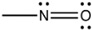Abstract
Fungi are the main contaminants of books and archival documents. In addition to their degrading power, offered by various types of lignolytic and cellulolytic enzymes, they can also hue the surface of the paper through the production of pigments. The fungi on paper release various types of pigments belonging mostly to two chemical groups (polyketides and carotenoids), which cause unpleasant anaesthetic stains. The paper surface can also be hued with several synthetic colors, which are part, for example, of stamps and inks. These synthetic colors could be degraded by lignin-modifying enzymes (LMEs) and also by dye-decolorizing peroxidases (DyPs). Therefore, the mechanism of action of LEMs and DyPs is illustrated. Moreover, we have examined the potentiality of LEMs and DyPs to remove the synthetic stains and also their hypothetical application in order to clean the fungal hues from the paper surface. Our review article, using the enzymatic removal parallelism between fungal and synthetic pigments, would like to show prospective solutions to this arduous problem.
1. Introduction
Many books, historical documents, sketches, paintings, music scores, architectural construction documents, postage stamps, and photographs are susceptible to destruction [1,2,3,4,5]. This destruction can be a result of the influence of endogenous factors such as acidity, metal ions, lignin, or paper degradation products, and exogenous factors such as heat, humidity, radiation (light, UV), oxygen, pollutants, or biodeteriogens [6,7].
Filamentous fungi have been considered to be the most important microorganisms responsible for the biodeterioration of collections in museums, archives, galleries, and libraries [6,8,9], causing severe material and information losses in libraries’ and archives’ collections [8].
Fungi possess the capacity to grow at average relative humidity levels, inducing several chemical and physical decomposing processes through the excretion of metabolites and enzymes that affect the structural organization of the paper [2,10]. These metabolites comprise a wide variety of substances such as colorants, enzymes, acids, chelating agents, etc. [11]. However, one of the major concerns regarding fungal colonization of paper is that it undergoes degradation that affects its mechanical and aesthetic properties [12] since the stains excreted by fungi are colored [13] (Figure 1). Several reviews and research papers can be found describing fungi, their colorants, and the foxing phenomenon causing biodeterioration of historical paper (Figure 1) [2,14,15,16,17], but very little data is presented about the acceptable methodology of how to treat and remove the stains from paper. To address these demands, first, it is important to collect available information about the fungal species that stain paper and the chemical nature of those stains. Another important demand is to develop new cleaning methods that would not bleach and destroy the aesthetics of the paper itself.

Figure 1.
Several types of stains on the paper surface. (A) red circle, probably black fungal stains; many black spots are spread on the book page. (B) red circles showing presumable yellowish and pinkish fungal spots. (C) the phenomenon of foxing. (D) colored stamps on the books’ pages.
Apart from fungal stains, another problem for the restorers is the colored stamps placed on a specific sheet (Figure 1), marking the book or the document as a property of the library. These stamps are usually composed of ink containing different kinds of synthetic dyes [18], and when restorers need to restore or digitalize a book or a document, they also need to remove the stamp from the sheet. Therefore, the ink of the stamp can also be considered as a potential colored stain necessary to be removed from the paper surface (personal communication, restorer Radka Benžová).
Generally, microorganisms are considered to be a threat to cultural heritage items in the form of biodeterioration. On the contrary, in the last few decades, these living organisms have been shown to be valuable restoration tools by means of their metabolic properties [19]. Many studies have reported the application of microorganisms and their enzymes in biocleaning, such as the removal of dirt, animal glue, and other organic residues from a variety of artworks [20,21,22,23,24,25,26].
For safety reasons, enzymatic cleaning is preferable in many cases over the application of living cells that can leave residues on the surfaces that are about to be restored and which can later serve as nutrients for dangerous microbes [19,27]. In the work of Jeszeová and colleagues [22], hydrolytic enzymes, in particular proteases, nucleases, and phosphatases, have been identified and assayed in biocleaning tests for degradation of animal glue performed on different kinds of surfaces such as wood, glass, and stone. These extracellular enzymes were excreted from Exiguobacterium undae and were used for the successful removal of different types of animal glue. Recently, the lipolytic abilities of the yeast Sporidiobolus metaroseus were also exploited to remove an oil-based patina from a historical painting from the XVIII Century [24]. Thus, it can be suggested that the isolated extracellular enzymes can be used for the restoration of different artworks and historical items without microbial contamination of the treated surface.
White-rot fungi, considered a physiological group of fungi exhibiting the activity of lignin degradation within lignocellulosic substrates, have been investigated intensively due to their great potential to degrade a wide variety of recalcitrant organic compounds and synthetic dyes [28,29,30,31,32]. These fungi, as well as brown-rot fungi, secrete various extracellular enzymes called lignin-modifying enzymes (LMEs), which include heme-dependent lignin peroxidases (LiP), manganese peroxidases (MnP), dye-decolorizing peroxidases (DyP), and copper-dependent laccases [33,34,35,36,37]. LMEs have found their use in environmental applications, including the transformation and degradation of compounds, such as different synthetic dyes, pharmaceuticals, and pesticides [38,39,40,41,42,43,44,45]. These enzymes are classified as Auxiliary Activity (AA) families in the Carbohydrate-Active enZYmes Database (CaZy database; http://www.cazy.org/ accessed on 22 September 2022), which provides an overview of cellulose- and lignin-degrading enzymes for biotechnical applications [46,47]. Some other enzymes such as vanillyl-alcohol oxidases, glyoxal oxidases, cellobiose/quinone oxidoreductase, and cellobiose dehydrogenase are correlated with lignin degradation through enhancement of the activity of laccases, MnPs, and LiPs [34,48,49]. Therefore, it could be useful to utilize these enzymes in order to remove the stains on archival documents and books. This review will illustrate several types of fungal pigments and synthetic dyes present in historical books and paper documents and the possible removal tools which can be represented by the ligninolytic enzymes and dye-decolorizing peroxidases, moreover, their function in known and potential decolorization applications.
2. Fungal Pigments Found on Paper
Colorants produced by microorganisms are the result of metabolic processes taking place in their cell structures. These secondary metabolites are complex chemical substances that can be incorporated into spores and mycelium or excreted into a substrate such as paper [50]. Fungi can produce a variety of stains with different colors ranging from brown, black, green, yellow, purple, or pink [2]. Colorants from fungi can be either pigments or dyes. The difference between the two is due to their physical characteristics, meaning that pigments are insoluble in the given medium, whereas dyes dissolve during the application, losing their crystal or particulate structure in the process [51].
Fungal colorants represent an organism’s response to light harvesting and processing, photo-protection, absorption, and neutralization of protons that could potentially damage the cell structures [52]. Some colorants are also acknowledged as enzyme inhibitors and antibiotics [53]. Dark pigments, melanins, have had several biological functions such as protection against radiation (e.g., UV), enzymatic lysis, high temperatures, or oxidizing agents; capacity for binding of metals, preventing the entry of toxic metals or concentrating essential metals; action as a virulence factor; or capacity for increasing resistance to fungicides [54].
Most of the fungal colorants are polyketide-based [55], but, in general, they can be classified into different groups, mainly: polyketides (and their derivatives), melanin, and carotenoids [56].
2.1. Polyketide Colorants
The important natural products encompassing a variety of toxins, antibiotics, therapeutic compounds, and colorants are represented by a group of chemical substances called polyketides. Due to their biosynthetic complexity and importance in the pharmaceutical industry, polyketides have drawn great attention among researchers, which has led to extensive molecular genetic studies carried out in actinomycetes and Gram-positive bacteria [57].
Polyketides are one of the largest and most structurally diverse classes of naturally occurring compounds with a variety of biological activities [58]. These compounds are biosynthesized from acyl CoA precursors catalyzed by polyketide synthases (PKSs) [59] that are members of a large enzyme family that are involved in the synthesis of different secondary metabolites such as pigments, toxins, antibiotics, and signaling molecules [60]. Pigments produced by fungi are mostly the result of the paucity of nutrients when mycelium produces secondary metabolites [61]. Fungal colorants are either polyketide- or carotenoid-based. Those that are polyketide-based are composed of tetraketides and octaketides with eight C2 units, creating a polyketide chain [62]. Polyketides comprise azaphilones and quinones, including anthraquinones, hydroxyanthraquinones, and naphthoquinones [55,62].
Azaphilones can be defined as a small but structurally diverse subset of the polyketide class that are excreted by fungi as second metabolites [55,63]. Due to their affinity for ammonia, the name azaphilones was given to these substances. In addition, azaphilones react with amines such as proteins, amino acids, and nucleic acids to construct red or purple vinylogous γ-pyridones due to the exchange of pyrane oxygen for nitrogen [55]. Fungi possessing these compounds show bright yellow, red, or green colors of mycelia. These polyketide derivatives are produced by a variety of representatives of ascomycetes, including the genera Aspergillus, Penicillium, Chaetomium, Talaromyces, Pestalotiopsis, Phomopsis, Eremicella, and Epicoccum, as well as Monascus and Hypoxylo [64], which may colonize paper and cause the appearance of colored spots [2].
Quinones are a class of quinoid compounds widely present in nature [65]. They are derived from aromatic compounds by the conversion of an even number of methane (-CH=) groups into carbonyl (C=O)groups with any necessary rearrangement of double bonds, resulting in a fully conjugated cyclic dione structure [66]. Those biosynthesized by the polyketide pathway have been isolated from fungi showing different colors, from yellow and orange to red. However, the color of the quinone isolated from a fungus does not reflect the color of that fungus. The reason for this may be that the quinone is accompanied by its reduction products, which lead to the formation of the quinhydrone complex [67]. The most produced quinones in fungi are Hydroxyanthraquinoid (HAQN) colourants. These colorants are derivatives of quinones, which are widely distributed in microorganisms. In particular, they have been produced mostly by the genera Penicillium and Aspregillus, with a different color tone [68]. Specifically, Aspergillus glaucus, Aspergillus versicolor, and Penicillium purpurogenum have been conspicuous anthraquinone producers [69]. The shade-like appearance of a specific anthraquinone called chrysophanols has been reported to be a consequence of the methabolic processes of Trichoderma with the species T. harzianum, T. polysporum, T. viridae, and T. aureoviride [70]. Chrysophanols are peculiar anthraquinones giving the shade-like appearance of different colors due to the presence of chromosphere for absorption of electronic spectra at diverse wavelengths [71]. Naphthoquinones, widespread quinones with naphthalene backbones, have been shown to possess biological activities such as antibacterial, antitumoral, antiviral, and antifungal activities [69,72,73,74]. These compounds have been synthesized in Fusarium and Candida species in response to stress [75,76]. Recently, novel naphthoquinones of a yellow color called Trypethelonamide A and a dark violet-red color called 5′-hydroxytrypethelone isolated from Trypethelium eluteriae have been shown to exhibit antioxidant and cytotoxic activities against cancer cell lines [77]. The fungal species Scytalidium cuboideum has been characterized as a producer of naturally occurring red/ping pigments that can crystalize [78]. These crystalline pigments have a naphthoquinone structure and were reported to require an easy methodology for their isolation, which is beneficial for different industries and green energy production [78,79,80].
2.2. Melanin
Melanin is an ubiquitous, widely distributed dark brown to black pigment in animals, plants, and microorganisms [81]. It is an insoluble and non-digestible, negatively charged polymer formed by oxidative polymerization of phenolic or indolic compounds [82,83,84]. There are a few forms of melanin existing in living organisms, including fungi: allomelanins, eumelanins, and pheomelanins. Eumelanins are characterized as black-brown insoluble melanin pigments partially derived from the oxidative polymerization of L-dopa via 5,6-dihydroxyindole intermediates. Pheomelanins represent a subgroup of melanins containing sulfur obtained by the oxidation of cysteinyldopa precursors via benzothiazine and benzothiazole intermediates. These pigments are alkali-soluble with a yellow to brown color [85]. Allomelanins are defined as melanins containing phenolic, catecholic, and 1,8-dihydroxynaphthalene (1,8-DHN) units [86].
Melanins were previously described as compounds resistant to degradation by hot acids, hot concentrated alkaline solutions, and bleaching by strong oxidizing agents, while nowadays, they stand for melanins, compounds insoluble in aqueous or organic solvents [87]. As a consequence of melanin’s recalcitrant nature, it is difficult to achieve a detailed chemical analysis of melanin. Additionally, the use of hazardous chemical methods for the purification of melanin makes it harder to chemically analyze them since the chemicals used in the purification process can modify their structure [87,88,89]. Microscopic analysis of fungi has suggested that the melanins, in general, have a granular shape and are enclosed in the fungal cell wall [60]. Many filamentous fungi, also including paper deteriogens [2,18], in the genera Alternaria, Aspergillus, Auricularia, Cladosporium, Epicoccum, Eurotium, Magnapothe, Penicillium, Phomopsis, Sporothri, Stachybotrys, and Wangiella, have been shown to synthesize melanin [90]. This pigment serves as a protection from disadvantageous conditions for fungal species, even though it is not essential for their development. It has been shown that fungi that produce melanin are more resistant to UV light-induced damage, damage caused by radioactive oxidative species, temperature extremes, different hydrolytic enzymes, antimicrobial drugs, and many other harmful factors [54,91,92]. Recently, it has been discovered that due to the production of melanin fungi, particularly Cladosporium and Alternaria, species could survive extreme conditions, such as those in industrial and roadside areas. For instance, Cladosporium spp., Alternaria alternata, and Aureobasidium pullulans were found to colonize the walls of the damaged reactor at Chernobyl, where they were exposed to extreme levels of radiation [93]. Melanin protects conidia against hydrolysis and digestion by different enzymes secreted by other microorganisms as well as against bacterial and fungicidal proteins of animal origin [83]. Thus, it can be concluded that melanin contributes to fungal pathogenesis and survival in unfavorable conditions.
2.3. Carotenoids
Despite the fact that fungi are non-photosynthetic organisms and carotenoids are not as widely distributed in them as in plants [2], some carotene hydrocarbons have been found in several fungal species [67,94]. Carotenoids are naturally occurring pigments that are produced in an isoprenoid pathway [95]. Thus, they are comprised of isoprene units that contain light-absorbing conjugated double bonds, forming an aliphatic polyene chain and providing characteristic yellow, orange, or reddish colors [94]. Their essential biological role is in the protection of an organism from oxidative stress and membrane stabilization [94]. It has been suggested that by growing a strain of Penicillium sp. under high oxidative stress and in the deficiency of antioxidants, higher carotenoid colorant yields and elevated sclerotial biomass would be obtained [96]. They are also important precursors to essential metabolites such as vitamin A [95].
It has been shown that β-carotene production, as the most abundant carotenoid in nature, seems to be present in species such as Aspergillus giganteus [97], Cercospora nicotianae [98], Penicillium sp. [96], and Aschersonia aleyroides [99].
In the review written by Melo et al. [2], it has been reported that of 80 different colorants produced by fungi identified as paper colonizers, approximately 96% were of a polyketide nature and only around 4% were carotenoids. They have found only three different carotenoids in the literature that were mentioned in the context of colorants produced by fungi identified on paper: neurosporaxanthin, β-carotene, and sporopollenins. Moreover, the most common stains produced by fungi on paper have been found to be brownish and black. It has been suggested that these stains can be a mixture of different colorants. Specifically, the hallmark of A. niger is the formation of black or dark brown conidia, which appears to be an outcome when dark brown melanins combine with hexahydroxyl pentacyclic quinoid green pigments [100]. Additionally, the color of the fungal stains on paper may be a result of the oxidation process of a colorant in the given medium, which can further lead to the darkening of the stain. This can be seen in the brown pigment sporopollenin, an oxidative polymer of yellow β-carotene [68].
For full insight into the chemical structure of the fungal pigments, refer to Melo et al. [2].
3. Synthetic Dyes
Synthetic dyes have been largely used in the textile, paper printing, food, pharmaceutical, and other industries due to their several effective characteristics, such as cost-effectiveness in synthesis, firmness, and high stability to different factors such as light, temperature, detergent, and microbial attack [101,102,103]. Nowadays, these dyes represent a big problem for the environment. The color impedes the effect of the sunlight on water and negatively affects photosynthesis, which consequently reduces the food source of aquatic organisms and inhibits their growth [101,103,104,105]. Additionally, many synthetic dyes have been considered toxic and carcinogenic [106].
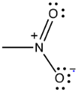
Dyes are organic compounds with the property of absorbing light in the visible spectrum (400–700 nm). This property is possible due to the presence of a chromophore or “color-bearing group”, described as a delocalized electron system with conjugated double bonds [107]. The chromophore is composed of groups of atoms, most commonly nitro (–NO2), azo (–N=N–), nitroso (–N=O), thiocarbonyl (–C=S), carbonyl (–C=O), and alkenes (–C=C–). Chemical classification of the dyes is based on these groups present in chromophores that are illustrated in Table 1 [108]. For a chromophore to absorb electromagnetic waves, an excitation of the electrons in a molecule is required. Once this requirement is fulfilled, the molecule becomes chromogenic. In addition, most of the dyes comprise groups called auxochromes or “color helpers” that can be acidic (COOH, SO3 and OH) or basic (NH2, NHR and NHR2).These groups are mostly responsible for dye solubility and color shifting [109].

Table 1.
Synthetic dyes: classification and examples of dyes according to the chromophore present.
Azo dyes have been by far the most utilized class of dyes in the industry. The azo chromophore is composed of at least one azo group (-N=N-), but sometimes two (diazo) and three (triazo) azo groups can be present in the structure. This azo group is bonded to two groups, of which at least one is aromatic [107].
The main compound of anthraquinone dyes is anthracene, composed of three fused benzene rings with two carbonyl groups on the central ring. After the substitution of the aromatic rings, the whole structure is given the color [110]. In the anthraquinone structure, the carbonyl group acts as an electron acceptor, which is not the case in the structure of azo dyes. In order to break the structure of anthraquinone dye, an electron donor is required [111,112] Moreover, anthraquinone dyes have a highly stabilized structure due to the resonance effect, which makes it harder to degrade them when compared with azo dyes [112,113].
Indigoid dyes are one of the oldest known classes of organic dyes. These dyes, also members of vat dyes (based on their application that takes place in a bucket or a vat), are insoluble in water and require oxygen for color development. They have an aromatic or heteroaromatic complex with an indigoid carbon skeleton, shown in Table 1 [108,109].
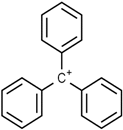
Another important class of dyes widely used in industries producing textiles, paper, leather, plastic, food, and pharmaceuticals is the triphenylmethane class [114,115,116]. These dyes are composed of a triphenylmethane backbone and are considered inexpensive colorants, but have shown cytotoxic effects on living organisms [117,118].
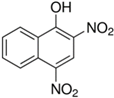
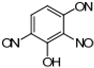
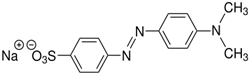
Stamp inks used in libraries have a complex composition containing dyes or pigments, hydrocarbons, plant or mineral oil as solvent, and many other additives [119]. Jasuja et al. [120] analyzed stamp pad inks by the TLC method and found that blue stamp ink contained Crystal Violet (CV) dye. Red stamp inks have been shown to be composed of alkyl-phenols, castor oil, and polyethylene glycol as volatile organic components [121]. Nowadays, the most commonly used are water-based stamp inks, usually containing colorants such as Acid Red R, Eosin Y, and Pigment Red 112 [18]. Eosin Y is a xanthene dye containing phenolic groups with hydroxyl groups and bromine.
4. Potential Decolorizing Enzymes and Their Characteristics
Many fungal pigments and synthetic dyes are composed of phenolic compounds [2,93], and their structure can be attacked by several oxidoreductive enzymes [122]. In this big group of enzymes, the LMEs and the DyPs are also included. Here, we describe the characteristics of the most promising candidates for decolorizing applications (laccases, LiPs, MnPs, and some DyPs) and their ways of action.
4.1. Laccases
Laccases (EC 1.10.3.2), also known as benzenediol: oxygen oxidoreductase or polyphenol oxidase, are members of the family of blue multicopper oxidases that catalyze single electron oxidation of four reducing-substrate molecules, accompanied by the four-electron reduction of molecular oxygen into the water. At the end of the nineteenth century, this enzyme was extracted from the Japanese lacquer Rhus vernicifera, after which it was named laccase [123,124]. Laccases have been considered to be ubiquitous in many white-rot fungi studied so far. The reason for their wide distribution is their function: degradation of the complex polyphenol structure that constitutes lignin in order to break the plant’s cell wall and uptake the host’s nutrients [125,126]. Fungal laccases also contribute to melanin formation in different fungal species [60,127]. This enzyme showed a specific function in some insects and plants [128]. Plant laccase, together with peroxidases, contributes to the biosynthesis of lignin during protoplast regeneration [129,130]. Laccases have also been found in some bacteria, having a function in melanin synthesis and showing stability at high temperatures and pH [131,132,133,134]. Thus, laccases are widely present in nature with a role in either synthetic or degradation processes.
Fungal laccases have shown large-scale applications for industrial and biotechnological processes because they are extracellular and inducible, and they do not require a cofactor and show low substrate specificity [128]. For example, they utilize oxygen as a cofactor [135] and oxidizing agent and showed the degradation of several compounds with a phenolic structure [136]. Thus, laccases have been utilized in many bioremediation processes as well as in the degradation of a wide number of textile dyes [39,128].
Laccase is a monomeric globular glycoprotein comprised of around 500 amino acid residues organized into three consecutive domains (A, B, C) with a Greek key β barrel topology [127].
The overall reaction that is catalyzed by laccase represents the oxidation of four electrons of a reducing substrate with a two-electron reduction of dioxygen to water [4RH + O2 → 4R + 2H2O]. The first step is a reduction of the reducing substrate carried out by the Cu2+ (Cu2+ to Cu+) at the T1 site, which is primarily an electron acceptor. The electrons extracted from the reducing substrate are transferred to the T2/T3 site, with three Cu2+ ions in total. This results in changing the enzyme state from fully oxidized to fully reduced [124].
Laccases catalyze the degradation of phenolic structures through the subtraction of one electron from phenolic hydroxyl groups to form phenoxy radicals that lead to the cleavage steps, which results in aromatic ring cleavage. Moreover, this enzyme can oxidize non-phenolic compounds but in the presence of a mediator such as 2,2′-azinobis-(3-ethylbenzthiazoline-6-sulfonate)(ABTS), 1-hydroxybenzotriazole (HBT), and 3-hyroxyanthranilic acid [124].
4.2. Lignin Peroxidases (LiPs)
LiP (E.C. 1.11.1.14) is one of the most significant ligninolytic enzymes that contains heme (protoporphyrin IX) as a prosthetic group. This enzyme was firstly discovered in nitrogen- and carbon-limited cultures of the white rot fungi Phanerochaete chrysosporium [137]. The presence of this enzyme was later detected in other white rot fungi such as Trametes versicolor [138]), Bjerkandera sp., Trametes trogii, and Phlebia tremellosa [139,140]. LiP has been purified in bacteria Acinetobacter calcoaceticus [140] and also has shown activity in Streptomyces viridosporus [141]. This enzyme is relatively non-specific to its substrate and its main function is the oxidation of different phenolic aromatic compounds as well as a broad range of non-phenolic lignin model compounds [124]. The aforementioned microorganisms secrete LiPs as a family of isozymes with relative composition and isoelectric points (pI), depending on the growth medium and nutrient conditions [142].
4.3. Manganese Peroxidases (MnPs)
Manganese peroxidase (EC 1.11.1.13) is another lignin-modifying enzyme firstly detected in P. chrysosporium by Kuwahara et al. [143]. It is secreted by many Basidiomycota species such as Panus tigrinus, Agaricus bisporus, Bjerkandera sp., and Nematoloma frowardii [142]. Additionally, this enzyme has been found in bacteria Bacillus pumilus, Paenibacillus sp. [144], Azospirillum brasilense and Actinobacterium S. psammoticus [142]. MnPs are glycoproteins with a molecular weight ranging between 38 and 62.5 kDa, comprised of approximately 350 amino acids. The overall structure is shown to be similar to that of LiPs, with two domains, a heme group in the middle, 10 major helices, and a minor helix. MnPs have five disulfide bonds unlike LiPs. Another major characteristic of MgP is the manganese-binding site. The additional disulfide bond participates in the Mn-binding site alongside Aspartic and Glutamic acid residues [145].
4.4. Dye-Decolorizing Peroxidases (DyPs)
Dye-decolorizing peroxidases (EC 1.11.1.19) represent a new family of heme peroxidases with one heme as a cofactor and a role in H2O2-dependent oxidation of a wide range of molecules [146,147]. Basidiomycete Thanatephorus cucumeris Dec1 was the first species suggested to be excreting these enzymes due to its ability to decolorize 18 types of reactive, acidic, and dispersive dyes, most of which were xenobiotics [148]. In 1999, the same author succeeded in the attempt to purify DyP for the first time. It is a glycoprotein with a molecular mass of 60 kD [149]. Later, it was reported that several bacteria have a role in lignin degradation, suggesting that these bacteria possess a unique class of dye decolorizing peroxidases [150,151]. Due to the low sequence identity shown between bacterial and fungal DyPs, they are divided into four classes, from A to D. The DyP from class D are the ones from Basidiomycota [146].
The catalytic cycle of the DYPs is composed of the resting state and transient intermediates comprising compound I and compound II (Figure 2) [152]. When the enzyme is in a resting state, it contains Fe3+. When this resting ferric enzyme is oxidized by H2O2, a high-valence compound I, [Fe4+ = O]+• is produced. This compound I reacts with 1 eq electrons and, from a reducing state, generates a compound II, [Fe4+ = O]+. Compound II is then reduced by a second substrate molecule and the resting state enzyme is produced again [152].
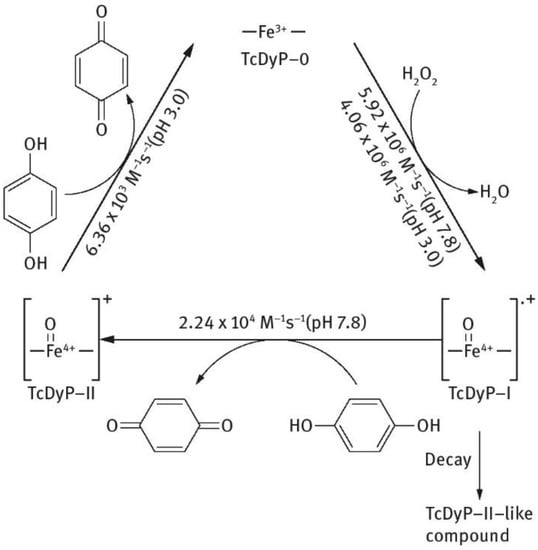
Figure 2.
Schematic description of catalytic cycle of Thermomonospora curvata DyPs.
Fernandez-Fueyo and colleagues [153] have shown that DyPs enzymes, Pleos-DyP1 and Pleos-DyP4, from fungi Pleurotus ostreatus have the ability to oxidize Mn2+ to Mn3+, a feature mostly present in MnPs and versatile peroxidases (VPs).
To compare the mechanisms of reaction of lignin modifying enzymes, including peroxidases and laccase, a schematic outline is shown (Figure 3) [142].
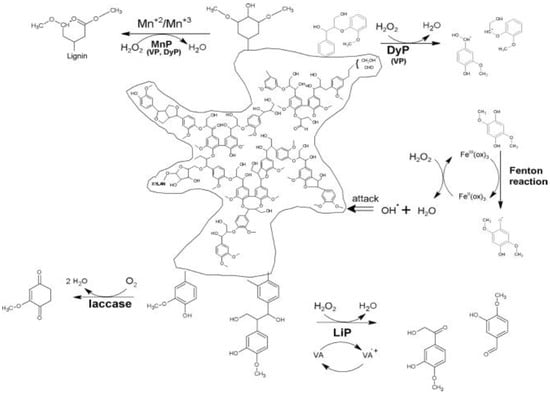
Figure 3.
Comparison of lignin degradation by peroxidases and laccase. MnP—manganese peroxidase, VP—versatile peroxidase, LiP—lignin peroxidase, DyP—dye-decolorizing peroxidase, ox—oxalate, VA—veratryl alcohol.
Many stains present on paper can be caused by past fungal contaminations but also by several types of synthetic dyes composing inks and stamp inks. As we saw in the previous paragraphs, there are some similarities between the structures of synthetic dyes and fungal pigments. Therefore, starting with the known properties of LMEs and DyPs to degrade synthetic dyes, we illustrate here the potential of these enzymes to also decolorize fungal pigments.
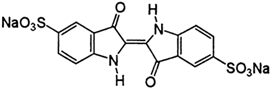
Osmanova and colleagues [55] were the first to present the transformation pathway of Remazol Brilliant Blue R (RBBR) dye by immobilizing laccase. Laccase was derived from the white-rot fungus Trametes pubescens and has shown a degradation activity of 44% in 42 h. The transformation pathway involved steps of deamination and oxidation and resulted in products that were less toxic than RBBR itself [55].
Levin and colleagues have demonstrated that two different strains of Trametes species produce different laccase isoforms capable of degrading Acid green 27 (AG27), another anthraquinone dye. Furthermore, they have shown that besides anthraquinone dyes, the culture fluids of the strain Trametes sp. LA1 also decolorized the azo dye Reactive Black 5 (RB5) and triphenylmethane CV, suggesting that this strain could be a new laccase source with dye-decolorization potential [154].
Another study showed that due to the presence of laccase, MnP, and LiP, the strain Aspergillus iizukae EAN60 was able to decolorize RBBR and two other azo dyes, methyl red (MR) and methyl orange (MO) [155]. After 9 days of monitoring, the maximum removal of RBBR was close to 95%, while the removal of MO and MR observed after 3 days was 84.90% and 47.65%, respectively.
Sing and colleagues [122] investigated the production of ligninolytic enzymes by fungi Marasmius cladophyllus during its application in the decolorization of RBBR, as well as two azo dyes, such as Orange G and Congo red (CR). By monitoring specific reactions characteristic of ligninolytic enzymes, it has been found that in the presence and absence of RBBR dye, this strain produced both laccase and LiP. After in vitro RBBR dye decolorization in the culture medium containing unpurified enzymes, it has been shown that laccase activity was in agreement with the dye decolorization. Moreover, the fresh dye has been successively added to the already decolorized culture medium. This step actually sped decolorization up in the subsequent round. After reconcentration of unpurified enzymes from decolorized culture medium, the protein precipitates contained laccase that decolorized 76% of RBBR dye in less than one day. The protein precipitates were also tested for in vitro decolorization of a mono-azo dye, Orange G, and a di-azo dye, CR. It has been shown that laccase decolorized 54% of Orange G and 33% of CR at the same time. On the other hand, the protein precipitates did not show the presence of LiP [122].
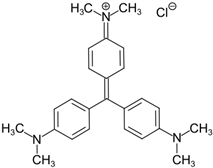
Another synthetic dye, Crystal Violet (CV), has been widely utilized in medicine and by the textile industry [44]. Several studies have shown that microorganisms can successfully degrade this dye and have proposed the hypothetical pathway of CV degradation [156,157]. In the first step of degradation, it has been suggested that laccase catalyzed reduction to its leuco derivative before reductively degrading into the next intermediates to the final phenol (Figure 4) [156]. An additional enzyme that appeared to be involved in the process was aminopyrine N-demethylase.
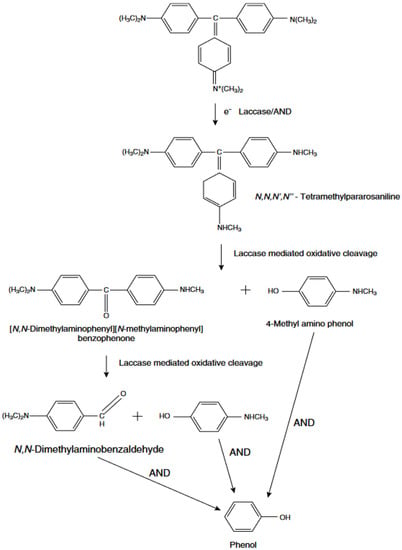
Figure 4.
Mechanism of Crystal Violet degradation by laccase.
Laccase catalytic activity in the degradation of Bisphenol A and decolorization of synthetic dyes was observed by Chairin et al. (2013) [40]. The purified enzyme from fungi Trametes polyzona WR710-1-induced oxidation of Bisphenol A within 3 h with redox mediator 1-hydroxybenzotriazole (HBT) present. Triphenylmethane dye (Bromophenol Blue; BRB), anthraquinone dye (RBBR), and azo dye (MO) were decolorized without the redox mediator present at a rate of 80%.
Another ascomycete, Leptosphaerulina sp., has been described as a producer of high amounts of laccase and MnP. These enzymes have shown the activity of decolorization of Remazol Black, Novacron Red, and Turquoise Blue.
To improve the production and decolorizing activity of Leptosphaerulina sp., Copete-Pertuz et al. [158] co-cultivated this species with other fungal species. Based on response surface methodology, they inferred that co-cultivation with Trichoderma viride and Aspergillus terreus was the best treatment for the production of ligninolytic enzymes, specifically laccase, MnP, and versatile peroxidase, as well as the degradation of RB5 [158].
Zhang et al. [159] cloned a novel MnP gene (mnp3) from the fungal strain Cerrena unicolor BBP6 and expressed it in the yeast Pichia pastoris. They have demonstrated that this recombinant enzyme, rMnP3-BBP6, is an effective decolorizing agent against different dyes. Additionally, it has been shown that rMnP3-BBP6 has great potential in denim bleaching. The decolorization potential of dyes such as RBBR, MO, BPB, and CV by recombinant MnP was higher than 50%.
Besides laccase and MnPs, LiP was confirmed to decolorize synthetic dyes. This enzyme showed degradative activity in a variety of substrates such as Mg2+, tryptophan, L-Dopa, hydroquinone, and synthetic dyes. Specifically, LiP isolated and purified from bacteria, Acinetobacter calcoaceticus decolorized azo-dyes such as MR and MO at a very high rate, 98% and 96% in one day, respectively. Other azo dyes were also decolorized by this enzyme, up to approximately 90% within two days [140]. The reaction mechanism of LiP in the decolorization of azo dyes was demonstrated by Jadhav and colleagues [160,161]. They have proposed that the cleavage of the dye is done asymmetrically, with an active site available for a LiP to excite the molecule. This asymmetric cleavage by LiP occurs between the nitrogen of the azo group (N=N) and the carbon of the aromatic ring and results in the production of different types of naphthalene-2-sulfonic acid, depending on the type of the dye that was subjected to degradation (Figure 5) [160,161].
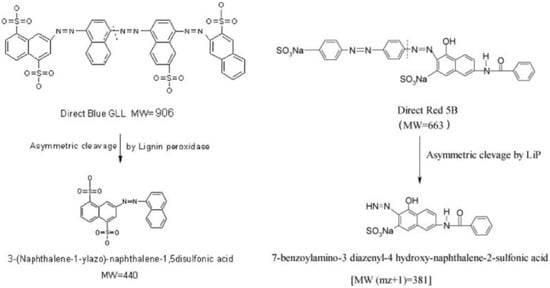
Figure 5.
Reaction mechanism of LiP in a degradation pathway of azo dyes.
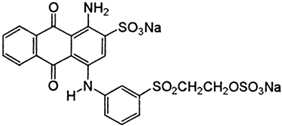
The anthraquinone dye degradation pathway has been reported by Sugano et al. (2009) [162]. The DyP from the basidiomycete Thanatephorus cucumeris was utilized in the process of degradation of RB5 dye. The pathway has the following intermediates: phthalic acid, Product 2, and Product 3 (Figure 6) [162]. These Product 2 and Product 3 were generated by the peroxidase reaction, while in the case of phthalic acid, it has been suggested that DyP acted as a hydrolase or oxygenase rather than a peroxidase [162].
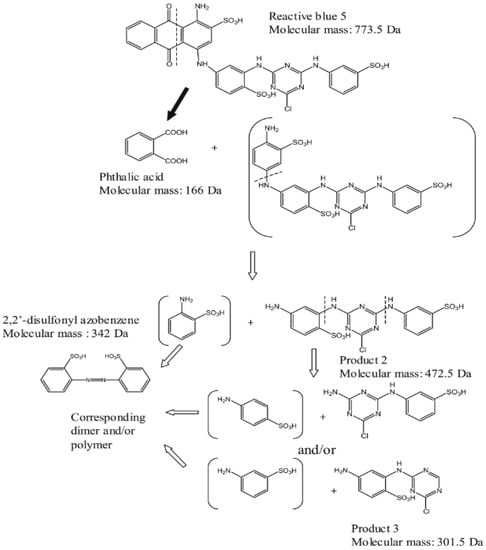
Figure 6.
Degradation pathway of RB5 by DyP.
Only a few attempts have been published, describing the degradation of uncharacterized fungal and melanin stains from various surfaces. Two different approaches were applied, a laccase-based method able to decolorize the fungal stains on paper and parchment [163] and a biomimetic copper-pyridine oxidation system able to remove fungal melanin from paper [164] and decolorize fungal stains from textile [165].
However, it has been shown that many quinone colorants are also synthesized by fungi are isolated from paper [2]. Quinones are a class of organic quinoid compounds widely distributed in nature [65]. A common basic structural pattern is an ortho or a para substituted dione. If this dione is conjugated to an aromatic nucleus, it is called benzene quinone, or to a condensed polycyclic aromatic system, then it can be naphthoquinones, anthraquinones, and anthracyclinones [65]. As was shown above, anthraquinone dyes used in textiles and in other industries can be degraded by LMEs synthesized by microorganisms, thus, we hypothesize that similar action can be applied to the quinoid colorants synthesized by different fungal species on historical paper.
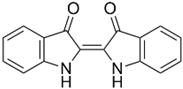
When one fungal pigment from the quinone class is compared with RBBR (Figure 7) [158,159] we can see this common cyclic dione with different functional groups. In the degradation of RBBR dye, laccase appears to be the first enzyme that cleaves hydrogen bonds between N-H groups due to the reduction of Cu2+ to Cu+, generating two compounds. In the case of fungal colorants, this cleavage would take place between the OH groups.
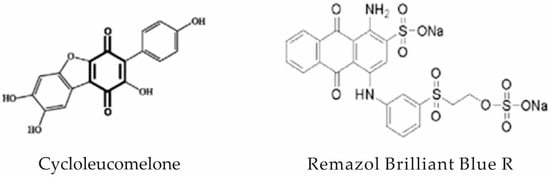
Figure 7.
Fungal pigment cycloleucomelone and anthraquinone dye RBBR.
If we take a look at the structures of pigments produced by fungi and synthetic dyes, we can see that most of them contain a phenolic ring bound to the hydroxy group (for a closer look, refer to [2]). For example, simple flavipin, a yellow pigment produced by Epicoccum purpurescens and Chaematomium globosum [2,166], has a phenyl group bonded to three hydroxyl groups (Figure 8b). Under the catalysis of MnPs, one of those hydroxyl groups can be oxidized, and Mn3+ is reduced to Mn2+. As the reaction goes further, with the series of oxidoreduction reactions, the phenolic ring would break down, causing the degradation of the pigment. Bilal et al. [39] have demonstrated a hypothetical degradation pathway of a phenol containing an organic compound (Figure 8a, [39]) as many fungal pigments and synthetic dyes are [39]. Based on the reactions shown by Bilal and colleagues [39], we proposed a similar degradation pathway of the fungal pigment flavipin (Figure 8b).
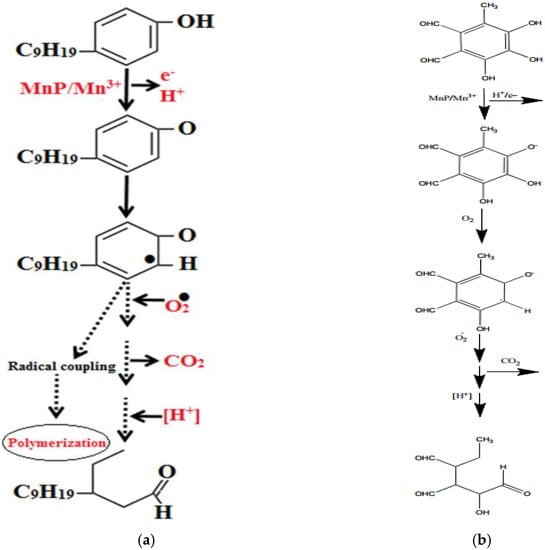
Figure 8.
(a) Hypothetical degradation of phenolic compounds (b) Hypothetical degradation of fungal pigment flavipin.
Due to the characteristic of DyP to act as a hydrolase/oxygenase, this enzyme can also be employed to degrade quinone structures that have been found abundantly in filamentous fungi, leaving different color hues on the paper. In Figure 9, we have demonstrated a hypothetical degradation pathway of a randomly chosen pigment, 3-acetyl-2,8-dihydroxy-6-methoxy-anthraquinone, found in Fusarium oxysporum. The chemical structure of the pigment itself is not as complicated as those of synthetic dyes; thus, it can be degraded by the cleavage of the –C(=O)– group. The same reaction is shown in Figure 6 [162] as the initial step of RB5 degradation. When it is broken down into simple phenolic compounds, these compounds can further be degraded by the MnP, as has previously been shown in Figure 8b.
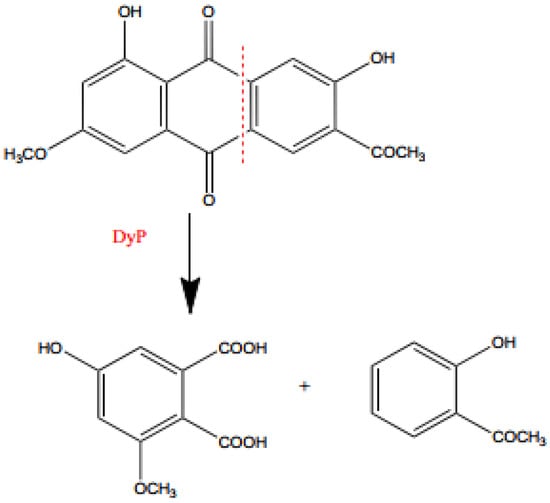
Figure 9.
Degradation of pigment 3-acetyl-2,8-dihydroxy-6-methoxy-anthraquinone.
Additional problems that libraries and book restorers are dealing with are stamp inks. Those inks are water-based and usually contain colorants such as Acid Red R, Eosin Y, and Pigment Red 112 [18]. Eosin Y is a xanthene dye containing phenolic groups with hydroxyl groups and bromine. Thus, this structure could also be degraded by MnPs. Acid Red R and Pigment Red 112 are typical azo dyes, so they should easily be degraded by laccase or LiP in a similar way as is shown in Figure 5 [160,161].
5. Concluding Remarks and Future Outlook
In summary, the data reviewed above illustrated information about the pigments produced by fungi that can colonize paper cultural heritage, synthetic dyes, ligninolytic and decolorizing enzymes, and their application. During the past few years, enzyme-based catalytic engineering has been used in the bioremediation of industrial pollutants. In this regard, the development of enzyme-based immobilized systems suggests a great potential for biotechnological applications for cultural heritage bioremediation purposes. Ligninolytic and dye-decolorizing enzymes have demonstrated features to degrade, decolorize, or detoxify several types of synthetic dyes and pollutants. When these dyes are compared with the fungal pigments found in the books, we hypothesized that the described enzymes have the ability to also degrade these structures. Depending on the side groups of the pigments’ structure, with the gathered action of laccase that can catalyze oxidation or demethylation, DyP that can cleave the –C(=O)– group and MnP that catalyzes oxidation-reduction reactions, specific fungal pigments could be degraded, which further leads to the cleaning of the stained paper. Similarly, water-based stamp inks can also be removed by the same principle since these inks are composed of azo dyes, previously shown to be removed by ligninolytic enzymes secreted by different microorganisms.
Even though the costs of biocleaning methods are comparable to those of chemical-physical techniques, the biological approach is more favorable in terms of both environmental and human health safety. Thus, future research should focus on the development of new protocols for the large-scale production of decolorizing enzymes and their application on paper and perhaps also on architectural cultural heritage since the direct application of living microorganisms can leave residues on the surfaces that are about to be restored and later serve as nutrients to other dangerous microbes. Additionally, the discovery of new chemical reactions catalyzed by ligninolytic and dye decolorizing enzymes represents a great rationale for their further utilization in bioremediation processes.
Author Contributions
Conceptualization, J.P. and D.P.; methodology, Z.F. and L.K.; resources, J.P., Z.F. and L.K.; data curation J.P.; writing—original draft preparation, J.P. and D.P.; writing—review and editing, J.P., Z.F., L.K. and D.P. All authors have read and agreed to the published version of the manuscript.
Funding
Authors acknowledge the projects APVV-19-0059 and VEGA 2/099/2021 which financed this study.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Karbowska-Berent, J.; Jarmiłko, J.; Czuczko, J. Fungi in Fox Spots of a Drawing by Leon Wyczółkowski. Restaur. Int. J. Preserv. Libr. Arch. Mater. 2014, 35, 159–179. [Google Scholar] [CrossRef]
- Melo, D.; Sequeira, S.O.; Lopes, J.A.; Macedo, M.F. Stains versus Colourants Produced by Fungi Colonising Paper Cultural Heritage: A Review. J. Cult. Herit. 2019, 35, 161–182. [Google Scholar] [CrossRef]
- Piñar, G.; Poyntner, C.; Lopandic, K.; Tafer, H.; Sterflinger, K. Rapid Diagnosis of Biological Colonization in Cultural Artefacts Using the MinION Nanopore Sequencing Technology. Int. Biodeterior. Biodegrad. 2020, 148, 104908. [Google Scholar] [CrossRef]
- Puškárová, A.; Bučková, M.; Habalová, B.; Kraková, L.; Maková, A.; Pangallo, D. Microbial Communities Affecting Albumen Photography Heritage: A Methodological Survey. Sci. Rep. 2016, 6, 20810. [Google Scholar] [CrossRef] [PubMed]
- Sclocchi, M.C.; Kraková, L.; Pinzari, F.; Colaizzi, P.; Bicchieri, M.; Šaková, N.; Pangallo, D. Microbial Life and Death in a Foxing Stain: A Suggested Mechanism of Photographic Prints Defacement. Microb. Ecol. 2017, 73, 815–826. [Google Scholar] [CrossRef]
- Roman, C.; Diaconescu, R.-M.; Scripcariu, L.; Grigoriu, A. Biocides Used in Preservation, Restoration and Conservation of the Paper. Eur. J. Sci. Theol. 2013, 9, 263–271. [Google Scholar]
- Strlič, M.; Kolar, J. Ageing and Stabilisation of Paper; National and University Library: Ljubljana, Slovenia, 2005; ISBN 978-961-6551-03-8. [Google Scholar]
- Michaelsen, A.; Piñar, G.; Pinzari, F. Molecular and Microscopical Investigation of the Microflora Inhabiting a Deteriorated Italian Manuscript Dated from the Thirteenth Century. Microb. Ecol. 2010, 60, 69–80. [Google Scholar] [CrossRef]
- Pangallo, D.; Šimonovičová, A.; Chovanová, K.; Ferianc, P. Wooden Art Objects and the Museum Environment: Identification and Biodegradative Characteristics of Isolated Microflora. Lett. Appl. Microbiol. 2007, 45, 87–94. [Google Scholar] [CrossRef]
- Sterflinger, K.; Pinzari, F. The Revenge of Time: Fungal Deterioration of Cultural Heritage with Particular Reference to Books, Paper and Parchment. Environ. Microbiol. 2012, 14, 559–566. [Google Scholar] [CrossRef]
- Viegas, C.; Pinheiro, A.C.; Sabino, R.; Viegas, S.; Brandão, J.; Veríssimo, C. Environmental Mycology in Public Health: Fungi and Mycotoxins Risk Assessment and Management; Academic Press: Cambridge, MA, USA, 2015; ISBN 978-0-12-411535-4. [Google Scholar]
- Arai, H. Foxing Caused by Fungi: Twenty-Five Years of Study. Int. Biodeterior. Biodegrad. 2000, 46, 181–188. [Google Scholar] [CrossRef]
- Mesquita, N.; Portugal, A.; Videira, S.; Rodríguez-Echeverría, S.; Bandeira, A.M.L.; Santos, M.J.A.; Freitas, H. Fungal Diversity in Ancient Documents. A Case Study on the Archive of the University of Coimbra. Int. Biodeterior. Biodegrad. 2009, 63, 626–629. [Google Scholar] [CrossRef]
- Kraková, L.; Chovanová, K.; Selim, S.A.; Šimonovičová, A.; Puškarová, A.; Maková, A.; Pangallo, D. A Multiphasic Approach for Investigation of the Microbial Diversity and Its Biodegradative Abilities in Historical Paper and Parchment Documents. Int. Biodeterior. Biodegrad. 2012, 70, 117–125. [Google Scholar] [CrossRef]
- Micheluz, A.; Manente, S.; Tigini, V.; Prigione, V.; Pinzari, F.; Ravagnan, G.; Varese, G.C. The Extreme Environment of a Library: Xerophilic Fungi Inhabiting Indoor Niches. Int. Biodeterior. Biodegrad. 2015, 99, 1–7. [Google Scholar] [CrossRef]
- Montanari, M.; Melloni, V.; Pinzari, F.; Innocenti, G. Fungal Biodeterioration of Historical Library Materials Stored in Compactus Movable Shelves. Int. Biodeterior. Biodegrad. 2012, 75, 83–88. [Google Scholar] [CrossRef]
- Sequeira, S.O.; de Carvalho, H.P.; Mesquita, N.; Portugal, A.; Macedo, M.F. Fungal Stains on Paper: Is What You See What You Get? Conserv. Património 2019, 32, 18–27. [Google Scholar] [CrossRef]
- Zhang, Q.; Zou, J.; Shi, G.; Zhang, L. [Analysis of major components in water based stamp pad inks and their imprints by ultra high performance liquid chromatography-mass spectrometry and gas chromatography-mass spectrometry]. Se Pu Chin. J. Chromatogr. 2010, 28, 1132–1136. [Google Scholar]
- Balloi, A.; Palla, F. Biocleaning. In Biotechnology and Conservation of Cultural Heritage; Palla, F., Barresi, G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 67–84. ISBN 978-3-319-46168-7. [Google Scholar]
- Barbabietola, N.; Tasso, F.; Alisi, C.; Marconi, P.; Perito, B.; Pasquariello, G.; Sprocati, A.R. A Safe Microbe-Based Procedure for a Gentle Removal of Aged Animal Glues from Ancient Paper. Int. Biodeterior. Biodegrad. 2016, 109, 53–60. [Google Scholar] [CrossRef]
- Bosch-Roig, P.; Ranalli, G. The Safety of Biocleaning Technologies for Cultural Heritage. Front. Microbiol. 2014, 5, 155. [Google Scholar] [CrossRef][Green Version]
- Jeszeová, L.; Bauerová-Hlinková, V.; Baráth, P.; Puškárová, A.; Bučková, M.; Kraková, L.; Pangallo, D. Biochemical and Proteomic Characterization of the Extracellular Enzymatic Preparate of Exiguobacterium undae, Suitable for Efficient Animal Glue Removal. Appl. Microbiol. Biotechnol. 2018, 102, 6525–6536. [Google Scholar] [CrossRef]
- Jeszeová, L.; Benžová, R.; Gluštíková, M.; Šišková, A.; Kisová, Z.; Planý, M.; Kraková, L.; Bauerová-Hlinková, V.; Pangallo, D. Biocleaning of Historical Documents: The Use and Characterization of Bacterial Enzymatic Resources. Int. Biodeterior. Biodegrad. 2019, 140, 106–112. [Google Scholar] [CrossRef]
- Kisová, Z.; Pavlović, J.; Šefčiková, L.; Bučková, M.; Puškárová, A.; Kraková, L.; Šišková, A.O.; Kleinová, A.; Machatová, Z.; Pangallo, D. Removal of Overpainting from an Historical Painting of the XVIII Century: A Yeast Enzymatic Approach. J. Biotechnol. 2021, 335, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Ranalli, G.; Alfano, G.; Belli, C.; Lustrato, G.; Colombini, M.P.; Bonaduce, I.; Zanardini, E.; Abbruscato, P.; Cappitelli, F.; Sorlini, C. Biotechnology Applied to Cultural Heritage: Biorestoration of Frescoes Using Viable Bacterial Cells and Enzymes. J. Appl. Microbiol. 2005, 98, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Valentini, F.; Diamanti, A.; Carbone, M.; Bauer, E.M.; Palleschi, G. New Cleaning Strategies Based on Carbon Nanomaterials Applied to the Deteriorated Marble Surfaces: A Comparative Study with Enzyme Based Treatments. Appl. Surf. Sci. 2012, 258, 5965–5980. [Google Scholar] [CrossRef]
- Mazzoli, R.; Giuffrida, M.G.; Pessione, E. Back to the past—Forever young: Cutting-edge biochemical and microbiological tools for cultural heritage conservation. Appl. Microbiol. Biotechnol. 2018, 102, 6815–6825. [Google Scholar] [CrossRef]
- Nyanhongo, G.S.; Gomes, J.; Gübitz, G.M.; Zvauya, R.; Read, J.; Steiner, W. Decolorization of Textile Dyes by Laccases from a Newly Isolated Strain of Trametes modesta. Water Res. 2002, 36, 1449–1456. [Google Scholar] [CrossRef]
- Ricotta, A.; Unz, R.F.; Bollag, J.-M. Role of a Laccase in the Degradation of Pentachlorophenol. Bull. Environ. Contam. Toxicol. 1996, 57, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Xu, F. Oxidation of Phenols, Anilines, and Benzenethiols by Fungal Laccases: Correlation between Activity and Redox Potentials as Well as Halide Inhibition. Biochemistry 1996, 35, 7608–7614. [Google Scholar] [CrossRef]
- Wong, Y.; Yu, J. Laccase-Catalyzed Decolorization of Synthetic Dyes. Water Res. 1999, 33, 3512–3520. [Google Scholar] [CrossRef]
- Bogan, B.W.; Lamar, R.T. Polycyclic Aromatic Hydrocarbon-Degrading Capabilities of Phanerochaete Laevis HHB-1625 and Its Extracellular Ligninolytic Enzymes. Appl. Environ. Microbiol. 1996, 62, 1597–1603. [Google Scholar] [CrossRef]
- Chi, Y.; Hatakka, A.; Maijala, P. Can Co-Culturing of Two White-Rot Fungi Increase Lignin Degradation and the Production of Lignin-Degrading Enzymes? Int. Biodeterior. Biodegrad. 2007, 59, 32–39. [Google Scholar] [CrossRef]
- Chowdhary, P.; More, N.; Yadav, A.; Bharagava, R.N. Chapter 12—Ligninolytic Enzymes: An Introduction and Applications in the Food Industry. In Enzymes in Food Biotechnology; Kuddus, M., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 181–195. ISBN 978-0-12-813280-7. [Google Scholar]
- Fackler, K.; Gradinger, C.; Hinterstoisser, B.; Messner, K.; Schwanninger, M. Lignin Degradation by White Rot Fungi on Spruce Wood Shavings during Short-Time Solid-State Fermentations Monitored by near Infrared Spectroscopy. Enzyme Microb. Technol. 2006, 39, 1476–1483. [Google Scholar] [CrossRef]
- Singh, P.; Sulaiman, O.; Hashim, R.; Rupani, P.F.; Peng, L.C. Biopulping of Lignocellulosic Material Using Different Fungal Species: A Review. Rev. Environ. Sci. Biotechnol. 2010, 9, 141–151. [Google Scholar] [CrossRef]
- Tuor, U.; Winterhalter, K.; Fiechter, A. Enzymes of White-Rot Fungi Involved in Lignin Degradation and Ecological Determinants for Wood Decay. J. Biotechnol. 1995, 41, 1–17. [Google Scholar] [CrossRef]
- Bagewadi, Z.K.; Mulla, S.I.; Ninnekar, H.Z. Purification and Immobilization of Laccase from Trichoderma harzianum Strain HZN10 and Its Application in Dye Decolorization. J. Genet. Eng. Biotechnol. 2017, 15, 139–150. [Google Scholar] [CrossRef]
- Bilal, M.; Asgher, M.; Iqbal, H.M.N.; Hu, H.; Wang, W.; Zhang, X. Bio-Catalytic Performance and Dye-Based Industrial Pollutants Degradation Potential of Agarose-Immobilized MnP Using a Packed Bed Reactor System. Int. J. Biol. Macromol. 2017, 102, 582–590. [Google Scholar] [CrossRef]
- Chairin, T.; Nitheranont, T.; Watanabe, A.; Asada, Y.; Khanongnuch, C.; Lumyong, S. Biodegradation of Bisphenol A and Decolorization of Synthetic Dyes by Laccase from White-Rot Fungus, Trametes polyzona. Appl. Biochem. Biotechnol. 2013, 169, 539–545. [Google Scholar] [CrossRef]
- Chen, S.H.; Yien Ting, A.S. Biodecolorization and Biodegradation Potential of Recalcitrant Triphenylmethane Dyes by Coriolopsis sp. Isolated from Compost. J. Environ. Manage. 2015, 150, 274–280. [Google Scholar] [CrossRef]
- Copete-Pertuz, L.S.; Plácido, J.; Serna-Galvis, E.A.; Torres-Palma, R.A.; Mora, A. Elimination of Isoxazolyl-Penicillins Antibiotics in Waters by the Ligninolytic Native Colombian Strain Leptosphaerulina sp. Considerations on Biodegradation Process and Antimicrobial Activity Removal. Sci. Total Environ. 2018, 630, 1195–1204. [Google Scholar] [CrossRef]
- Lundell, T.K.; Mäkelä, M.R.; Hildén, K. Lignin-Modifying Enzymes in Filamentous Basidiomycetes – Ecological, Functional and Phylogenetic Review. J. Basic Microbiol. 2010, 50, 5–20. [Google Scholar] [CrossRef]
- Mani, S.; Bharagava, R.N. Exposure to Crystal Violet, Its Toxic, Genotoxic and Carcinogenic Effects on Environment and Its Degradation and Detoxification for Environmental Safety. In Reviews of Environmental Contamination and Toxicology Volume 237; de Voogt, W.P., Ed.; Reviews of Environmental Contamination and Toxicology; Springer International Publishing: Cham, Switzerland, 2016; pp. 71–104. ISBN 978-3-319-23573-8. [Google Scholar]
- Zeng, S.; Qin, X.; Xia, L. Degradation of the Herbicide Isoproturon by Laccase-Mediator Systems. Biochem. Eng. J. 2017, 119, 92–100. [Google Scholar] [CrossRef]
- Daniel, G. Chapter 8—Fungal Degradation of Wood Cell Walls. In Secondary Xylem Biology; Kim, Y.S., Funada, R., Singh, A.P., Eds.; Academic Press: Boston, MA, USA, 2016; pp. 131–167. ISBN 978-0-12-802185-9. [Google Scholar]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the Enzymatic Repertoire of the CAZy Database to Integrate Auxiliary Redox Enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Daou, M.; Faulds, C.B. Glyoxal Oxidases: Their Nature and Properties. World J. Microbiol. Biotechnol. 2017, 33, 87. [Google Scholar] [CrossRef]
- Gygli, G.; de Vries, R.P.; van Berkel, W.J.H. On the Origin of Vanillyl Alcohol Oxidases. Fungal Genet. Biol. 2018, 116, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Caneva, G. Biology in the Conservation of Works of Art; ICCROM: Rome, Italy, 1991; ISBN 978-92-9077-101-2.
- Torres, F.A.E.; Zaccarim, B.R.; de Lencastre Novaes, L.C.; Jozala, A.F.; dos Santos, C.A.; Teixeira, M.F.S.; Santos-Ebinuma, V.C. Natural Colorants from Filamentous Fungi. Appl. Microbiol. Biotechnol. 2016, 100, 2511–2521. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, B.; Martínez-Morales, F.; Tinoco-Valencia, R.; Rojas, S.; Acosta-Urdapilleta, L.; Trejo-Hernández, M.R. Biochemical and Molecular Characterization of Laccase Isoforms Produced by the White-Rot Fungus Trametes versicolor under Submerged Culture Conditions. J. Mol. Catal. B Enzym. 2015, 122, 339–347. [Google Scholar] [CrossRef]
- Szcepanowska, H.; Mathia, T.G.; Belin, P. Morphology of Fungal Stains on Paper Characterized with Multi-Scale and Multi-Sensory Surface Metrology. Scanning 2014, 36, 76–85. [Google Scholar] [CrossRef]
- Butler, M.J.; Day, A.W. Fungal Melanins: A Review. Can. J. Microbiol. 1998, 44, 1115–1136. [Google Scholar] [CrossRef]
- Osmanova, N.; Schultze, W.; Ayoub, N. Azaphilones: A Class of Fungal Metabolites with Diverse Biological Activities. Phytochem. Rev. 2010, 9, 315–342. [Google Scholar] [CrossRef]
- Lin, L.; Xu, J. Fungal Pigments and Their Roles Associated with Human Health. J. Fungi 2020, 6, 280. [Google Scholar] [CrossRef]
- Tsai, H.-F.; Fujii, I.; Watanabe, A.; Wheeler, M.H.; Chang, Y.C.; Yasuoka, Y.; Ebizuka, Y.; Kwon-Chung, K.J. Pentaketide Melanin Biosynthesis in Aspergillus fumigatus Requires Chain-Length Shortening of a Heptaketide Precursor. J. Biol. Chem. 2001, 276, 29292–29298. [Google Scholar] [CrossRef]
- Dutta, S.; Whicher, J.R.; Hansen, D.A.; Hale, W.A.; Chemler, J.A.; Congdon, G.R.; Narayan, A.R.H.; Håkansson, K.; Sherman, D.H.; Smith, J.L.; et al. Structure of a Modular Polyketide Synthase. Nature 2014, 510, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Shen, B. Polyketide Biosynthesis beyond the Type I, II and III Polyketide Synthase Paradigms. Curr. Opin. Chem. Biol. 2003, 7, 285–295. [Google Scholar] [CrossRef]
- Eisenman, H.C.; Casadevall, A. Synthesis and Assembly of Fungal Melanin. Appl. Microbiol. Biotechnol. 2012, 93, 931–940. [Google Scholar] [CrossRef]
- Mukherjee, G.; Mishra, T.; Deshmukh, S.K. Fungal Pigments: An Overview. In Developments in Fungal Biology and Applied Mycology; Satyanarayana, T., Deshmukh, S.K., Johri, B.N., Eds.; Springer: Singapore, 2017; pp. 525–541. ISBN 978-981-10-4768-8. [Google Scholar]
- Mapari, S.A.S.; Thrane, U.; Meyer, A.S. Fungal Polyketide Azaphilone Pigments as Future Natural Food Colorants? Trends Biotechnol. 2010, 28, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.-M.; Yang, S.-X.; Qin, J.-C. Azaphilones: Chemistry and Biology. Chem. Rev. 2013, 113, 4755–4811. [Google Scholar] [CrossRef]
- Gao, Y.; Badejo, A.A.; Sawa, Y.; Ishikawa, T. Analysis of Two L-Galactono-1,4-Lactone-Responsive Genes with Complementary Expression during the Development of Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 592–601. [Google Scholar] [CrossRef] [PubMed]
- El-Najjar, N.; Gali-Muhtasib, H.; Ketola, R.A.; Vuorela, P.; Urtti, A.; Vuorela, H. The Chemical and Biological Activities of Quinones: Overview and Implications in Analytical Detection. Phytochem. Rev. 2011, 10, 353. [Google Scholar] [CrossRef]
- Moss, H.E.; Ostrin, R.K.; Tyler, L.K.; Marslen-Wilson, W.D. Accessing Different Types of Lexical Semantic Information: Evidence from Priming. J. Exp. Psychol. Learn. Mem. Cogn. 1995, 21, 863–883. [Google Scholar] [CrossRef]
- Hanson, J.R. The Chemistry of Fungi; The Royal Society of Chemistry: Cambridge, UK, 2008; ISBN 978-0-85404-136-7. [Google Scholar]
- Dufossé, L.; Fouillaud, M.; Caro, Y.; Mapari, S.A.S.; Sutthiwong, N. Filamentous Fungi Are Large-Scale Producers of Pigments and Colorants for the Food Industry. Curr. Opin. Biotechnol. 2014, 26, 56–61. [Google Scholar] [CrossRef]
- Meruvu, H.; dos Santos, J.C. Colors of Life: A Review on Fungal Pigments. Crit. Rev. Biotechnol. 2021, 41, 1153–1177. [Google Scholar] [CrossRef]
- Caro, Y.; Anamale, L.; Fouillaud, M.; Laurent, P.; Petit, T.; Dufosse, L. Natural Hydroxyanthraquinoid Pigments as Potent Food Grade Colorants: An Overview. Nat. Prod. Bioprospecting 2012, 2, 174–193. [Google Scholar] [CrossRef]
- Fouillaud, M.; Venkatachalam, M.; Girard-Valenciennes, E.; Caro, Y.; Dufossé, L. Anthraquinones and Derivatives from Marine-Derived Fungi: Structural Diversity and Selected Biological Activities. Mar. Drugs 2016, 14, 64. [Google Scholar] [CrossRef] [PubMed]
- Rahmoun, N.M.; Boucherit-Otmani, Z.; Boucherit, K.; Benabdallah, M.; Villemin, D.; Choukchou-Braham, N. Antibacterial and Antifungal Activity of Lawsone and Novel Naphthoquinone Derivatives. Med. Mal. Infect. 2012, 42, 270–275. [Google Scholar] [CrossRef]
- Ravichandiran, P.; Masłyk, M.; Sheet, S.; Janeczko, M.; Premnath, D.; Kim, A.R.; Park, B.-H.; Han, M.-K.; Yoo, D.J. Synthesis and Antimicrobial Evaluation of 1,4-Naphthoquinone Derivatives as Potential Antibacterial Agents. ChemistryOpen 2019, 8, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Abe, H.; Yoshizaki, F. In Vitro Antifungal Activity of Naphthoquinone Derivatives. Biol. Pharm. Bull. 2002, 25, 669–670. [Google Scholar] [CrossRef]
- Lebeau, J.; Petit, T.; Clerc, P.; Dufossé, L.; Caro, Y. Isolation of Two Novel Purple Naphthoquinone Pigments Concomitant with the Bioactive Red Bikaverin and Derivates Thereof Produced by Fusarium oxysporum. Biotechnol. Prog. 2019, 35, e2738. [Google Scholar] [CrossRef] [PubMed]
- Futuro, D.O.; Ferreira, P.G.; Nicoletti, C.D.; Borba-Santos, L.P.; Silva, F.C.D.; Rozental, S.; Ferreira, V.F. The Antifungal Activity of Naphthoquinones: An Integrative Review. An. Acad. Bras. Cienc. 2018, 90, 1187–1214. [Google Scholar] [CrossRef]
- Basnet, B.B.; Liu, L.; Zhao, W.; Liu, R.; Ma, K.; Bao, L.; Ren, J.; Wei, X.; Yu, H.; Wei, J.; et al. New 1, 2-Naphthoquinone-Derived Pigments from the Mycobiont of Lichen Trypethelium eluteriae Sprengel. Nat. Prod. Res. 2019, 33, 2044–2050. [Google Scholar] [CrossRef]
- Gutierrez, S.M.V.; Hazell, K.K.; Simonsen, J.; Robinson, S.C. Description of a Naphthoquinonic Crystal Produced by the Fungus Scytalidium cuboideum. Molecules 2018, 23, 1905. [Google Scholar] [CrossRef]
- Palomino Agurto, M.E.; Vega Gutierrez, S.M.; Van Court, R.C.; Chen, H.-L.; Robinson, S.C. Oil-Based Fungal Pigment from Scytalidium cuboideum as a Textile Dye. J. Fungi 2020, 6, 53. [Google Scholar] [CrossRef]
- Stone, D.W.; Vega Gutierrez, S.M.; Walsh, Z.M.; Robinson, S.C. Preliminary Exploration of the Red Pigment from Scytalidium cuboideum as a Cellulosic Pulp Colorant. Challenges 2022, 13, 15. [Google Scholar] [CrossRef]
- Huang, L.; Liu, M.; Huang, H.; Wen, Y.; Zhang, X.; Wei, Y. Recent Advances and Progress on Melanin-like Materials and Their Biomedical Applications. Biomacromolecules 2018, 19, 1858–1868. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Liebmann, B. Aspergillus fumigatus Conidial Pigment and CAMP Signal Transduction: Significance for Virulence. Med. Mycol. 2005, 43 (Suppl. 1), S75–S82. [Google Scholar] [CrossRef] [PubMed]
- Nosanchuk, J.D.; Stark, R.E.; Casadevall, A. Fungal Melanin: What Do We Know About Structure? Front. Microbiol. 2015, 6, 1463. [Google Scholar] [CrossRef] [PubMed]
- White, L.P. Melanin: A Naturally Occurring Cation Exchange Material. Nature 1958, 182, 1427–1428. [Google Scholar] [CrossRef]
- d’Ischia, M.; Wakamatsu, K.; Napolitano, A.; Briganti, S.; Garcia-Borron, J.-C.; Kovacs, D.; Meredith, P.; Pezzella, A.; Picardo, M.; Sarna, T.; et al. Melanins and Melanogenesis: Methods, Standards, Protocols. Pigment Cell Melanoma Res. 2013, 26, 616–633. [Google Scholar] [CrossRef] [PubMed]
- Solano, F. Melanins: Skin Pigments and Much More—Types, Structural Models, Biological Functions, and Formation Routes. New J. Sci. 2014, 2014, e498276. [Google Scholar] [CrossRef]
- Casadevall, A.; Rosas, A.L.; Nosanchuk, J.D. Melanin and Virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 2000, 3, 354–358. [Google Scholar] [CrossRef]
- Pralea, I.E.; Moldovan, R.C.; Petrache, A.M.; Ilieș, M.; Hegheș, S.C.; Ielciu, I.; Nicoară, R.; Moldovan, M.; Ene, M.; Radu, M.; et al. From extraction to advanced analytical methods: The challenges of melanin analysis. Int. J. Mol. Sci. 2019, 20, 3943. [Google Scholar] [CrossRef]
- Gonçalves, R.C.R.; Lisboa, H.C.F.; Pombeiro-Sponchiado, S.R. Characterization of Melanin Pigment Produced by Aspergillus nidulans. World J. Microbiol. Biotechnol. 2012, 28, 1467–1474. [Google Scholar] [CrossRef]
- Suwannarach, N.; Kumla, J.; Watanabe, B.; Matsui, K.; Lumyong, S. Characterization of Melanin and Optimal Conditions for Pigment Production by an Endophytic Fungus, Spissiomyces endophytica SDBR-CMU319. PLOS ONE 2019, 14, e0222187. [Google Scholar] [CrossRef] [PubMed]
- Dadachova, E.; Bryan, R.A.; Howell, R.C.; Schweitzer, A.D.; Aisen, P.; Nosanchuk, J.D.; Casadevall, A. The Radioprotective Properties of Fungal Melanin Are a Function of Its Chemical Composition, Stable Radical Presence and Spatial Arrangement. Pigment Cell Melanoma Res. 2008, 21, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Fogarty, R.V.; Tobin, J.M. Fungal Melanins and Their Interactions with Metals. Enzyme Microb. Technol. 1996, 19, 311–317. [Google Scholar] [CrossRef]
- Gessler, N.N.; Egorova, A.S.; Belozerskaya, T.A. Melanin Pigments of Fungi under Extreme Environmental Conditions (Review). Appl. Biochem. Microbiol. 2014, 50, 105–113. [Google Scholar] [CrossRef]
- Avalos, J.; Carmen Limón, M. Biological Roles of Fungal Carotenoids. Curr. Genet. 2015, 61, 309–324. [Google Scholar] [CrossRef]
- Echavarri-Erasun, C.; Johnson, E.A. Fungal Carotenoids. In Applied Mycology and Biotechnology; Khachatourians, G.G., Arora, D.K., Eds.; Agriculture and Food Production; Elsevier: Amsterdam, The Netherlands, 2002; Volume 2, pp. 45–85. [Google Scholar]
- Han, J.R.; Zhao, W.J.; Gao, Y.Y.; Yuan, J.M. Effect of Oxidative Stress and Exogenous β-Carotene on Sclerotial Differentiation and Carotenoid Yield of Penicillium sp. PT95. Lett. Appl. Microbiol. 2005, 40, 412–417. [Google Scholar] [CrossRef]
- El-Jack, M.; Mackenzie, A.; Bramley, P.M. The Photoregulation of Carotenoid Biosynthesis in Aspergillus giganteus mut. alba. Planta 1988, 174, 59–66. [Google Scholar] [CrossRef]
- Chung, K.-R.; Jenns, A.E.; Ehrenshaft, M.; Daub, M.E. A Novel Gene Required for Cercosporin Toxin Resistance in the Fungus Cercospora nicotianae. Mol. Gen. Genet. MGG 1999, 262, 382–389. [Google Scholar] [CrossRef]
- van Eijk, G.W.; Mummery, R.S.; Roeymans, H.J.; Valadon, L.R.G. A Comparative Study of Carotenoids of Aschersonia aleyroides and Aspergillus giganteus. Antonie Van Leeuwenhoek 1979, 45, 417–422. [Google Scholar] [CrossRef]
- Jørgensen, T.R.; Park, J.; Arentshorst, M.; van Welzen, A.M.; Lamers, G.; vanKuyk, P.A.; Damveld, R.A.; van den Hondel, C.A.M.; Nielsen, K.F.; Frisvad, J.C.; et al. The Molecular and Genetic Basis of Conidial Pigmentation in Aspergillus niger. Fungal Genet. Biol. 2011, 48, 544–553. [Google Scholar] [CrossRef]
- Ali, H. Biodegradation of Synthetic Dyes—A Review. Water. Air. Soil Pollut. 2010, 213, 251–273. [Google Scholar] [CrossRef]
- Kuhad, R.C.; Sood, N.; Tripathi, K.K.; Singh, A.; Ward, O.P. Developments in Microbial Methods for the Treatment of Dye Effluents. In Advances in Applied Microbiology; Academic Press: Cambridge, MA, USA, 2004; Volume 56, pp. 185–213. [Google Scholar]
- Rodríguez Couto, S. Dye Removal by Immobilised Fungi. Biotechnol. Adv. 2009, 27, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Annuar, M.S.M.; Adnan, S.; Vikineswary, S.; Chisti, Y. Kinetics and Energetics of Azo Dye Decolorization by Pycnoporus sanguineus. Water. Air. Soil Pollut. 2009, 202, 179–188. [Google Scholar] [CrossRef]
- Banat, I.M.; Nigam, P.; Singh, D.; Marchant, R. Microbial Decolorization of Textile-Dyecontaining Effluents: A Review. Bioresour. Technol. 1996, 58, 217–227. [Google Scholar] [CrossRef]
- Singh, H. Fungal Decolorization and Degradation of Dyes. In Mycoremediation; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2006; pp. 420–483. ISBN 978-0-470-05059-0. [Google Scholar]
- Knackmuss, H.-J. Basic Knowledge and Perspectives of Bioelimination of Xenobiotic Compounds. J. Biotechnol. 1996, 51, 287–295. [Google Scholar] [CrossRef]
- El-Sikaily, A.; Khaled, A.; El Nemr, A. Textile Dyes Xenobiotic and Their Harmful Effect. In Non-Conventional Textile Waste Water Treatment; NOVA Publishers: Hauppauge, NY, USA, 2012; pp. 31–64. ISBN 978-1-62100-079-2. [Google Scholar]
- Benkhaya, S.; M’ rabet, S.; El Harfi, A. A Review on Classifications, Recent Synthesis and Applications of Textile Dyes. Inorg. Chem. Commun. 2020, 115, 107891. [Google Scholar] [CrossRef]
- Hunger, K.; Gregory, P.; Miederer, P.; Berneth, H.; Heid, C.; Mennicke, W. Important Chemical Chromophores of Dye Classes. In Industrial Dyes; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2004; pp. 13–112. ISBN 978-3-527-60201-8. [Google Scholar]
- Pereira, L.; Alves, M. Dyes—Environmental Impact and Remediation. In Environmental Protection Strategies for Sustainable Development; Malik, A., Grohmann, E., Eds.; Strategies for Sustainability; Springer: Dordrecht, The Netherlands, 2012; pp. 111–162. ISBN 978-94-007-1591-2. [Google Scholar]
- Routoula, E.; Patwardhan, S.V. Degradation of Anthraquinone Dyes from Effluents: A Review Focusing on Enzymatic Dye Degradation with Industrial Potential. Environ. Sci. Technol. 2020, 54, 647–664. [Google Scholar] [CrossRef]
- Lizama, C.; Freer, J.; Baeza, J.; Mansilla, H.D. Optimized Photodegradation of Reactive Blue 19 on TiO2 and ZnO Suspensions. Catal. Today 2002, 76, 235–246. [Google Scholar] [CrossRef]
- Azmi, W.; Sani, R.K.; Banerjee, U.C. Biodegradation of Triphenylmethane Dyes. Enzyme Microb. Technol. 1998, 22, 185–191. [Google Scholar] [CrossRef]
- Bumpus, J.A.; Brock, B.J. Biodegradation of Crystal Violet by the White Rot Fungus Phanerochaete Chrysosporium. Appl. Environ. Microbiol. 1988, 54, 1143–1150. [Google Scholar] [CrossRef]
- Malik, R.; Ramteke, D.S.; Wate, S.R. Adsorption of Malachite Green on Groundnut Shell Waste Based Powdered Activated Carbon. Waste Manag. 2007, 27, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Ting, A.S.Y. Microfungi for the Removal of Toxic Triphenylmethane Dyes. Min. Microb. Wealth MetaGenomics 2017, 405–429. [Google Scholar] [CrossRef]
- Lellis, B.; Fávaro-Polonio, C.Z.; Pamphile, J.A.; Polonio, J.C. Effects of Textile Dyes on Health and the Environment and Bioremediation Potential of Living Organisms. Biotechnol. Res. Innov. 2019, 3, 275–290. [Google Scholar] [CrossRef]
- Raza, A.; Saha, B. Application of Raman Spectroscopy in Forensic Investigation of Questioned Documents Involving Stamp Inks. Sci. Justice 2013, 53, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Jasuja, O.P.; Singla, A.K.; Seema, B.L. Thin-Layer Chromatographic Analysis of Indian Stamp Pad Inks. Forensic Sci. Int. 1989, 42, 255–262. [Google Scholar] [CrossRef]
- Yao, Y.-T.; Song, J.; Yu, J.; Wang, X.-F.; Hou, F.; Zhang, A.-L.; Liu, Y.; Han, J.; Xie, M.-X. Differentiation and Dating of Red Ink Entries of Seals on Documents by HPLC and GC/MS. J. Sep. Sci. 2009, 32, 2919–2927. [Google Scholar] [CrossRef]
- Sing, N.N.; Husaini, A.; Zulkharnain, A.; Roslan, H.A. Decolourisation Capabilities of Ligninolytic Enzymes Produced by Marasmius Cladophyllus UMAS MS8 on Remazol Brilliant Blue R and Other Azo Dyes. BioMed Res. Int. 2017, 2017, 1325754. [Google Scholar] [CrossRef] [PubMed]
- Piontek, K.; Antorini, M.; Choinowski, T. Crystal Structure of a Laccase from the Fungus Trametes versicolor at 1.90-Å Resolution Containing a Full Complement of Coppers. J. Biol. Chem. 2002, 277, 37663–37669. [Google Scholar] [CrossRef]
- Wong, D.W.S. Structure and Action Mechanism of Ligninolytic Enzymes. Appl. Biochem. Biotechnol. 2009, 157, 174–209. [Google Scholar] [CrossRef]
- Eggert, C.; Temp, U.; Eriksson, K.-E.L. Laccase Is Essential for Lignin Degradation by the White-Rot Fungus Pycnoporus cinnabarinus. FEBS Lett. 1997, 407, 89–92. [Google Scholar] [CrossRef]
- Youn, H.-D.; Hah, Y.C.; Kang, S.-O. Role of Laccase in Lignin Degradation by White-Rot Fungi. FEMS Microbiol. Lett. 1995, 132, 183–188. [Google Scholar] [CrossRef]
- Upadhyay, S.; Torres, G.; Lin, X. Laccases Involved in 1,8-Dihydroxynaphthalene Melanin Biosynthesis in Aspergillus fumigatus Are Regulated by Developmental Factors and Copper Homeostasis. Eukaryot. Cell 2013, 12, 1641–1652. [Google Scholar] [CrossRef] [PubMed]
- Plácido, J.; Capareda, S. Ligninolytic Enzymes: A Biotechnological Alternative for Bioethanol Production. Bioresour. Bioprocess. 2015, 2, 23. [Google Scholar] [CrossRef]
- de Marco, A.; Roubelakis-Angelakis, K.A. Laccase Activity Could Contribute to Cell-Wall Reconstitution in Regenerating Protoplasts. Phytochemistry 1997, 46, 421–425. [Google Scholar] [CrossRef]
- Sterjiades, R.; Dean, J.F.D.; Gamble, G.; Himmelsbach, D.S.; Eriksson, K.-E.L. Extracellular Laccases and Peroxidases from Sycamore Maple (Acer pseudoplatanus) Cell-Suspension Cultures. Planta 1993, 190, 75–87. [Google Scholar] [CrossRef]
- Castro-Sowinski, S.; Martinez-Drets, G.; Okon, Y. Laccase Activity in Melanin-Producing Strains of Sinorhizobium meliloti. FEMS Microbiol. Lett. 2002, 209, 119–125. [Google Scholar] [CrossRef]
- Chauhan, P.S.; Goradia, B.; Saxena, A. Bacterial Laccase: Recent Update on Production, Properties and Industrial Applications. 3 Biotech 2017, 7, 323. [Google Scholar] [CrossRef]
- Forootanfar, H.; Faramarzi, M.A. Insights into Laccase Producing Organisms, Fermentation States, Purification Strategies, and Biotechnological Applications. Biotechnol. Prog. 2015, 31, 1443–1463. [Google Scholar] [CrossRef]
- Hullo, M.-F.; Moszer, I.; Danchin, A.; Martin-Verstraete, I. CotA of Bacillus subtilis Is a Copper-Dependent Laccase. J. Bacteriol. 2001, 183, 5426–5430. [Google Scholar] [CrossRef]
- Matera, I.; Gullotto, A.; Tilli, S.; Ferraroni, M.; Scozzafava, A.; Briganti, F. Crystal Structure of the Blue Multicopper Oxidase from the White-Rot Fungus Trametes trogii Complexed with p-Toluate. Inorganica Chim. Acta 2008, 361, 4129–4137. [Google Scholar] [CrossRef]
- Zouari-Mechichi, H.; Mechichi, T.; Dhouib, A.; Sayadi, S.; Martínez, A.T.; Martínez, M.J. Laccase Purification and Characterization from Trametes trogii Isolated in Tunisia: Decolorization of Textile Dyes by the Purified Enzyme. Enzyme Microb. Technol. 2006, 39, 141–148. [Google Scholar] [CrossRef]
- Dias, A.A.; Sampaio, A.; Bezerra, R.M. Environmental Applications of Fungal and Plant Systems: Decolourisation of Textile Wastewater and Related Dyestuffs. In Environmental Bioremediation Technologies; Singh, S.N., Tripathi, R.D., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 445–463. ISBN 978-3-540-34793-4. [Google Scholar]
- Johansson, T.; Nyman, P.O. Isozymes of Lignin Peroxidase and Manganese(II) Peroxidase from the White-Rot Basidiomycete Trametes versicolor. I. Isolation of Enzyme Forms and Characterization of Physical and Catalytic Properties. Arch. Biochem. Biophys. 1993, 300, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Aarti, C.; Valan Arasu, M.; Agastian, P. Lignin Degradation:A Microbial Approach. South Indian J. Biol. Sci. 2015, 1, 119. [Google Scholar] [CrossRef]
- Ghodake, G.S.; Kalme, S.D.; Jadhav, J.P.; Govindwar, S.P. Purification and Partial Characterization of Lignin Peroxidase from Acinetobacter calcoaceticus NCIM 2890 and Its Application in Decolorization of Textile Dyes. Appl. Biochem. Biotechnol. 2009, 152, 6–14. [Google Scholar] [CrossRef]
- Gottschalk, L.M.F.; Bon, E.P.S.; Nobrega, R. Lignin Peroxidase from Streptomyces viridosporus T7A: Enzyme Concentration Using Ultrafiltration. Appl. Biochem. Biotechnol. 2008, 147, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Janusz, G.; Pawlik, A.; Sulej, J.; Świderska-Burek, U.; Jarosz-Wilkołazka, A.; Paszczyński, A. Lignin Degradation: Microorganisms, Enzymes Involved, Genomes Analysis and Evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef]
- Kuwahara, M.; Glenn, J.K.; Morgan, M.A.; Gold, M.H. Separation and Characterization of Two Extracelluar H2O2-Dependent Oxidases from Ligninolytic Cultures of Phanerochaete chrysosporium. FEBS Lett. 1984, 169, 247–250. [Google Scholar] [CrossRef]
- de Oliveira, P.L.; Duarte, M.C.T.; Ponezi, A.N.; Durrant, L.R. Purification and Partial Characterization of Manganese Peroxidase from Bacillus Pumilus AND Paenibacillus sp. Braz. J. Microbiol. Publ. Braz. Soc. Microbiol. 2009, 40, 818–826. [Google Scholar] [CrossRef]
- Sundaramoorthy, M.; Kishi, K.; Gold, M.H.; Poulos, T.L. The Crystal Structure of Manganese Peroxidase from Phanerochaete chrysosporium at 2.06-A Resolution. J. Biol. Chem. 1994, 269, 32759–32767. [Google Scholar] [CrossRef]
- Chen, C.; Li, T. Bacterial Dye-Decolorizing Peroxidases: Biochemical Properties and Biotechnological Opportunities. Phys. Sci. Rev. 2016, 1, 20160051. [Google Scholar] [CrossRef]
- Sugano, Y.; Muramatsu, R.; Ichiyanagi, A.; Sato, T.; Shoda, M. DyP, a Unique Dye-Decolorizing Peroxidase, Represents a Novel Heme Peroxidase Family: ASP171 Replaces the Distal Histidine of Classical Peroxidases. J. Biol. Chem. 2007, 282, 36652–36658. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Ishikawa, K.; Hirai, M.; Shoda, M. Characteristics of a Newly Isolated Fungus, Geotrichum candidum Dec 1, Which Decolorizes Various Dyes. J. Ferment. Bioeng. 1995, 79, 601–607. [Google Scholar] [CrossRef]
- Kim, S.J.; Shoda, M. Purification and Characterization of a Novel Peroxidase from Geotrichum candidum Dec 1 Involved in Decolorization of Dyes. Appl. Environ. Microbiol. 1999, 65, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Roberts, J.N.; Hardiman, E.M.; Singh, R.; Eltis, L.D.; Bugg, T.D.H. Identification of DypB from Rhodococcus jostii RHA1 as a Lignin Peroxidase. Biochemistry 2011, 50, 5096–5107. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.E.; Barros, T.; Chang, M.C.Y. Identification and Characterization of a Multifunctional Dye Peroxidase from a Lignin-Reactive Bacterium. ACS Chem. Biol. 2012, 7, 2074–2081. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Shrestha, R.; Jia, K.; Gao, P.F.; Geisbrecht, B.V.; Bossmann, S.H.; Shi, J.; Li, P. Characterization of Dye-Decolorizing Peroxidase (DyP) from Thermomonospora curvata Reveals Unique Catalytic Properties of A-Type DyPs. J. Biol. Chem. 2015, 290, 23447–23463. [Google Scholar] [CrossRef]
- Fernández-Fueyo, E.; Linde, D.; Almendral, D.; López-Lucendo, M.F.; Ruiz-Dueñas, F.J.; Martínez, A.T. Description of the First Fungal Dye-Decolorizing Peroxidase Oxidizing Manganese(II). Appl. Microbiol. Biotechnol. 2015, 99, 8927–8942. [Google Scholar] [CrossRef]
- Levin, L.N.; Hernández-Luna, C.E.; Niño-Medina, G.; García-Rodríguez, J.P.; López-Sadin, I.; Méndez-Zamora, G.; Gutiérrez-Soto, G. Decolorization and Detoxification of Synthetic Dyes by Mexican Strains of Trametes sp. Int. J. Environ. Res. Public Health 2019, 16, 4610. [Google Scholar] [CrossRef]
- Noman, E.; Al-Gheethi, A.; Talip, B.A.; Mohamed, R.; Kassim, A.H. Decolourization of Dye Wastewater by A Malaysian Isolate of Aspergillus iizukae 605EAN Strain: A Biokinetic, Mechanism and Microstructure Study. Int. J. Environ. Anal. Chem. 2020, 101, 1592–1615. [Google Scholar] [CrossRef]
- Parshetti, G.K.; Parshetti, S.G.; Telke, A.A.; Kalyani, D.C.; Doong, R.A.; Govindwar, S.P. Biodegradation of Crystal Violet by Agrobacterium radiobacter. J. Environ. Sci. 2011, 23, 1384–1393. [Google Scholar] [CrossRef]
- Yan, K.; Wang, H.; Zhang, X. Biodegradation of Crystal Violet by Low Molecular Mass Fraction Secreted by Fungus. J. Biosci. Bioeng. 2009, 108, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Copete-Pertuz, L.S.; Alandete-Novoa, F.; Plácido, J.; Correa-Londoño, G.A.; Mora-Martínez, A.L. Enhancement of Ligninolytic Enzymes Production and Decolourising Activity in Leptosphaerulina sp. by Co–Cultivation with Trichoderma viride and Aspergillus terreus. Sci. Total Environ. 2019, 646, 1536–1545. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.; Geng, A. Expression of a Novel Manganese Peroxidase from Cerrena unicolor BBP6 in Pichia pastoris and Its Application in Dye Decolorization and PAH Degradation. Biochem. Eng. J. 2020, 153, 107402. [Google Scholar] [CrossRef]
- Jadhav, U.U.; Dawkar, V.V.; Ghodake, G.S.; Govindwar, S.P. Biodegradation of Direct Red 5B, a Textile Dye by Newly Isolated Comamonas sp. UVS. J. Hazard. Mater. 2008, 158, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, U.U.; Dawkar, V.V.; Telke, A.A.; Govindwar, S.P. Decolorization of Direct Blue GLL with Enhanced Lignin Peroxidase Enzyme Production in Comamonas sp. UVS. J. Chem. Technol. Biotechnol. 2009, 84, 126–132. [Google Scholar] [CrossRef]
- Sugano, Y.; Matsushima, Y.; Tsuchiya, K.; Aoki, H.; Hirai, M.; Shoda, M. Degradation Pathway of an Anthraquinone Dye Catalyzed by a Unique Peroxidase DyP from Thanatephorus cucumeris Dec 1. Biodegradation 2009, 20, 433–440. [Google Scholar] [CrossRef]
- Abd El Monssef, R.A.; Hassan, E.A.; Ramadan, E.M. Production of Laccase Enzyme for Their Potential Application to Decolorize Fungal Pigments on Aging Paper and Parchment. Ann. Agric. Sci. 2016, 61, 145–154. [Google Scholar] [CrossRef]
- Tavzes, Č.; Palčič, J.; Fackler, K.; Pohleven, F.; Koestler, R.J. Biomimetic System for Removal of Fungal Melanin Staining on Paper. Int. Biodeterior. Biodegrad. 2013, 84, 307–313. [Google Scholar] [CrossRef]
- Kavkler, K.; Demšar, A.; Tavzes, Č.; Gostinčar, C.; Zalar, P. Discolouration of Fungal Stains on Cotton Textiles. Int. Biodeterior. Biodegrad. 2022, 172, 105427. [Google Scholar] [CrossRef]
- Ye, Y.; Xiao, Y.; Ma, L.; Li, H.; Xie, Z.; Wang, M.; Ma, H.; Tang, H.; Liu, J. Flavipin in Chaetomium globosum CDW7, an Endophytic Fungus from Ginkgo biloba, Contributes to Antioxidant Activity. Appl. Microbiol. Biotechnol. 2013, 97, 7131–7139. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).