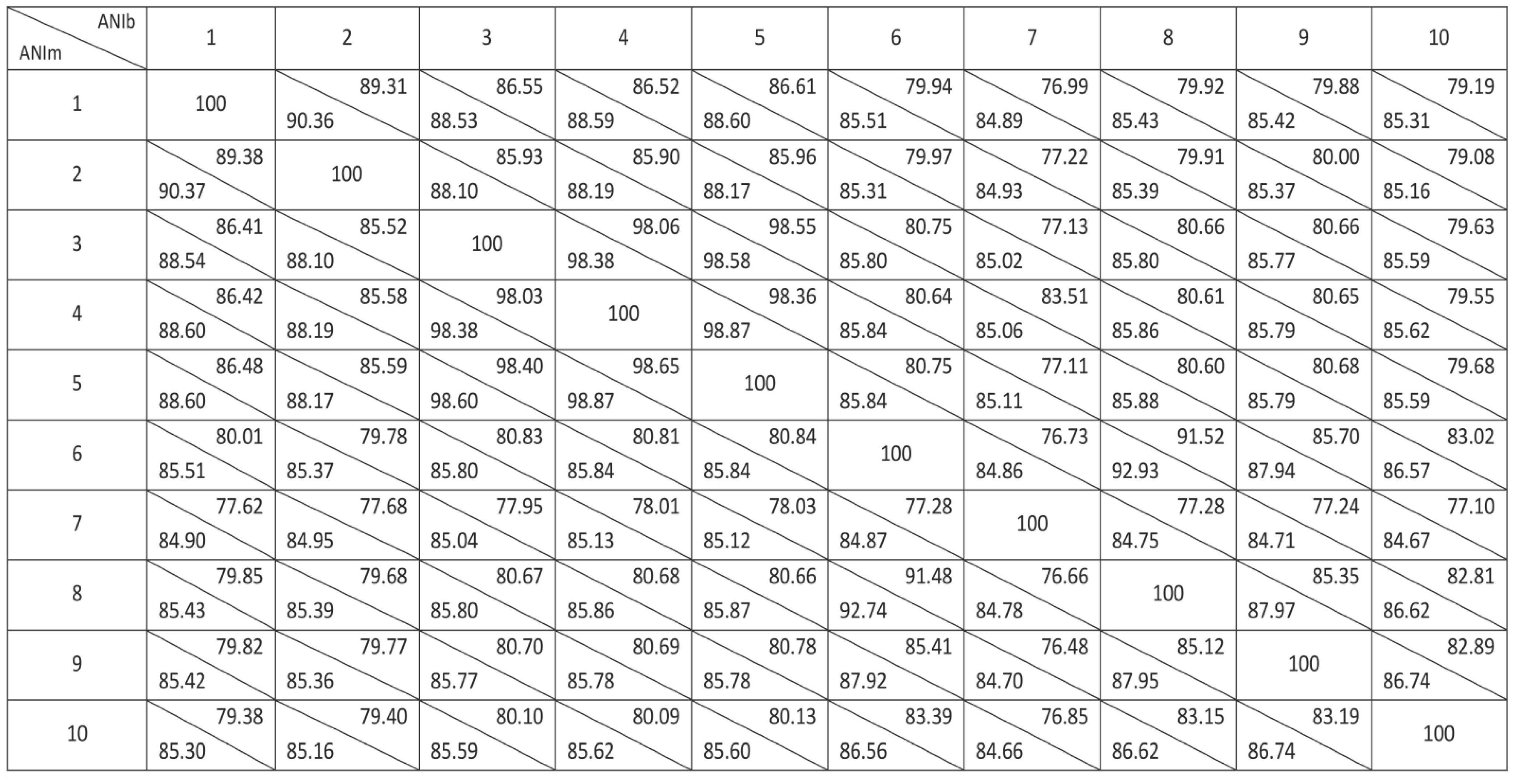
3.3. Genomics Analyses
3.4. Functional Annotations
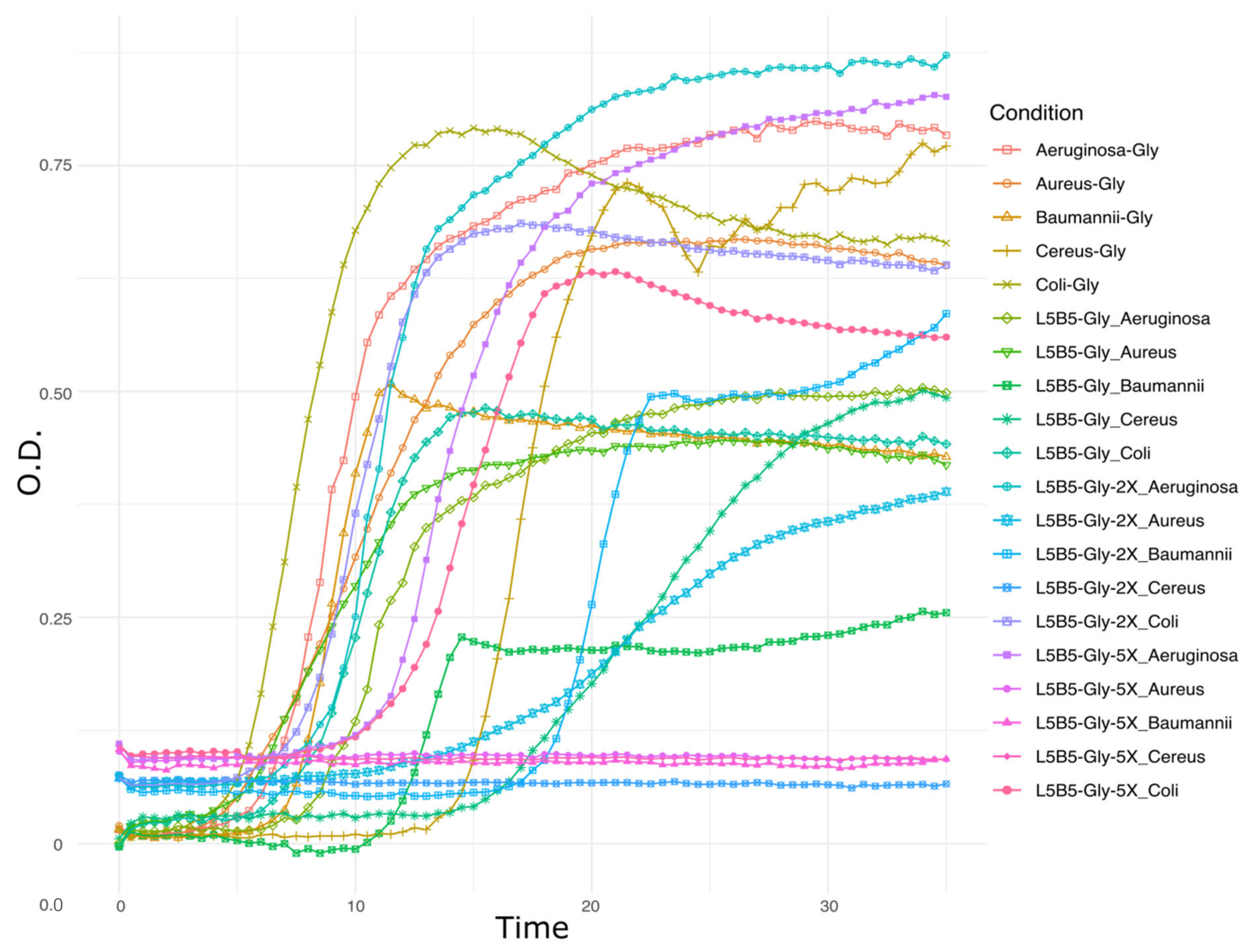
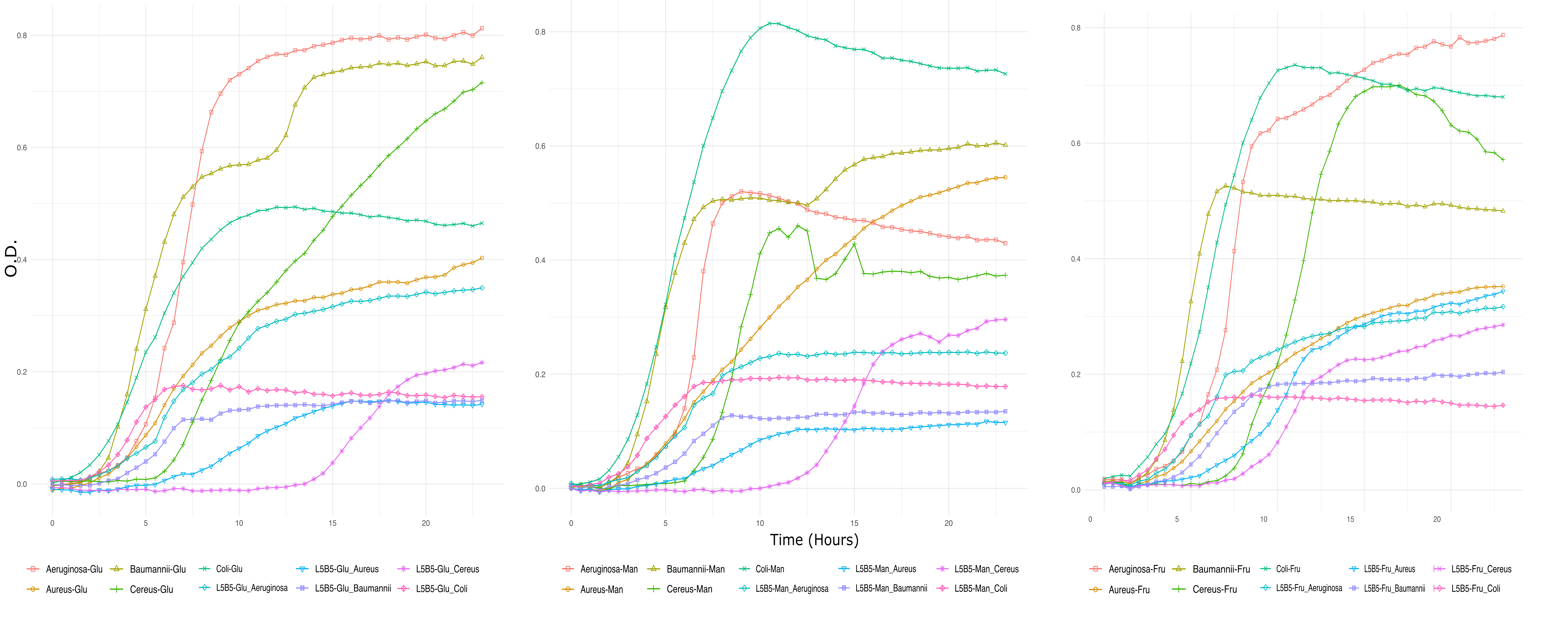
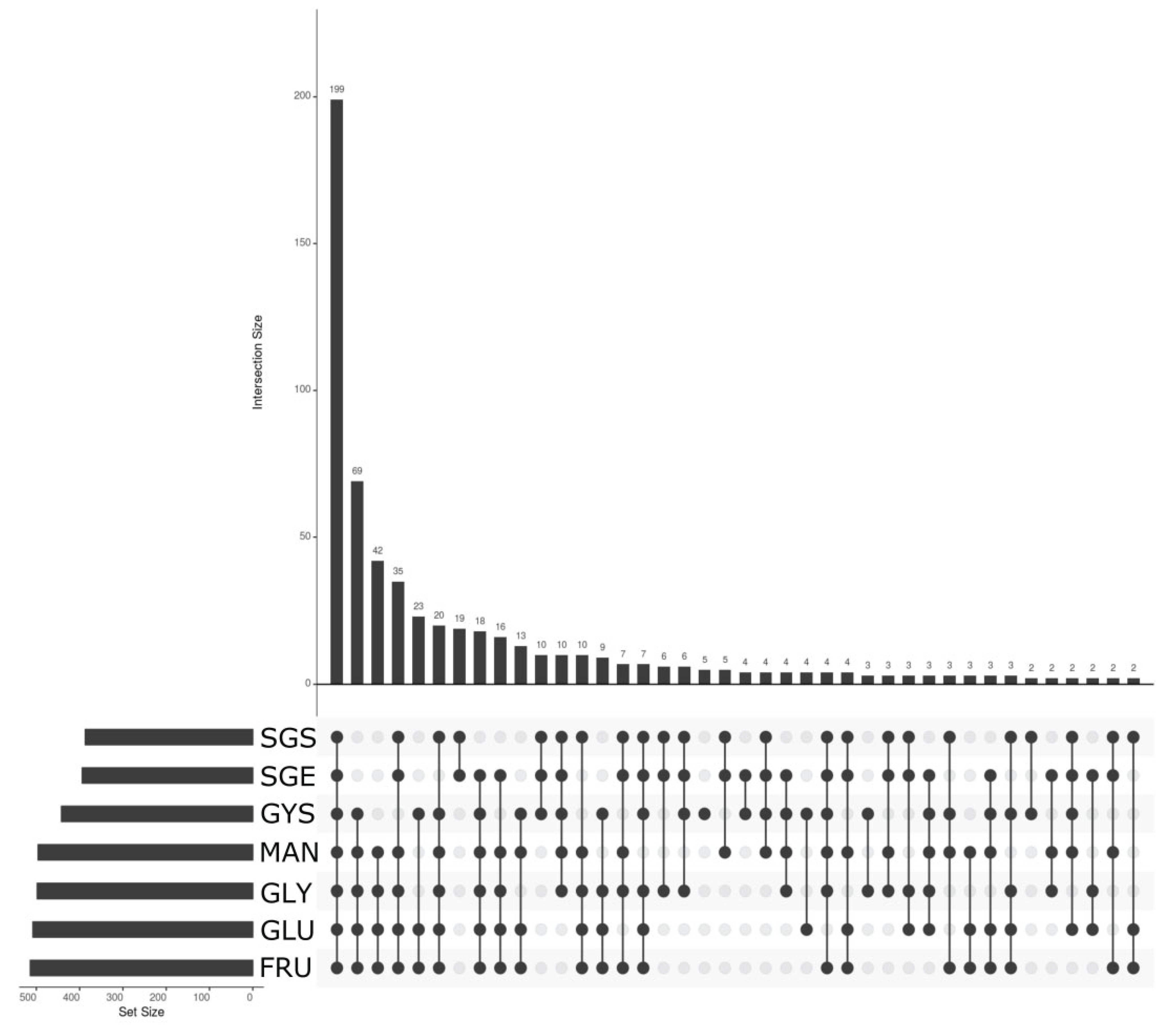
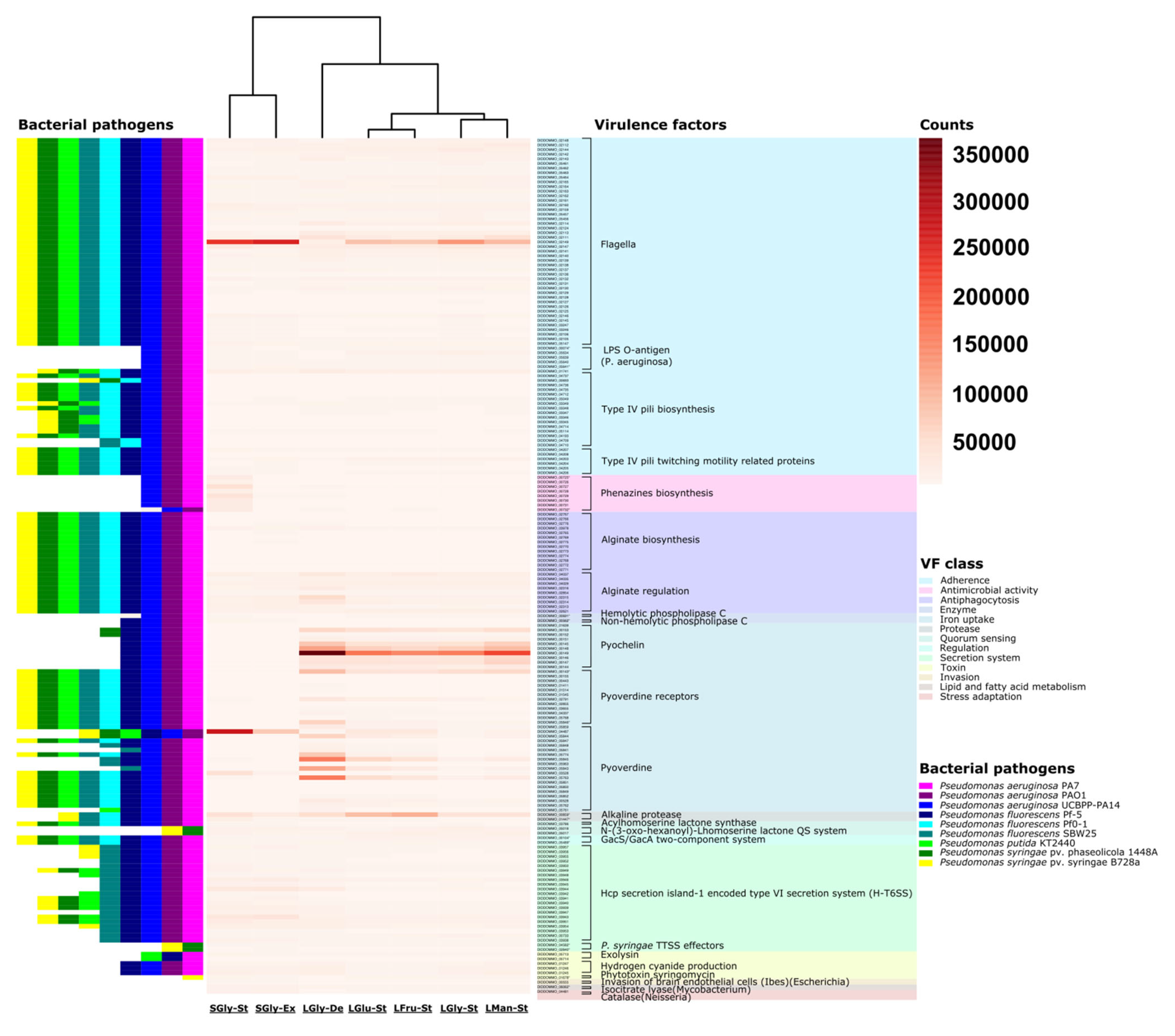
3.5. Transcriptomic Analysis—Pre-Treatment and Statistics
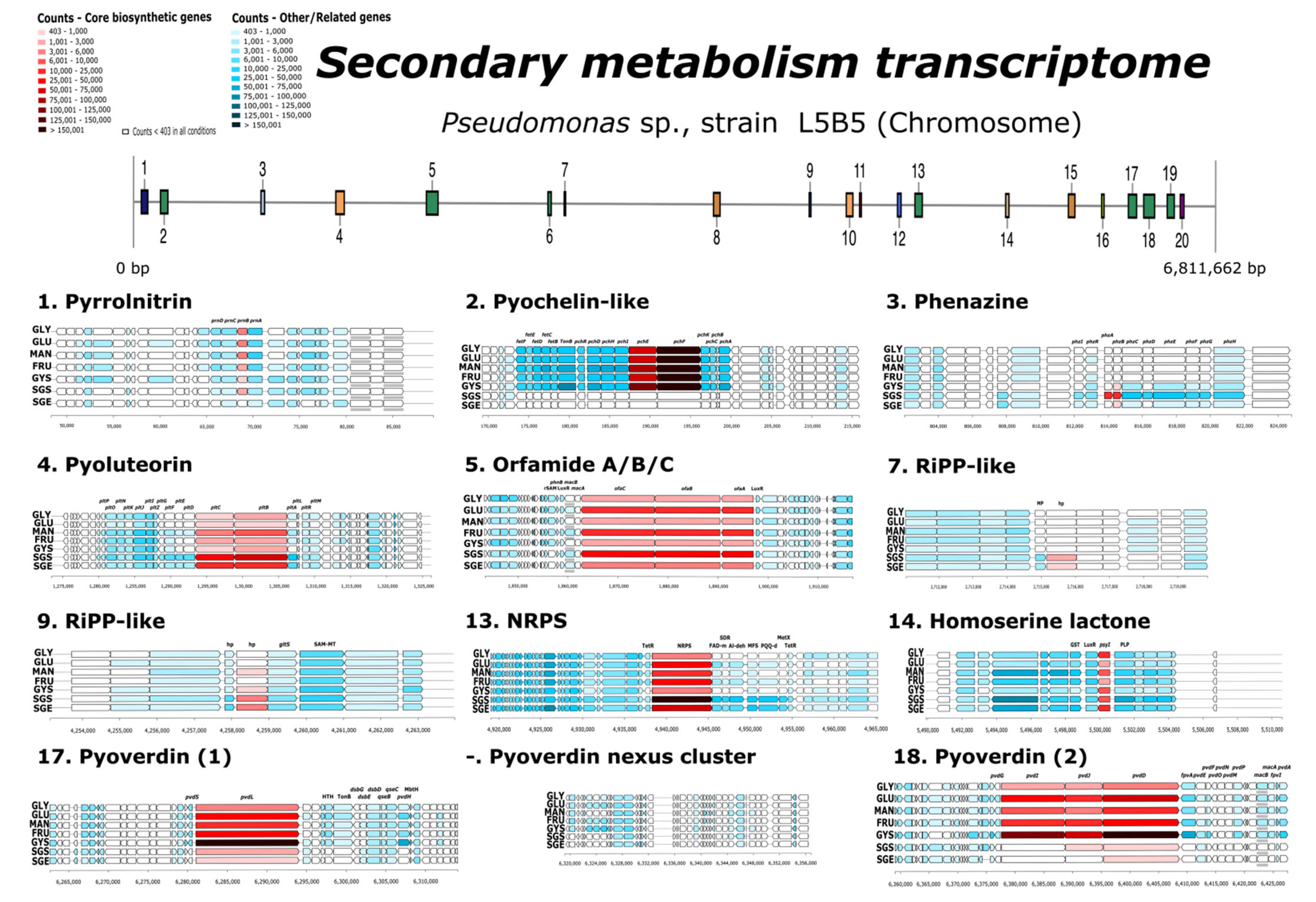
3.6. Overexpression in Secondary Metabolism
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choudhary, S.; Sar, P. Uranium biomineralization by a metal resistant Pseudomonas aeruginosa strain isolated from contaminated mine waste. J. Hazard Mater. 2011, 186, 336–343. [Google Scholar] [CrossRef]
- Choudhary, S.; Sar, P. Identification and characterization of uranium accumulation potential of a uranium mine isolated Pseudomonas strain. World J. Microbiol. Biotechnol. 2011, 27, 1795–1801. [Google Scholar] [CrossRef]
- Thavamani, P.; Samkumar, R.A.; Satheesh, V.; Subashchandrabose, S.R.; Ramadass, K.; Naidu, R.; Venkateswarlu, K.; Megharaj, M. Microbes from mined sites: Harnessing their potential for reclamation of derelict mine sites. Environ. Pollut. 2017, 230, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.M.; Escudero, C.; Leandro, T.; Mateos, G.; Amils, R. Draft genome sequence of Pseudomonas sp. strain T2.31D-1, isolated from a drilling core sample obtained 414 meters below surface in the Iberian Pyrite Belt. Microbiol. Resour. Announc. 2021, 10, e01165-20. [Google Scholar] [CrossRef]
- Bastian, F.; Alabouvette, C.; Saiz-Jimenez, C. Impact of biocide treatments on the bacterial communities of the Lascaux Cave. Naturwissenschaften 2009, 96, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Busquet, A.; Mulet, M.; Gomila, M.; García-Valdés, E. Pseudomonas lalucatii sp. nov. isolated from Vallgornera, a karstic cave in Mallorca, Western Mediterranean. Syst. Appl. Microbiol. 2021, 44, 126205. [Google Scholar] [CrossRef] [PubMed]
- Švec, P.; Kosina, M.; Zeman, M.; Holochová, P.; Králová, S.; Němcová, E.; Micenková, L.; Gupta, V.; Sood, U.; Lal, R.; et al. Pseudomonas karstica sp. nov. and Pseudomonas spelaei sp. nov., isolated from calcite moonmilk deposits from caves. Int. J. Syst. Evol. Microbiol. 2020, 70, 5131–5140. [Google Scholar] [CrossRef] [PubMed]
- Relvas, J.M.R.S.; Pinto, A.; Fernandes, C.; Matos, J.X.; Vieira, A.; Mendonça, A.; Malha, C.; Albuquerque, F.; Alegre, L.; Abrunhosa, M.; et al. Lousal: An old mine, a recent dream, a new reality. Comm. Geol. 2014, 101, 1345–1347. [Google Scholar]
- Garcia-Anton, E.; Cuezva, S.; Jurado, V.; Porca, E.; Miller, A.Z.; Fernandez-Cortes, A.; Sáiz-Jiménez, C.; Sanchez-Moral, S. Combining stable isotope (δ13C) of trace gases and aerobiological data to monitor the entry and dispersion of microorganisms in caves. Environ. Sci. Pollut. Res. 2014, 21, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Martin-Sanchez, P.M.; Jurado, V.; Porca, E.; Bastian, F.; Lacanette, D.; Alabouvette, C.; Saiz-Jimenez, C. Aerobiology of Lascaux Cave (France). Int. J. Speleol. 2014, 43, 295–303. [Google Scholar] [CrossRef]
- Dominguez-Moñino, I.; Jurado, V.; Rogerio-Candelera, M.A.; Hermosin, B.; Saiz-Jimenez, C. Airborne bacteria in show caves from Southern Spain. Microb. Cell 2021, 8, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Özen, A.I.; Ussery, D.W. Defining the Pseudomonas genus: Where do we draw the line with Azotobacter? Microb. Ecol. 2012, 63, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Andreolli, M.; Zapparoli, G.; Angelini, E.; Lucchetta, G.; Lampis, S.; Vallini, G. Pseudomonas protegens MP12: A plant growth-promoting endophytic bacterium with broad-spectrum antifungal activity against grapevine phytopathogens. Microbiol. Res. 2019, 219, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Shinde, S.; Zerbs, S.; Collart, F.R.; Cumming, J.R.; Noirot, P.; Larsen, P.E. Pseudomonas fluorescens increases mycorrhization and modulates expression of antifungal defense response genes in roots of aspen seedlings. BMC Plant Biol. 2019, 19, 4. [Google Scholar] [CrossRef]
- Peix, A.; Ramírez-Bahena, M.H.; Velázquez, E. The current status on the taxonomy of Pseudomonas revisited: An update. Infect. Genet. Evol. 2018, 57, 106–116. [Google Scholar] [CrossRef]
- Thomas, C.; Hothersall, J.; Willis, C.L.; Simpson, T.J. Resistance to and synthesis of the antibiotic mupirocin. Nat. Rev. Microbiol. 2010, 8, 281–289. [Google Scholar] [CrossRef]
- Pierson, L.S., 3rd; Thomashow, L.S. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30-84. Mol. Plant Microbe Interact. 1992, 5, 330–339. [Google Scholar] [CrossRef]
- Gross, H.; Stockwell, V.O.; Henkels, M.D.; Nowak-Thompson, B.; Loper, J.E.; Gerwick, W.H. The genomisotopic approach: A systematic method to isolate products of orphan biosynthetic gene clusters. Chem. Biol. 2007, 14, 53–63. [Google Scholar] [CrossRef]
- Ma, Z.; Geudens, N.; Kieu, N.P.; Sinnaeve, D.; Ongena, M.; Martins, J.C.; Höfte, M. Biosynthesis, chemical structure, and structure-activity relationship of orfamide lipopeptides produced by Pseudomonas protegens and related species. Front. Microbiol. 2016, 7, 382. [Google Scholar] [CrossRef]
- Cornelis, P.; Dingemans, J. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell. Infect. Microbiol. 2013, 3, 75. [Google Scholar] [CrossRef]
- Youard, Z.A.; Mislin, G.L.A.; Majcherczyk, P.A.; Schalk, I.J.; Reimmann, C. Pseudomonas fluorescens CHA0 produces enantio-pyochelin, the optical antipode of the Pseudomonas aeruginosa siderophore pyochelin. J. Biol. Chem. 2007, 282, 35546–35553. [Google Scholar] [CrossRef] [PubMed]
- Hammer, P.E.; Hill, D.S.; Lam, S.T.; Van Pée, K.H.; Ligon, J.M. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Microbiol. 1997, 63, 2147–2154. [Google Scholar] [CrossRef] [PubMed]
- Nowak-Thompson, B.; Chaney, N.; Wing, J.; Gould, S.J.; Loper, J.E. Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J. Bacteriol. 1999, 181, 2166–2174. [Google Scholar] [CrossRef] [PubMed]
- Poblete-Castro, I.; Wittmann, C.; Nikel, P.I. Biochemistry, genetics and biotechnology of glycerol utilization in Pseudomonas species. Microb. Biotechnol. 2020, 13, 32–53. [Google Scholar] [CrossRef]
- Martin-Sanchez, P.M.; Nováková, A.; Bastian, F.; Alabouvette, C.; Saiz-Jimenez, C. Two new species of the genus Ochroconis, O. lascauxensis and O. anomala isolated from black stains in Lascaux Cave, France. Fungal Biol. 2012, 116, 574–589. [Google Scholar] [CrossRef]
- Dominguez-Moñino, I.; Jurado, V.; Rogerio-Candelera, M.A.; Hermosin, B.; Saiz-Jimenez, C. Airborne fungi in show caves from Southern Spain. Appl. Sci. 2021, 11, 5027. [Google Scholar] [CrossRef]
- Zimmermann, J.; Gonzalez, J.M.; Saiz-Jimenez, C.; Ludwig, W. Detection and phylogenetic relationships of highly diverse uncultured acidobacterial communities in Altamira Cave using 23S rRNA sequence analyses. Geomicrobiol. J. 2005, 22, 379–388. [Google Scholar] [CrossRef]
- Muyzer, G.; Teske, A.; Wirsen, C.O.; Jannasch, H.W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel-electrophoresis of 16S rDNA fragments. Arch. Microbiol. 1995, 164, 165–172. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Pimentel, J.L.; Dominguez-Moñino, I.; Jurado, V.; Caldeira, A.T.; Saiz-Jimenez, C. Complete genome sequence of the airborne Pseudomonas sp. strain L5B5, isolated from an inactive pyrite mine. Microbiol. Resour. Announc. 2021, 10, e0102921. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Hunt, M.; Silva, N.D.; Otto, T.D.; Parkhill, J.; Keane, J.A.; Harris, S.R. Circlator: Automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015, 16, 294. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome project data processing subgroup. The sequence alignment/map format and SAM tools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.D.; Jin, Q.; Chen, L.H.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Stothard, P.; Wishart, D.S. Circular genome visualization and exploration using CGView. Bioinformatics 2005, 21, 537–539. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Tarazona, S.; Furio-Tari, P.; Turra, D.; Pietro, A.D.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.J.; Bolstad, B.M.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. UpSet: Visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Arima, K.; Imanaka, H.; Kousaka, M.; Fukuta, A.; Tamura, G. Pyrrolnitrin, a new antibiotic substance produced by Pseudomonas. Agric. Biol. Chem. 1964, 28, 575–576. [Google Scholar] [CrossRef]
- Pawar, S.; Chaudhari, A.; Prabha, R.; Shukla, R.; Singh, D.P. Microbial pyrrolnitrin: Natural metabolite with immense practical utility. Biomolecules 2019, 9, 443. [Google Scholar] [CrossRef] [PubMed]
- Reimmann, C. Inner-membrane transporters for the siderophores pyochelin in Pseudomonas aeruginosa and enantio-pyochelin in Pseudomonas fluorescens display different enantioselectivities. Microbiology 2012, 158, 1317–1324. [Google Scholar] [CrossRef]
- Mentel, M.; Ahuja, E.G.; Mavrodi, D.V.; Breinbauer, R.; Thomashow, L.S.; Blankenfeldt, W. Of two make one: The biosynthesis of phenazines. Chembiochem 2009, 10, 2295–2304. [Google Scholar] [CrossRef] [PubMed]
- Mavrodi, D.V.; Blankenfeldt, W.; Thomashow, L.S. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 2006, 44, 417–445. [Google Scholar] [CrossRef]
- Chin-A-Woeng, T.F.C.; Thomas-Oates, J.E.; Lugtenberg, B.J.J.; Bloemberg, G.V. Introduction of the phzH gene of Pseudomonas chlororaphis PCL 1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol. Plant Microbe. Interact. 2001, 14, 1006–1015. [Google Scholar] [CrossRef]
- Brodhagen, M.; Paulsen, I.; Loper, J.E. Reciprocal regulation of pyoluteorin production with membrane transporter gene expression in Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 2005, 271, 6900–6909. [Google Scholar] [CrossRef]
- Li, S.; Huang, X.; Wang, G.; Xu, Y. Transcriptional activation of pyoluteorin operon mediated by the LysR-type regulator PltR bound at a 22 bp lys box in Pseudomonas aeruginosa M18. PLoS ONE 2012, 7, e39538. [Google Scholar] [CrossRef]
- D’aes, J.; Kieu, N.P.; Léclère, V.; Tokarski, C.; Olorunleke, F.E.; De Maeyer, K.; Jacques, P.; Höfte, M.; Ongena, M. To settle or to move? The interplay between two classes of cyclic lipopeptides in the biocontrol strain Pseudomonas CMR12a. Environ. Microbiol. 2014, 16, 2282–2300. [Google Scholar] [CrossRef]
- Ma, Z.; Zhang, S.; Liang, J.; Sun, K.; Hu, J. Isolation and characterization of a new cyclic lipopeptide orfamide H from Pseudomonas protegens CHA0. J. Antibiot. 2020, 73, 179–183. [Google Scholar] [CrossRef]
- Olorunleke, F.E.; Kieu, N.P.; De Waele, E.; Timmerman, M.; Ongena, M.; Höfte, M. Coregulation of the cyclic lipopeptides orfamide and sessilin in the biocontrol strain Pseudomonas sp. CMR12a. MicrobiologyOpen 2017, 6, e00499. [Google Scholar] [CrossRef] [PubMed]
- Hetrick, K.J.; van der Donk, W.A. Ribosomally synthesized and post-translationally modified peptide natural product discovery in the genomic era. Curr. Opin. Chem. Biol. 2017, 38, 36–44. [Google Scholar] [CrossRef]
- Carreño-López, R.; Alatorre-Cruz, J.M.; Marín-Cevada, V. Pyrroloquinoline quinone (PQQ): Role in plant-microbe interactions. In Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms; Singh, H., Keswani, C., Reddy, M., Sansinenea, E., García-Estrada, C., Eds.; Springer: Singapore, 2019; pp. 169–184. [Google Scholar]
- Biessy, A.; Novinscak, A.; Blom, J.; Léger, G.; Thomashow, L.S.; Cazorla, F.M.; Josic, D.; Filion, M. Diversity of phytobeneficial traits revealed by whole-genome analysis of worldwide-isolated phenazine-producing Pseudomonas spp. Environ. Microbiol. 2019, 21, 437–455. [Google Scholar] [CrossRef] [PubMed]
- Lucena, T.; Arahal, D.R.; Ruvira, M.A.; Navarro-Torre, S.; Mesa, J.; Pajuelo, E.; Rodriguez-Llorente, I.D.; Rodrigo-Torres, L.; Pinar, M.J.; Pujalte, M.J. Vibrio palustris sp. nov. and Vibrio spartinae sp. nov., two novel members of the Gazogenes clade, isolated from salt-marsh plants (Arthrocnemum macrostachyum and Spartina maritima). Int. J. Syst. Evol. Microbiol. 2017, 67, 3506–3512. [Google Scholar] [CrossRef]
- Sugar, D.R.; Murfin, K.E.; Chaston, J.M.; Andersen, A.W.; Richards, G.R.; deLéon, L.; Baum, J.A.; Clinton, W.P.; Forst, S.; Goldman, B.S.; et al. Phenotypic variation and host interactions of Xenorhabdus bovienii SS-2004, the entomopathogenic symbiont of Steinernema jollieti nematodes. Environ. Microbiol. 2012, 14, 924–939. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L. Induction of microbial secondary metabolism. Int. Microbiol. 1998, 1, 259–264. [Google Scholar] [PubMed]
- Scarpellini, M.; Franzetti, L.; Galli, A. Development of PCR assay to identify Pseudomonas fluorescens and its biotype. FEMS Microbiol. Lett. 2004, 236, 257–260. [Google Scholar] [CrossRef]
- Merriman, T.R.; Merriman, M.E.; Lamont, I.L. Nucleotide sequence of pvdD, a pyoverdine biosynthetic gene from Pseudomonas aeruginosa: PvdD has similarity to peptide synthetases. J. Bacteriol. 1995, 177, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Lehoux, D.E.; Sanschagrin, F.; Levesque, R.C. Genomics of the 35-kb pvd locus and analysis of novel pvdIJK genes implicated in pyoverdine biosynthesis in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2000, 190, 141–146. [Google Scholar] [CrossRef]
- Mossialos, D.; Ochsner, U.; Baysse, C.; Chablain, P.; Pirnay, J.P.; Koedam, N.; Budzikiewicz, H.; Fernández, D.U.; Schäfer, M.; Ravel, J.; et al. Identification of new, conserved, non-ribosomal peptide synthetases from fluorescent pseudomonads involved in the biosynthesis of the siderophore pyoverdine. Mol. Microbiol. 2002, 45, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Ackerley, D.F.; Caradoc-Davies, T.T.; Lamont, I.L. Substrate specificity of the nonribosomal peptide synthetase PvdD from Pseudomonas aeruginosa. J. Bacteriol. 2003, 185, 2848–2855. [Google Scholar] [CrossRef]
- Nadal-Jimenez, P.; Koch, G.; Reis, C.R.; Muntendam, R.; Raj, H.; Jeronimus-Stratingh, C.M.; Cool, R.H.; Quax, W.J. PvdP is a tyrosinase that drives maturation of the pyoverdine chromophore in Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 2681–2690. [Google Scholar] [CrossRef] [PubMed]
- Ringel, M.T.; Dräger, G.; Brüser, T. PvdO is required for the oxidation of dihydropyoverdine as the last step of fluorophore formation in Pseudomonas fluorescens. J. Biol. Chem. 2018, 293, 2330–2341. [Google Scholar] [CrossRef] [PubMed]
- Lamont, I.L.; Beare, P.A.; Ochsner, U.; Vasil, A.I.; Vasil, M.L. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2002, 99, 7072–7077. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhou, R.; Sauder, J.M.; Tonge, P.J.; Burley, S.K.; Swaminathan, S. Structural and functional studies of fatty acyl adenylate ligases from E. coli and L. pneumophila. J. Mol. Biol. 2011, 406, 313–324. [Google Scholar] [CrossRef]
- Miller, B.R.; Gulick, A.M. Structural biology of nonribosomal peptide synthetases. Methods Mol. Biol. 2016, 1401, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Cianfanelli, F.R.; Monlezun, L.; Coulthurst, S.J. Aim, load, fire: The type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 2016, 24, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Fakhr, M.K.; Peñaloza-Vázquez, A.; Chakrabarty, A.M.; Bender, C.L. Regulation of alginate biosynthesis in Pseudomonas syringae pv. syringae. J. Bacteriol. 1999, 181, 3478–3485. [Google Scholar] [CrossRef] [PubMed]
- Ertesvåg, H.; Sletta, H.; Senneset, M.; Sun, Y.Q.; Klinkenberg, G.; Konradsen, T.A.; Ellingsen, T.E.; Valla, S. Identification of genes affecting alginate biosynthesis in Pseudomonas fluorescens by screening a transposon insertion library. BMC Genomics 2017, 18, 11. [Google Scholar] [CrossRef]
- Liyama, K.; Takahashi, E.; Lee, J.M.; Mon, H.; Morishita, M.; Kusakabe, T.; Yasunaga-Aoki, C. Alkaline protease contributes to pyocyanin production in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2017, 364, fnx051. [Google Scholar] [CrossRef]
- Keshavarz-Tohid, V.; Vacheron, J.; Dubost, A.; Prigent-Combaret, C.; Taheri, P.; Tarighi, S.; Taghavi, S.M.; Moenne-Loccoz, Y.; Muller, D. Genomic, phylogenetic and catabolic re-assessment of the Pseudomonas putida clade supports the delineation of Pseudomonas alloputida sp. nov., Pseudomonas inefficax sp. nov., Pseudomonas persica sp. nov., and Pseudomonas shirazica sp. nov. Syst. Appl. Microbiol. 2019, 42, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Tohya, M.; Watanabe, S.; Teramoto, K.; Shimojima, M.; Tada, T.; Kuwahara-Arai, K.; War, M.W.; Mya, S.; Tin, H.H.; Kirikae, T. Pseudomonas juntendi sp. nov., isolated from patients in Japan and Myanmar. Int. J. Syst. Evol. Microbiol. 2019, 69, 3377–3384. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Rao, Q.; Blom, J.; Lin, Q.; Luo, T. Pseudomonas piscis sp. nov., isolated from the profound head ulcers of farmed Murray cod (Maccullochella peelii peelii). Int. J. Syst. Evol. Microbiol. 2020, 70, 2732–2739. [Google Scholar] [CrossRef] [PubMed]
- Radlinski, L.; Rowe, S.E.; Kartchner, L.B.; Maile, R.; Cairns, B.A.; Vitko, N.P.; Gode, C.J.; Lachiewicz, A.M.; Wolfgang, M.C.; Conlon, B.P. Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol. 2017, 15, e2003981. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.W.; Lin, Y.S.; Huang, W.C.; Lai, C.C.; Chien, H.J.; Hu, N.-J.; Chen, J.-H. Inhibition of the clinical isolates of Acinetobacter baumannii by Pseudomonas aeruginosa: In vitro assessment of a case-based study. J. Microbiol. Immunol. Infect. 2022, 55, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Jurado, V.; Sanchez-Moral, S.; Saiz-Jimenez, C. Entomogenous fungi and the conservation of the cultural heritage: A review. Int. Biodeterior. Biodegr. 2008, 62, 325–330. [Google Scholar] [CrossRef]
- Zhang, Z.F.; Liu, F.; Zhou, X.; Liu, X.Z.; Liu, S.J.; Cai, L. Culturable mycobiota from karst caves in China, with descriptions of 20 new species. Persoonia 2017, 39, 1–31. [Google Scholar] [CrossRef]
- Howell, C.R.; Stipanovic, R.D. Suppression of Pythium ultimum-induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic, pyoluteorin. Phytopathology 1980, 70, 712–715. [Google Scholar] [CrossRef]
- Gorantla, J.N.; Kumar, S.N.; Nisha, G.V.; Sumandu, A.S.; Dileep, C.; Sudaresan, A.; Kumar, M.S.; Lankalapalli, R.; Kumar, B.D. Purification and characterization of antifungal phenazines from a fluorescent Pseudomonas strain FPO4 against medically important fungi. J. Mycol. Med. 2014, 24, 185–192. [Google Scholar] [CrossRef]
- Hoegy, F.; Celia, H.; Mislin, G.L.; Vincent, M.; Gallay, J.; Schalk, I.J. Binding of iron-free siderophore, a common feature of siderophore outer membrane transporters of Escherichia coli and Pseudomonas aeruginosa. J. Biol. Chem. 2005, 280, 20222–20230. [Google Scholar] [CrossRef]
- Moynié, L.; Milenkovic, S.; Mislin, G.L.A.; Gasser, V.; Malloci, G.; Baco, E.; McCaughan, R.P.; Page, M.G.P.; Schalk, I.J.; Ceccarelli, M.; et al. The complex of ferric-enterobactin with its transporter from Pseudomonas aeruginosa suggests a two-site model. Nat. Commun. 2019, 10, 3673. [Google Scholar] [CrossRef]
- Filkins, L.M.; Graber, J.A.; Olson, D.G.; Dolben, E.L.; Lynd, L.R.; Bhuju, S.; O’Toole, G.A. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J. Bacteriol. 2015, 197, 2252–2264. [Google Scholar] [CrossRef] [PubMed]
- Moynié, L.; Luscher, A.; Rolo, D.; Pletzer, D.; Tortajada, A.; Weingart, H.; Braun, Y.; Page, M.G.P.; Naismith, J.H.; Köhler, T. Structure and function of the PiuA and PirA siderophore-drug receptors from Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob. Agents Chemother. 2017, 61, e02531-16. [Google Scholar] [CrossRef] [PubMed]
- Tanii, A.; Miyajima, K.; Akita, T. The sheath brown rot disease of rice plant and its causal bacterium, Pseudomonas fuscovaginae. Ann. Phytopathol. Jpn. 1976, 42, 540–548. [Google Scholar] [CrossRef]
- Zada, S.; Sajjad, W.; Rafiq, M.; Ali, S.; Hu, Z.; Wang, H.; Cai, R. Cave microbes as a potential source of drugs development in the modern era. Microb. Ecol. 2021. [Google Scholar] [CrossRef]
- Gonzalez-Pimentel, J.L.; Dominguez-Moñino, I.; Jurado, V.; Laiz, L.; Caldeira, A.T.; Saiz-Jimenez, C. The rare actinobacterium Crossiella sp. is a potential source of new bioactive compounds with activity against bacteria and fungi. Microorganisms 2022, 10, 1575. [Google Scholar] [CrossRef] [PubMed]
| Pseudomonas sp., strain L5B5 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| Metabolite-Region (R) | UniProtKb Similarity (Core Biosynthetic Genes) | ||||
| Other-R1 (Pyrrolnitrin) | 95% Pseudomonas sp. CMR5c (prnB) | + | + | + | + |
| NRPS-R2 (Enantio/Iso-pyochelin) | 90%–95% Pseudomonas sp. CMR5c (pchEF) | − | + | + | + |
| Phenazine-R3 | 84.7%–85.3%–98.2% P. piscis-P. yamanorum-Pseudomonas sp. FW507-12TSA (phzI-phzAB) | + | − | − | − |
| T1PKS-R4 (Pyoluteorin) | 84.1%–77% Pseudomonas sp. MSSRFD41 (PltBC-like PKSynthase) | − | + | + | + |
| NRPS-R5 (Orfamides) | 89.3%–90.3%–90% Pseudomonas sp. CMR5c (ofaABC) | + | + | + | + |
| CDPS-R6 | 97.6% P. protegens CHA0 (Cyclodipeptide synthase) | + | + | + | + |
| RiPP-like-R7 | 86.3% Pseudomonas sp. FW507-12TSA (Uncharacterized protein) | + | + | + | + |
| Arylpolyene-R8 | 89.7%–97.3% P. protegens CHA0-Pseudomonas sp. CMR5c (Beta-ketoacyl-ACP synthases) | + | + | + | + |
| RiPP-like-R9 | 93.2% Pseudomonas sp. CMR12a (Uncharacterized protein) | + | + | + | + |
| T3PKS-R10 (2,4-diacetylphloroglucinol) | 94.6% P. protegens Pf-5 (phlD) | + | + | + | + |
| Siderophore-R11 | 80.7% Pseudomonas sp. CMR12a (Uncharacterized protein) | − | − | − | − |
| Redox-cofactor-R12 (Lankacidin C) | 98.8%–96.7%–97.2% Pseudomonas sp. CMR12a-P. protegens Pf-5 (pqqABCDEF) | + | + | + | + |
| NRPS-R13 (Putative NRPS) | 88.7% P. juntendi/P. putida (Thioesterase) | − | − | − | − |
| Hserlactone-R14 | 80.5% Pseudomonas sp. FW507-12TSA (psyL) | + | − | − | − |
| Arylpolyene-R15 | 90.6% Pseudomonas sp. CMR5c (Thiolase) | − | − | − | − |
| NAGGN-R16 | 98.7%–96%–97.6% Pseudomonas sp. BIOMIG1BAC (M42 family peptidase- N-acetylglutaminylglutamine synthetase | + | + | + | + |
| NRPS-R17-18 (Pyoverdine) | 96.4%–96%–94.3%–95.2% P. protegens CHA0 (pvdLIJD) | + | + | + | + |
| NRPS-R19 | 85.4% Pseudomonas sp. CMR5c (Thioester reductase-like domain) | − | − | − | − |
| Betalactone-R20 | 94.8%–95% P. piscis-Pseudomonas sp. CMR12a (Acyl-CoA synthase-pyruvate carboxylase) | + | + | + | + |
| RNA-Sample | Raw Reads | Reads after rRNA Removing | Filtered Reads | Passed Reads | Mapped Reads |
|---|---|---|---|---|---|
| SGE | 40,589,012 | 40,333,005 | 38,932,275 | 95.92% | 94.46% |
| SGS | 31,304,717 | 31,221,230 | 30,076,663 | 96.08% | 94.17% |
| GYS | 31,931,209 | 31,583,884 | 30,443,705 | 95.34% | 90.74% |
| GLY | 35,637,786 | 35,492,545 | 34,789,606 | 97.62% | 94.06% |
| GLU | 33,422,936 | 32,805,879 | 32,587,198 | 97.50% | 93.38% |
| MAN | 32,739,745 | 32,624,884 | 31,908,361 | 97.46% | 94.83% |
| FRU | 33,195,686 | 33,098,880 | 32,344,406 | 97.44% | 93.90% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez-Pimentel, J.L.; Dominguez-Moñino, I.; Jurado, V.; Caldeira, A.T.; Saiz-Jimenez, C. Pseudomonas sp., Strain L5B5: A Genomic and Transcriptomic Insight into an Airborne Mine Bacterium. Appl. Sci. 2022, 12, 10854. https://doi.org/10.3390/app122110854
Gonzalez-Pimentel JL, Dominguez-Moñino I, Jurado V, Caldeira AT, Saiz-Jimenez C. Pseudomonas sp., Strain L5B5: A Genomic and Transcriptomic Insight into an Airborne Mine Bacterium. Applied Sciences. 2022; 12(21):10854. https://doi.org/10.3390/app122110854
Chicago/Turabian StyleGonzalez-Pimentel, Jose Luis, Irene Dominguez-Moñino, Valme Jurado, Ana Teresa Caldeira, and Cesareo Saiz-Jimenez. 2022. "Pseudomonas sp., Strain L5B5: A Genomic and Transcriptomic Insight into an Airborne Mine Bacterium" Applied Sciences 12, no. 21: 10854. https://doi.org/10.3390/app122110854
APA StyleGonzalez-Pimentel, J. L., Dominguez-Moñino, I., Jurado, V., Caldeira, A. T., & Saiz-Jimenez, C. (2022). Pseudomonas sp., Strain L5B5: A Genomic and Transcriptomic Insight into an Airborne Mine Bacterium. Applied Sciences, 12(21), 10854. https://doi.org/10.3390/app122110854







